Abstract
The well-established killing of bacteria by copper surfaces, also called contact killing, is currently believed to be a combined effect of bacterial contact with the copper surface and the dissolution of copper, resulting in lethal bacterial damage. Iron can similarly be released in ionic form from iron surfaces and would thus be expected to also exhibit contact killing, although essentially no contact killing is observed by iron surfaces. However, we show here that the exposure of bacteria to iron surfaces in the presence of copper ions results in efficient contact killing. The process involves reduction of Cu2+ to Cu+ by iron; Cu+ has been shown to be considerably more toxic to cells than Cu2+. The specific Cu+ chelator, bicinchoninic acid, suppresses contact killing by chelating the Cu+ ions. These findings underline the importance of Cu+ ions in the contact killing process and infer that iron-based alloys containing copper could provide novel antimicrobial materials.
INTRODUCTION
The killing of bacteria by metallic copper surfaces, so-called “contact killing,” is now well established and has explicitly been shown for many species (1). Bacteria are killed within minutes on surfaces of copper or copper alloys containing at least 60% copper. In contrast, cells can survive for days on surfaces of stainless steel, glass, or plastics. Copper and copper alloys have attracted attention as a means of creating self-sanitizing surfaces in the light of increasing nosocomial infections in Western hospitals. In a number of hospital trials, rooms have been fitted with copper alloy table tops, bedrails, door handles, light switches, bathroom fixtures, etc., in an effort to curb nosocomial infections (2–5; K. Laitinen et al., unpublished data). These copper surfaces resulted in a 2- to 3-log reduction of the microbial burden on a continuous basis. However, further data are needed to convincingly demonstrate that these measures also lead to a lasting reduction of nosocomial infections. Nevertheless, it appears clear that copper-containing materials can contribute to hospital hygiene and lower the bacterial burden also in other facilities where clean or aseptic working procedures are required (6).
The mechanism of contact killing of bacteria by copper-containing materials is of interest not only in connection to its use in hospitals but also from a purely scientific point of view. Laboratory studies have shown that bacteria on copper surfaces suffer rapid membrane damage and DNA degradation, in addition to other less well-defined cellular damage (7–12). The importance and the order of the different processes leading to cell death may depend on the type of microorganism (9). One key element required for contact killing is the release of copper ions from the metal surface. Bacterial copper resistance systems appear unable to cope with the released copper (13–15). The second important requirement for contact killing is bacterial contact with the metal surface (16). Recently, we showed that bacteria are also killed effectively on iron surfaces if ionic copper ions are present (16). In the present study, we also show that the reduction of Cu2+ to Cu+ by the iron surface plays a key role in the killing process. These findings underline the greater toxicity of Cu+ compared to Cu2+ and suggest novel antimicrobial materials based on iron alloys able to release copper.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type strain Enterococcus hirae ATCC 9790 was grown anaerobically by inoculating 10 ml of air-saturated N-medium (17), followed by growth in sealed tubes to stationary phase at 37°C. Cells were collected by centrifugation for 5 min at 5,000 × g, washed twice with 20 ml of 100 mM Na-HEPES (pH 7) or, where indicated, with 100 mM Tris-Cl (pH 7), and resuspended in 10 ml of the same buffer. The average cell density was 2 × 108 to 8 × 108 CFU/ml. All handling of cells was performed aerobically.
Preparation of iron coupons.
Iron plates (10 by 20 by 0.5 mm), called “coupons,” were composed of >99% iron, and <1% carbon and were cleaned by ultrasonication in chloroform and ethanol for 10 min each, followed by air-drying. After cleaning, all coupons used in the present study were stored under nitrogen until used.
Measurement of contact killing.
To assess contact killing, a wet plating technique was used, essentially as previously described (14). Briefly, 40 μl of cells suspended in 100 mM Tris-Cl or Na-HEPES (pH 7) and supplemented with 2 mM bicinchoninic acid (BCA) and/or 4 mM CuSO4 was applied to the coupons. After incubation at 25°C for 0 to 300 min in a water-saturated atmosphere, 10-μl samples were withdrawn, and serial dilutions in phosphate-buffered saline were spread on N agar plates. After growth for 24 h, survival was calculated from plate counts and expressed in CFU.
Copper and iron determinations.
Copper or iron release from coupons during wet plating was assessed by removing 20-μl aliquots at 0 to 300 min, diluting them 50-fold with 0.065% HNO3, and measuring the copper content by inductively coupled plasma atomic emission (ICP-AE) spectroscopy on a Jobin Yvon JY 24 instrument (Horiba; Jobin Yvon GmbH, Munich, Germany) at 324.754 nm for Cu or 259.940 nm for Fe.
Measurement of copper reduction.
Cell suspensions used to measure contact killing with 4 mM CuSO4 (40 μl) were applied to iron coupons, and at 0 to 300 min 10-μl aliquots were withdrawn and mixed with 990 μl of 100 μM BCA in 0.1 M Tris-Cl (pH 7). Formation of Cu+ was determined by measuring the concentration of the formed Cu(BCA)2 complex at 354.5 nm, using an extinction coefficient ε = 4.6 × 104 M−1 cm−1 (18).
RESULTS
Contact killing on iron coupons.
Iron has redox properties similar to those of copper, yet it does not exhibit contact killing of bacteria. When 2 × 107 Enterococcus hirae cells were applied to an iron surface, there was no significant reduction (<1 log) in the number of live bacteria after 300 min, under both anaerobic and aerobic conditions (Fig. 1A). However, if 4 mM CuSO4 is added to the cells, no survivors could be recovered after 300 min under both anaerobic and aerobic conditions. After 100 min of exposure, a difference between anaerobic and aerobic conditions can be observed, with 3 logs of killing under aerobic and nearly 6 logs of killing under anaerobic conditions. The concentration of 4 mM CuSO4 was chosen on the basis of previous findings, which had shown that the rate of killing on iron is proportional to the copper concentration (16); 4 mM CuSO4 provided an ideal concentration for the present studies.
FIG 1.
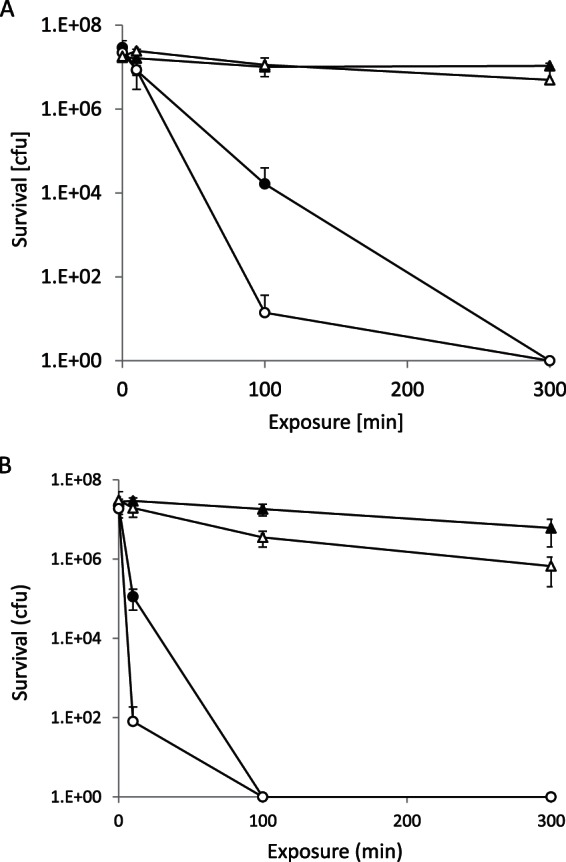
(A) Contact killing of E. hirae on iron. Cells suspended in Na-HEPES buffer and 4 mM CuSO4 were incubated on iron coupons under either aerobic (●) or anaerobic (○) conditions. The results for controls without copper under anaerobic (▲) or anaerobic (△) conditions are also indicated. (B) Same as in panel A, but the cells suspended in Tris-Cl buffer. The error bars indicate the standard deviations of three independent experiments.
These experiments were conducted in Na-HEPES buffer, which exhibits negligible complex formation with copper. If these experiments were conducted in Tris-Cl buffer, which is known to complex copper strongly (19), contact killing was even more rapid, with complete killing observed already after 100 min, compared to 300 min in Na-HEPES, under both aerobic and anaerobic conditions (Fig. 1B). There was also a significant decline in viability in Tris-Cl buffer in the absence of copper. The cause of this loss in viability remains unknown but may be connected to the membrane permeability of Tris-Cl in its nondissociated state. Taken together, these experiments show that the addition of copper to cells on metallic iron induces contact killing and that the effect is accentuated by anaerobic conditions.
Role of copper reduction.
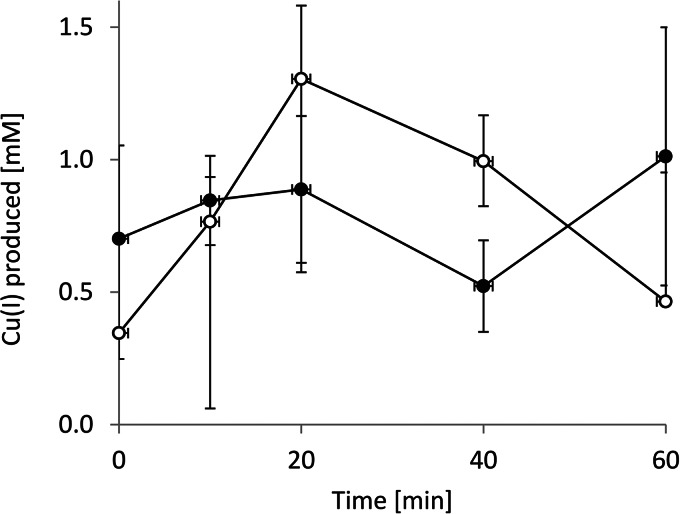
The enhancement of copper-induced contact killing by anaerobic conditions led us to conclude that the oxidation state of the copper ions is important to the process. We thus determined the presence of Cu+ in the course of contact killing experiments on iron. When cells were exposed to metallic iron in the presence of CuSO4, there was significant generation of Cu+ ions, even at the shortest times measurable (1 to 2 s; Fig. 2). Overall, Cu+ generation did not differ significantly between aerobic and anaerobic conditions and, on average, remained at ∼0.8 mM throughout the experiment. It has previously been shown that Cu+ is considerably more toxic to cells than Cu2+ (20). The faster killing of E. hirae on iron in the presence of copper thus appears to be related to the generation of Cu+.
FIG 2.
Measurement of Cu+ production. Iron coupons were incubated with cell suspensions containing 4 mM CuSO4 under either aerobic (●) or anaerobic (○) conditions. At the times indicated, samples were withdrawn, and the Cu+ formed was complexed with BCA, followed by spectrophotometric determination of Cu+ as described in Materials and Methods. The error bars indicate the standard deviations of three independent experiments.
Copper reduction by iron.
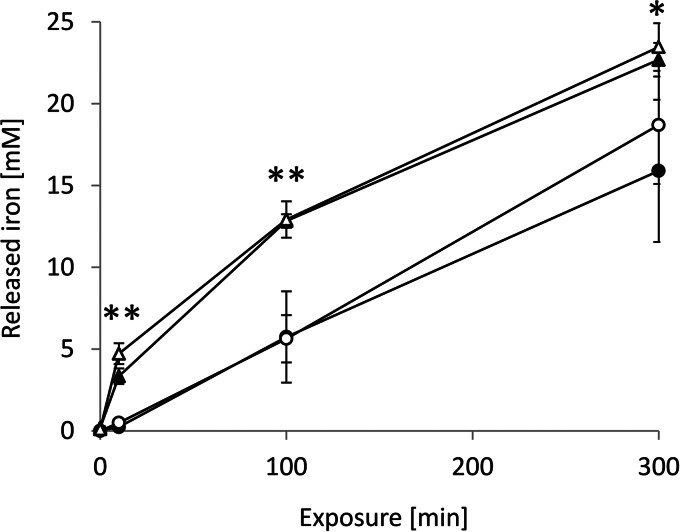
The obvious source of electrons for copper reduction is the Fe(0) of the coupons. We therefore looked at iron release from the coupons in the presence of copper. There was substantial release of iron into the aqueous medium, regardless of the presence of either copper or cells (Fig. 3). After 300 min, 15 mM iron was released into the aqueous phase in the absence of copper. When 4 mM CuSO4 was present, iron release was enhanced by ca. 30%. Iron is not very soluble under aerobic conditions at pH 7, and it must be assumed that most of the iron was present in the hydroxide form. In fact, the formation of a visible film, presumably of iron hydroxide, on the surface of the aqueous phase could be observed. Iron release was unexpectedly high, exceeding copper reduction almost 20-fold, and this release was unlikely to play an important role in contact killing by metallic iron in the presence of copper. The generation of Cu+ appears to be the key toxicity mechanism.
FIG 3.
Determination of iron release. Iron coupons were incubated with cell suspensions under the following conditions: minus cells, minus copper (○); plus cells, minus copper (●); plus cells, plus copper (▲); and minus cells, plus copper(△). The copper concentration was always 4 mM CuSO4. At the times indicated, samples were withdrawn, and the iron content was determined by ICP-AE spectroscopy as described in Materials and Methods. The error bars indicate the standard deviations of three independent experiments. *, P < 0.006; **, P < 0.03 (Student t test).
Cu+ chelation suppresses copper toxicity on iron.
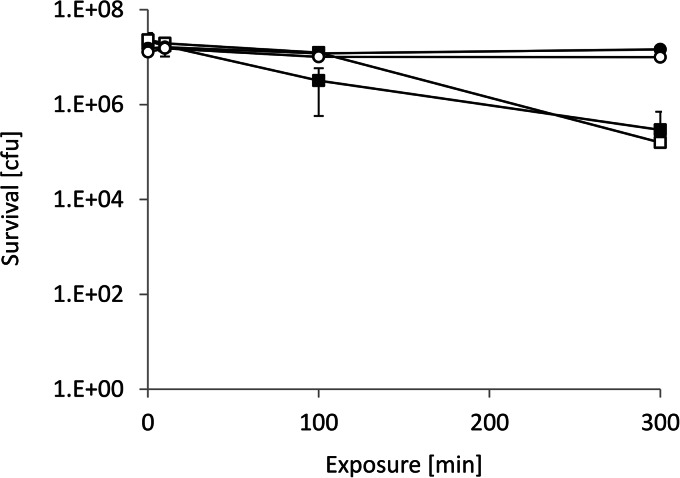
Since the generation of Cu+ appears to be the toxicity mechanism of contact killing on iron in the presence of copper, specific chelation of Cu+ by BCA was investigated. Figure 4 shows that contact killing in the presence of copper was massively reduced by BCA. After 300 min of exposure, only a 2-log (99%) reduction in cell survival was observed in the presence of 4 mM CuSO4 and 2 mM BCA compared to experiments without chelation, where survival was reduced by >7 logs. BCA by itself had no significant effect on the survival of bacteria on iron or glass. This further supports that contact killing on iron surfaces in the presence of copper is due to the reduction of Cu2+ to the more toxic Cu+.
FIG 4.
Contact killing in the presence of BCA. Survival of cells in suspension was determined in the presence of both 4 mM CuSO4 and 2 mM BCA on either iron (■) or glass (□) or in the presence of only BCA on iron (●) or glass (○). The experiment was conducted as described in the legend to Fig. 1. The error bars show the standard deviations of three independent experiments.
Redox-inactive metal ions do not promote contact killing on iron.
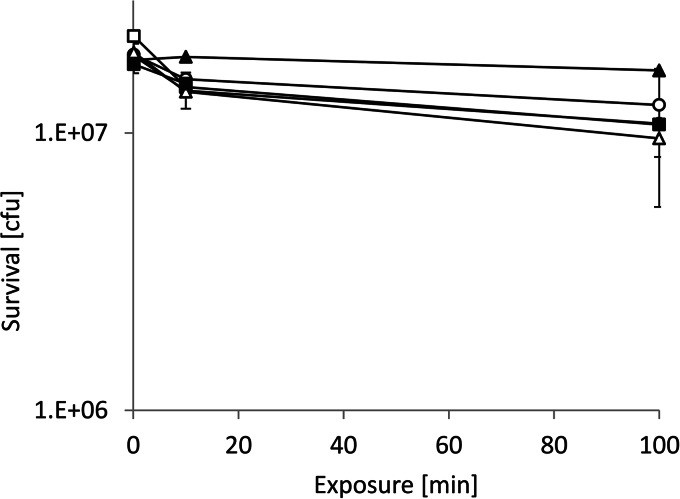
As a further test of the concept of copper reduction as the active principle in contact killing on iron plus copper ions, metal ions which are not redox-active were tested as to their effect on bacterial survival on iron. As shown in Fig. 5, neither Zn2+ nor Cd2+ had a significant effect on bacterial survival on iron (note that the ordinate of this figure is greatly expanded for clarity). Clearly, redox reactions between iron and copper are the underlying mechanism of contact killing of bacteria on iron in the presence of copper ions.
FIG 5.
Exposure to iron in the presence of Zn2+ or Cd2+. Cells in the presence of 4 mM ZnSO4 were exposed to either iron (■) or glass (□) or were exposed in the presence of 4 mM CdSO4 to either iron (▲) or glass (△). Also shown are controls without metal ions on iron (●) or glass (○). Other details of the experiment are as described in the legend to Fig. 4. The error bars show the standard deviations of three independent experiments.
DISCUSSION
We previously reported the augmentation of contact killing of bacteria on iron surfaces by copper ions (16). The present study extends those observations to gain further insight into the biocidal mechanism. Solid iron by itself only marginally impairs bacterial survival. However, when cell suspensions are supplemented with Cu2+, rapid bacterial killing is triggered. Key events in the process appear to be iron solubilization and copper reduction. Reduction of Cu2+ to Cu+ can be driven by metallic iron or Fe2+ serving as reductants. This process has been thermodynamically analyzed in detail by Matocha et al. (21). Cu+ can then form insoluble cuprite (Cu2O), even under anaerobic conditions (21). Cu+ is considerably more toxic to bacteria than Cu2+, due presumably to its greater membrane permeability than Cu2+ (20, 22). Cu2O was also found to be as toxic to bacteria as metallic copper (23). The copper-induced contact killing on iron shown here is more rapid than contact killing by copper surfaces, presumably due to the greater toxicity of Cu+ versus Cu2+. However, there may also be synergistic effects of the simultaneous presence of Fe2+, Fe3+, Cu+, Cu2+, and Cu2O (16). A copper-sensitive E. hirae mutant with both copper ATPases deleted and thus unable to expel cytoplasmic copper was completely killed on iron in the presence of copper in 100 min (>6 logs) compared to 300 min for wild-type E. hirae under the same conditions (data not shown). This further underlines the importance of copper ions in the contact killing process.
The buffer used to suspend cells had a significant effect on the rate of killing, with Tris-Cl buffer mediating faster killing than Na-HEPES. Tris is known to form complexes with copper, which could be more membrane-permeable than free copper ions (19). Tris was also shown to permeabilize the outer membrane of Escherichia coli and may damage the cell membrane of Gram-positive organisms such as E. hirae, but this will require further investigation (24). Furthermore, chloride ions at the concentration present in Tris-Cl buffer stabilize the more toxic Cu+ ions (21).
Contact killing of bacteria by copper surfaces involves the following steps: damage of the outer and/or inner bacterial membrane, accumulation of copper ions in the cell, and degradation of the bacterial DNA (1). The order in which these events lead to cell death is an issue of debate and may vary with the organism (7–10, 25). Copper can lead to the production of reactive oxygen species (ROS) in both Gram-positive and Gram-negative organisms by Fenton-type reactions. However, cell death can only partially be suppressed by ROS quenchers such as superoxide dismutase or catalase (13). ROS generation and lipid peroxidation by copper ions was also shown to occur in E. coli and Salmonella exposed to solid copper (9, 26). A mutant strain with higher levels of unsaturated fatty acids and thus more sensitive to ROS exhibited an earlier rise in lipid peroxidation, higher sensitivity to contact killing, and an earlier onset of DNA degradation (26). Evidence for oxidative damage was also apparent from the proteome of E. coli exposed to metallic copper by the increased presence of oxidatively modified proteins (27). Although ROS clearly cause cell damage in contact killing, it is probably an accompanying effect rather than the primary cause of cell death.
In the experiments presented here, Cu+ is produced by the reduction of Cu2+ via oxidation of iron. Anaerobic conditions lead to more rapid killing than aerobic conditions, suggesting that ROS production as not a primary cause of cell death. Rather, anaerobic conditions stabilize Cu+, leading to higher transient concentrations (cf. Fig. 2) and favor oxidation to Cu2O rather than to CuO; Cu2O has been shown to be as toxic to cells as unoxidized, metallic copper, whereas CuO is less toxic (23). Substantial amounts of iron are released from the coupons and copper stimulates this release by ca. 30%. Ionic iron can also induce killing of bacteria, as previously shown (28).
That Cu+ is a key player in contact killing in the experiments reported here is evident by the protective effect of the specific Cu+-chelator BCA. How copper kills cells in contact killing is clearly different from the mechanism in growing cells. In culture, the toxic effect of copper on E. coli was shown to be the displacement of iron from [4Fe-4S] clusters of dehydratases (29–31). Destruction of [4Fe-4S] clusters was also shown for Ag+, Hg2+, Cd2+, and Zn2+ at concentrations that only marginally inhibited growth (31). In line with an attack of iron-sulfur clusters by these ions, their cytotoxicity was related to their thiophilicity. In our experiments, the redox inactive metals Cd2+ and Zn2+ were unable to elicit significant cell death on iron coupons. Also, zinc has previously been shown to display a death rate constant of contact killing <1/20 of that of copper or silver (32), whereas cadmium has never been tested for contact killing. Taken together, these observations suggest that displacement of iron from [4Fe-4S] clusters does not play a significant role in contact killing or, for that matter, the killing of cells on iron in the presence of copper.
Killing of various species of streptococci in solution by Fe2+ or Cu+ ions, but not by Fe3+ or Cu2+ ions, has previously been reported by Dunning et al. (28). These experiments were conducted under anaerobic conditions to prevent ROS production, but in the absence of metallic surfaces. More than 4 logs of killing in 50 min by 5 to 10 mM Fe2+ or Cu+ was observed for streptococci, but considerably slower killing occurred with E. hirae ATCC 9790, the strain used in the present study. Dunning et al. concluded that killing was primarily due to inhibition of F-ATPase by Fe2+ or Cu+, but we do not share this interpretation. Although inhibition of cellular functions impairs growth, it does not necessarily lead to cell death. Killing by the simultaneous presence of iron and copper, as in the experiments reported here, was not investigated by Dunning et al. Also, no measurements of residual oxygen or ROS production were reported, but our findings here underline the importance of ionic species of iron and copper in the antibacterial activity of these metals.
In an atomic force microscopy study, it was found that the outer membrane of bacteria in contact with antibacterial stainless steel that contains 3.8% copper undergoes substantial changes, and this suggests that membrane damage is a major event in contact killing (33). There was also release of copper ions by the copper-containing antibacterial stainless steel, thus providing an additional toxic component. It would be interesting to know if there was also generation of Cu+ under these conditions, but this question was not addressed. The study does, however, highlight the importance of bacterium-metal contact, as previously reported for copper, and thus supports the current model of contact killing (16). Taken together, these and our findings suggest novel design criteria for antimicrobial, functional materials, based on combinations of iron and copper, and show the greater potency of Cu+ compared to Cu2+ in contact killing.
ACKNOWLEDGMENTS
We thank Thomas Weber for expert technical assistance and Ilya Kublanov for suggesting the use of copper ions on iron coupons.
This study was supported by a Russian Federation Government grant to leading scientists (contract 14.Z50.31.0011).
REFERENCES
- 1.Grass G, Rensing C, Solioz M. 2011. Metallic copper as an antimicrobial surface. Appl Environ Microbiol 77:1541–1547. doi: 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rai S, Hirsch BE, Attaway HH, Nadan R, Fairey S, Hardy J, Miller G, Armellino D, Moran WR, Sharpe P, Estelle A, Michel JH, Michels HT, Schmidt MG. 2012. Evaluation of the antimicrobial properties of copper surfaces in an outpatient infectious disease practice. Infect Control Hosp Epidemiol 33:200–201. doi: 10.1086/663701. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt MG, Attaway HH, Sharpe PA, John J Jr, Sepkowitz KA, Morgan A, Fairey SE, Singh S, Steed LL, Cantey JR, Freeman KD, Michels HT, Salgado CD. 2012. Sustained reduction of microbial burden on common hospital surfaces through the introduction of copper. J Clin Microbiol 50:2217–2223. doi: 10.1128/JCM.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey AL, Adams D, Karpanen TJ, Lambert PA, Cookson BD, Nightingale P, Miruszenko L, Shillam R, Christian P, Elliott TS. 2010. Role of copper in reducing hospital environment contamination. J Hosp Infect 74:72–77. doi: 10.1016/j.jhin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Mikolay A, Huggett S, Tikana L, Grass G, Braun J, Nies DH. 2010. Survival of bacteria on metallic copper surfaces in a hospital trial. Appl Microbiol Biotechnol 87:1875–1879. doi: 10.1007/s00253-010-2640-1. [DOI] [PubMed] [Google Scholar]
- 6.O'Gorman J, Humphreys H. 2012. Application of copper to prevent and control infection. Where are we now? J Hosp Infect 81:217–223. doi: 10.1016/j.jhin.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Espirito Santo C, Lam EW, Elowsky CG, Quaranta D, Domaille DW, Chang CJ, Grass G. 2011. Bacterial killing by dry metallic copper surfaces. Appl Environ Microbiol 77:794–802. doi: 10.1128/AEM.01599-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espirito Santo C, Quaranta D, Grass G. 2012. Antimicrobial metallic copper surfaces kill Staphylococcus haemolyticus via membrane damage. Microbiologyopen 1:46–52. doi: 10.1002/mbo3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by lower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol 14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 10.Warnes SL, Keevil CW. 2011. Mechanism of copper surface toxicity in vancomycin-resistant enterococci following ‘wet’ or ‘dry’ contact. Appl Environ Microbiol 77:6049–6059. doi: 10.1128/AEM.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnes SL, Green SM, Michels HT, Keevil CW. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNA. Appl Environ Microbiol 76:5390–5401. doi: 10.1128/AEM.03050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weaver L, Noyce JO, Michels HT, Keevil CW. 2010. Potential action of copper surfaces on methicillin-resistant Staphylococcus aureus. J Appl Microbiol 109:2200–2205. doi: 10.1111/j.1365-2672.2010.04852.x. [DOI] [PubMed] [Google Scholar]
- 13.Espirito Santo C, Taudte N, Nies DH, Grass G. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl Environ Microbiol 74:977–986. doi: 10.1128/AEM.01938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molteni C, Abicht HK, Solioz M. 2010. Killing of bacteria by copper surfaces involves dissolved copper. Appl Environ Microbiol 76:4099–4101. doi: 10.1128/AEM.00424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elguindi J, Wagner J, Rensing C. 2009. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J Appl Microbiol 106:1448–1455. doi: 10.1111/j.1365-2672.2009.04148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews S, Hans M, Mücklich F, Solioz M. 2013. Contact killing of bacteria on copper is suppressed if bacteria-metal contact is prevented and is induced on iron by copper ions. Appl Environ Microbiol 79:2605–2611. doi: 10.1128/AEM.03608-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol 29:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner AJ, Harris ED. 1995. A quantitative test for copper using bicinchoninic acid. Anal Biochem 226:80–84. doi: 10.1006/abio.1995.1194. [DOI] [PubMed] [Google Scholar]
- 19.McPhail DB, Goodman BA. 1984. Tris buffer–a case for caution in its use in copper-containing systems. Biochem J 221:559–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abicht HK, Gonskikh Y, Gerber SD, Solioz M. 2013. Non-enzymatic copper reduction by menaquinone enhances copper toxicity in Lactococcus lactis IL1403. Microbiology 159:1190–1197. doi: 10.1099/mic.0.066928-0. [DOI] [PubMed] [Google Scholar]
- 21.Matocha CJ, Karathanasis AD, Rakshit S, Wagner KM. 2005. Reduction of copper(II) by iron(II). J Environ Qual 34:1539–1546. doi: 10.2134/jeq2005.0002. [DOI] [PubMed] [Google Scholar]
- 22.Chaturvedi KS, Henderson JP. 2014. Pathogenic adaptations to host-derived antibacterial copper. Front Cell Infect Microbiol 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hans M, Erbe A, Mathews S, Chen Y, Solioz M, Mücklich F. 2013. Role of copper oxides in contact killing of bacteria. Langmuir 29:16160–16166. doi: 10.1021/la404091z. [DOI] [PubMed] [Google Scholar]
- 24.Irvin RT, MacAlister TJ, Costerton JW. 1981. Tris(hydroxymethyl)aminomethane buffer modification of Escherichia coli outer membrane permeability. J Bacteriol 145:1397–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian WX, Yu S, Ibrahim M, Almonaofy AW, He L, Hui Q, Bo Z, Li B, Xie GL. 2012. Copper as an antimicrobial agent against opportunistic pathogenic and multidrug resistant Enterobacter bacteria. J Microbiol 50:586–593. doi: 10.1007/s12275-012-2067-8. [DOI] [PubMed] [Google Scholar]
- 26.Hong R, Kang TY, Michels CA, Gadura N. 2012. Membrane lipid peroxidation in copper alloy mediated contact killing of Escherichia coli. Appl Environ Microbiol 78:1776–1784. doi: 10.1128/AEM.07068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandakumar R, Espirito Santo C, Madayiputhiya N, Grass G. 2011. Quantitative proteomic profiling of the Escherichia coli response to metallic copper surfaces. Biometals 24:429–444. doi: 10.1007/s10534-011-9434-5. [DOI] [PubMed] [Google Scholar]
- 28.Dunning JC, Ma Y, Marquis RE. 1998. Anaerobic killing of oral streptococci by reduced, transition metal cations. Appl Environ Microbiol 64:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu FF, Imlay JA. 2012. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol 78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami H, Yoshida K, Nishida Y, Kikuchi Y, Sato Y. 2008. Antibacterial properties of metallic elements for alloying evaluated with application of JIS Z 2801:2000. ISIJ Intl 48:1299–1304. doi: 10.2355/isijinternational.48.1299. [DOI] [Google Scholar]
- 33.Nan L, Liu Y, Lu M, Yang K. 2008. Study on antibacterial mechanism of copper-bearing austenitic antibacterial stainless steel by atomic force microscopy. J Mater Sci Mater Med 19:3057–3062. doi: 10.1007/s10856-008-3444-z. [DOI] [PubMed] [Google Scholar]