Abstract
The human pathogen Vibrio vulnificus is the leading cause of seafood-related deaths in the United States. Strains are genotyped on the basis of alleles that correlate with isolation source, with clinical (C)-genotype strains being more often implicated in disease and environmental (E)-genotype strains being more frequently isolated from oysters and estuarine waters. Previously, we have shown that the ecologically distinct C- and E-genotype strains of V. vulnificus display different degrees of chitin attachment, with C-genotype strains exhibiting reduced attachment relative to their E-genotype strain counterparts. We identified type IV pili to be part of the molecular basis for this observed genotypic variance, as E-genotype strains exhibit higher levels of expression of these genes than C-genotype strains. Here, we used a C-genotype quorum-sensing (QS) mutant to demonstrate that quorum sensing is a negative regulator of type IV pilus expression, which results in decreased chitin attachment. Furthermore, calcium depletion reduced E-genotype strain attachment to chitin, which suggests that calcium is necessary for proper functioning of the type IV pili in E-genotype strains. We also found that starvation or dormancy can alter the efficiency of chitin attachment, which has significant implications for the environmental persistence of V. vulnificus. With the increasing incidence of wound infections caused by V. vulnificus, we investigated a subset of E-genotype strains isolated from human wound infections and discovered that they attached to chitin in a manner more similar to that of C-genotype strains. This study enhances our understanding of the molecular and physical factors that mediate chitin attachment in V. vulnificus, providing insight into the mechanisms that facilitate the persistence of this pathogen in its native environment.
INTRODUCTION
The opportunistic human pathogen Vibrio vulnificus is part of the normal microflora of the estuarine environment and is found concentrated within bivalves and other organisms that inhabit these waters. V. vulnificus can infect individuals who consume raw or undercooked shellfish harboring this bacterium, and it has the potential to cause fulminating septicemia and organ failure, particularly in individuals with liver disease or immunodeficiency. With a case fatality rate of up to 50%, the majority (95%) of deaths associated with seafood in the United States are caused by this pathogen (1–3).
V. vulnificus is an exceptional pathogen in that it has more than one portal of entry. In addition to septicemia caused by ingestion of raw or undercooked seafood, this bacterium causes grievous wound infections and necrotizing fasciitis via entry into preexisting cuts or wounds. In the latter case, infection can result from exposure to seawater or shellfish harboring sufficient quantities of the pathogen, which can occur during fishing, swimming, or other coastal activities. The incubation period postexposure is relatively short, averaging only 16 h, and wound infection often requires tissue debridement, skin grafts, and even amputation of the affected limbs (4). Approximately 25% of wound infections result in the death of the patient. Unlike patients who suffer septicemia from ingestion, victims suffering from wound infections often do not have underlying diseases (4, 5). Interestingly, the incidence of wound infections in the United States has been increasing over the last few decades, and wound infections are now the predominant form of infection caused by this pathogen (6).
Genetic polymorphisms within specific genes have been found to correlate highly with the source of isolation and pathogenicity (1, 7–11). Currently, we use an allele referred to as the virulence-correlated gene (vcg) to distinguish between strains that are more often implicated in human disease (clinical [C]-genotype strains) from those that are more frequently isolated from the natural environment (environmental [E]-genotype strains) (7–9). Previous studies have provided further distinctions between these genotypes. Specifically, we have found that C-genotype strains demonstrate superior survival in human serum relative to E-genotype strains (12–14), and genome comparisons have allowed the identification of several putative virulence factors that could potentially aid this bacterium in disease progression (15, 16).
Despite these studies, no single, causal virulence factor has been found to be exclusively associated with clinical genotype strains (17). While the C/E genotyping scheme strongly correlates with virulence potential, this method does not strictly predict virulence (17). For example, some strains with the vcgE allele have been isolated from clinical cases, often from wound infections. Previously, we analyzed a subset of E-genotype wound isolates and demonstrated their ability to resist the bactericidal effects of human serum in a manner similar to that of C-genotype blood isolates (13). Thus, it appears that a subset of E-genotype strains has the ability to cause human disease, highlighting the need for better predictors of virulence potential within E-genotype strains.
The environmental occurrence of V. vulnificus is favored by high temperatures (>20°C) and intermediate salinities (15 to 25 ppt); hence, this bacterium can be readily isolated in environments with these physiochemical parameters (18). However, this organism can tolerate wide ranges of salinities and temperatures, which likely contributes to its seasonal and geographical recurrence (18–20). For example, when water temperatures drop below 4°C for extended periods of time, V. vulnificus has been documented to enter a state of dormancy referred to as the viable but nonculturable (VBNC) state (4, 21). This process of overwintering is thought to explain the observed seasonality of this organism in its native environment.
While it is well-known how salinity and seasonality influence the diversity of V. vulnificus in the environment (22, 23), the population structure of C- and E-genotype strains within a given environment appears to be more complex. In their natural environment, C- and E-genotype strains are routinely found to have different environmental distributions, with E-genotype strains often predominating (ca. 85%) in oysters and the water column relative to the prevalence of C-genotype strains (ca. 15%) (24). This distribution anomaly highlights the need for a better understanding of the biotic and abiotic factors that influence the spatial and temporal distribution of C- and E-genotype strains.
Deeper investigation of ecological niches, such as microenvironments, has provided considerable insight into the niche differentiation of C- and E-genotype strains. Marine aggregates, conglomerates of organic debris and inorganic matter, form in the upper layers of the ocean and serve as a carbon and nitrogen hot spot for planktonic microorganisms. A study investigating the ability of V. vulnificus C- and E-genotype strains to integrate into marine aggregates found that they do so with significantly different efficiencies (25). That study found that E-genotype strains incorporate into marine aggregates much more efficiently than their C-genotype counterparts, and this increased attachment results in the preferential uptake and retention of E-genotype strains by oysters feeding on these marine aggregates. That study offered insight into why E-genotype strains have previously been documented to predominate within the oyster environment; however, the physiological and genetic causalities for this demonstrated genotypic heterogeneity remain unexplained.
Chitin is a major constituent of marine aggregates, and chitinous substrates are considered to play a pivotal role in the survival and persistence of vibrios, serving as a critical reservoir for pathogens such as Vibrio cholerae (26). Importantly, a number of Vibrio spp. have been shown to adhere to chitinous substrates, and these associations with chitin are thought to influence the organism's overall metabolism and physiology (26–29). This substrate provides the organism with a number of advantages, including food availability, tolerance to stress, and protection from predators (26, 30, 31). Given the abundance of chitin in marine aggregates and the significance of chitin in the ecology of vibrios, we subsequently investigated the efficiency of attachment of C- and E-genotype strains to chitin particles (32). Our study revealed that E-genotype strains attach to chitin with a significantly greater efficiency than C-genotype strains, and surface-associated proteins, including type IV pilus proteins (PilA pilus and mannose-sensitive hemagglutinin [MSHA]), were implicated in the observed adherence to chitin. Interestingly, even in the absence of chitin, the expression levels of these attachment genes were significantly higher in E-genotype strains than C-genotype strains, suggesting that E-genotype strains have an inherent predisposition for adherence to chitin. From these findings, we proposed that C- and E-genotype strains have intrinsically divergent physiological programs which may help to explain the observed genotypic differences in oyster colonization and possibly pathogenic potential.
Collectively, these studies provided significant insight into the genotypic differences in environmental abundance and oyster colonization that have been documented. Here, we further elaborate on the molecular mechanisms that facilitate the differential chitin attachment of C- and E-genotype strains by revealing a role for quorum sensing (QS) in this process. Additionally, we explore how fluctuations in environmental conditions can influence chitin attachment and show that starvation and cold temperatures can alter the efficiency of attachment by V. vulnificus. Lastly, given the increasing prevalence of E-genotype wound isolates, we compare the chitin attachment efficiency of representative isolates and reveal that they behave more like C-genotype strains. Overall, these studies contribute to our understanding of the behavior of C- and E-genotype strains in the environment and reveal how environmental conditions can influence bacterial interactions with chitinous surfaces.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains of V. vulnificus used in this study are summarized in Table 1. Cultures were stored at −80°C in Bacto Luria broth (LB; BD Difco, NJ) containing 20% glycerol. All strains were grown in Bacto heart infusion (HI) broth for 24 h at 30°C with aeration. Strain JDO1 is a quorum-sensing, luxS mutant derived from parent strain C7184 and is unable to make 4,5-dihydroxy-2,3-pentanedione, an essential precursor to autoinducer-2 (AI-2). JDO1 was grown with 2 μg/ml chloramphenicol. JDO1 carries an insertional mutation in the luxS gene. The mutation was created via single-crossover homologous recombination using a plasmid that carries a truncated copy of the luxS gene as well as the gene encoding chloramphenicol resistance (33).
TABLE 1.
Strains used in this study
| Strain name | Source | Genotypesa | Lineageb | Genomic region XIIc |
|---|---|---|---|---|
| JY1701 | Environmental | A, E | II | − |
| JY1305 | Environmental | A, E | II | − |
| E64MW | Human wound | A, E | UNKd | − |
| LSU1657 | Human wound | A, E | UNK | UNK |
| CMCP6 | Human blood | B, C | I | + |
| MO6-24 | Human blood | B, C | I | + |
| C7184e | Clinical | B, C | I | + |
| JDO1 | luxS mutant | B, C | I | + |
Genotype groupings according to polymorphisms within the 16S rRNA gene (58) and the virulence-correlated gene (9), respectively.
Lineage groupings based on multilocus sequence typing analysis of six housekeeping genes (59).
+ and −, presence and absence of the 33-kb genomic island (region XII), respectively (59).
UNK, unknown.
Parental wild-type strain of the luxS mutant.
Chitin attachment assay.
Chitin attachment was performed as previously described (32). Briefly, chitin magnetic beads (New England BioLabs, MA) were washed twice by vortexing in 1/2-strength artificial seawater (ASW; 15 ppt). Bacterial cultures were washed twice in 1/2-strength ASW by centrifugation and added to the washed chitin beads at a final concentration of 5 × 107 cells/ml (final volume, 1 ml). The mixture was allowed to incubate at 20°C for 1 h on a rotisserie rotating at 8 rpm. The supernatant, which contained the unattached cells, was removed by placing the tube onto a 1.5-ml microcentrifuge magnetic stand (Life Technologies) and gently washing the beads three times with 1/2-strength ASW. The chitin beads, which harbored the attached cells, were suspended in 1 ml 1/2-strength ASW and prepared for quantification. To quantify the cells, 0.2 g of 0.5-mm ZR BashingBead lysis matrix was added to the washed chitin beads suspended in 1 ml 1/2-strength ASW, and the tube was vortexed vigorously for 60 s to detach the bound bacteria. The chitin beads were separated from the supernatant by using the magnetic stand, and the cell suspension containing the detached cells was serially diluted and plated onto HI agar.
Effect of quorum sensing on chitin attachment.
To test the role of quorum sensing on chitin attachment, the ability of the luxS mutant to attach to chitin was evaluated as described above. To test whether the addition of AI-2 to the luxS mutant would allow the mutant to attach to chitin similarly to the parent strain, the mutant was grown overnight in HI broth supplemented with 250 nM AI-2 prior to performing the chitin attachment assay. AI-2 was prepared as previously described (34). Chitin attachment was performed in 1/2-strength ASW supplemented with 250 nM AI-2. To test whether the addition of a quorum-sensing inhibitor (cinnamaldehyde) would cause the parent strain to attach to chitin at levels similar to those for the luxS mutant, the parent strain was grown overnight in HI broth supplemented with 150 μM cinnamaldehyde prior to performing the chitin attachment assay. Chitin attachment was performed in 1/2-strength ASW supplemented with 150 μM cinnamaldehyde.
Expression of type IV pilin genes by the luxS mutant.
To analyze the gene expression of the quorum-sensing luxS mutant relative to that of the wild-type strain, we performed relative quantitative reverse transcription-PCR (qRT-PCR) using methods previously described (32). Briefly, V. vulnificus strains C7184 and JDO1 were exposed to 15 ppt ASW for 1 h, after which the cells were subjected to RNA extraction using Qiagen's RNAprotect bacterial reagent with an RNeasy minikit and on-column DNase I treatment (Qiagen). After RNA extraction, the cells were subjected to a second DNase treatment using a Turbo DNA-free kit (Ambion). Endpoint PCR was performed on the RNA samples to confirm the complete removal of DNA (32).
Primers (Table 2) were designed, analyzed, and validated as previously described (32), and the PCR amplification efficiency of each primer set was validated by generating standard curves and evaluating the slope. To measure gene expression, 1 μg of total RNA was reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences), and 50 ng of cDNA template was used for quantitative PCR (qPCR). qRT-PCR was performed on three technical and two biological replicates for each sample using PerfeCTa SYBR green FastMix, Low ROX (carboxy-X-rhodamine; Quanta Biosciences). Water as a negative control and no-reverse-transcription controls were employed to rule out the influence of contaminants and residual genomic DNA, respectively. The expression levels of each gene were normalized using an endogenous control gene (DNA gyrase subunit B [gyrB]) to correct for sampling errors. Fold changes in the levels of gene expression for the luxS mutant relative to levels of expression for the wild-type strain were measured using the Pfaffl equation (35), taking into account the differences in PCR efficiencies between the primer sets.
TABLE 2.
Primers used in this study
| Gene | Primer target (orientationa) | Sequence (5′–3′) | Size of expected product (bp) |
|---|---|---|---|
| DNA gyrase subunit B | gyrB (F) | CTGAAGGGTCTGGATGCGG | 97 |
| gyrB (R) | GTGCCATCATCTGTGTCCCC | ||
| Type IV pilin protein subunit | pilA (F) | GCACAGCTCCAACCAGTAGT | 57 |
| pilA (R) | TTGGCGGCACTTCAACAATG | ||
| Type IV pilin protein prepilin peptidase | pilD (F) | TTGGCTTACTGGTAGGCAGC | 128 |
| pilD (R) | GGTTTCTGTCGGTGGTGTGA |
F, forward; R, reverse.
Effect of calcium on attachment to chitin.
To test the effect of calcium on the attachment efficiencies of C- and E-genotype V. vulnificus strains, strains were grown overnight in HI broth, and chitin attachment was performed as described above. To test whether calcium has an effect on attachment, we incubated cells with chitin in either normal 1/2-strength ASW (containing 0.018% Ca2+) or 1/2-strength ASW lacking Ca2+. Both ASW types were maintained at 15 ppt salinity. To test whether calcium has an effect on the chitin attachment of the luxS mutant, chitin attachment was performed for the mutant using ASW with and without calcium chloride.
Effect of starvation and low temperature on chitin attachment.
Cultures of strain C7184 were grown overnight in HI broth, washed, resuspended in 1/2-strength ASW, and allowed to attach to chitin as described above (control). Separate aliquots of this culture were left in ASW at 20°C and 4°C for 18 days to induce starvation and the VBNC state, respectively. The ability of each of these cultures to attach to chitin was assessed after 2 and 18 days of incubation under these conditions. Cells were determined to be in the VBNC state using methods previously described (36). Briefly, cell cultures were quantified every other day until V. vulnificus was no longer culturable on HI agar (when <10 CFU/ml was detectable). Cells were confirmed to be VBNC by successful resuscitation after the cultures were returned to 20°C for 24 h. For cultures that entered the VBNC state due to 4°C incubation (18 days), the chitin attachment assay was modified to allow the detection of VBNC cells, since they are not detectable on HI agar. Chitin attachment was performed as described above; however, after bead bashing to detach the cells that had adhered to the chitin beads, cultures were incubated in ASW (nutrient-deplete medium) at 20°C for 24 h to allow VBNC cells to resuscitate (to allow their detection on HI agar). Cells were then quantified by serial dilution and plating on HI agar.
Statistical analysis.
For culture-based quantification, attachment was expressed as a percentage, calculated from the total number of cells attached to the chitin beads (output) divided by the total number of cells added to the system (input) multiplied by 100. Data were analyzed by using unpaired Student's t test or one-way analysis of variance (ANOVA) followed by Tukey's post hoc test for multiple comparisons. Significance was determined by using a 95% confidence interval. All data were analyzed by using GraphPad Prism (version 5.0) software (GraphPad Software Inc.).
RESULTS AND DISCUSSION
QS reduces chitin attachment by negatively regulating expression of type IV pili.
Quorum sensing (QS) is the production and transduction of autoinducers, which are generated in an effort to gauge the population density in a certain environment or to evaluate the cells' environmental location (37). This leads to global changes in gene expression and has been shown to orchestrate multicellular behaviors in participating communities. Vibrios rely on quorum-sensing regulators to control stages of biofilm formation (38). In V. cholerae, cells initiate biofilm formation when they are in a low-cell-density state and allow quorum-sensing regulators to mediate natural maturation during biofilm development (38, 39). A similar phenomenon has been documented in V. vulnificus, in which the biofilm of a quorum-sensing mutant was initially robust but ultimately there was an early collapse of the biofilm architecture, illustrating that the QS system regulates proper biofilm development and maintenance (40).
Pili, such as type IV pili (MSHA and PilA), have repeatedly been implicated in facilitating cell-surface and cell-cell interactions in several Vibrio species, including V. vulnificus, and these cell surface structures mediate critical steps of attachment and biofilm formation (41–46). Previously, an inverse relationship between pili and quorum sensing has been suggested in V. vulnificus, since a QS mutant was shown to phenotypically exhibit significantly more pili and fimbriae than the wild-type strain (40).
Considering the suggested role of QS in pilus production and biofilm formation, we examined the ability of a V. vulnificus C-genotype QS mutant (deficient in the ability to produce AI-2) to attach to chitin particles and found a significant increase in attachment relative to that of the parental C-genotype strain (Fig. 1). Addition of synthetic AI-2 to the QS mutant restored the level of attachment to the wild-type level. Further, addition of an AI-2 inhibitor (cinnamaldehyde) to the wild-type strain significantly enhanced attachment to levels similar to the level for the QS mutant. Collectively, these results strongly suggest that the presence of AI-2 negatively affects chitin attachment for C-genotype strains. Further studies should be performed to compare what effect that AI-2 production has on E-genotype strains.
FIG 1.
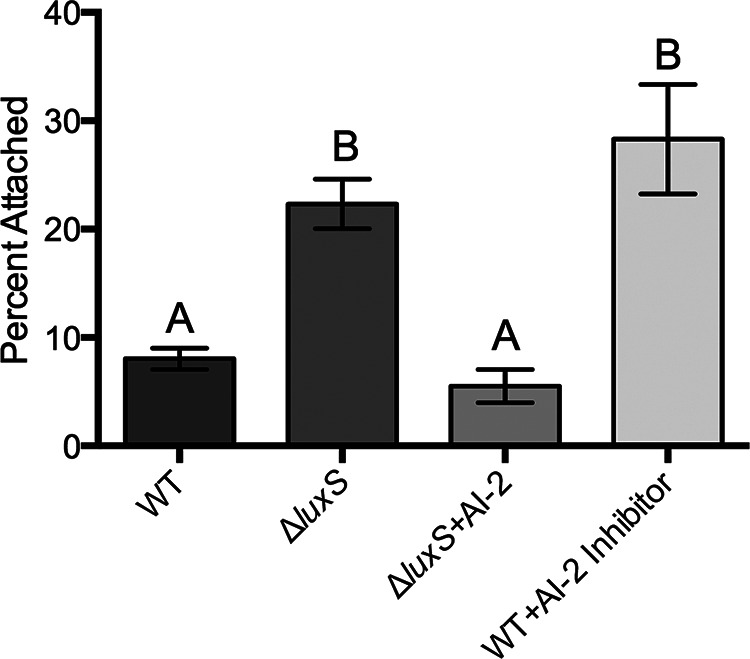
Effect of quorum sensing on chitin attachment. The attachment efficiencies of a C-genotype wild-type (WT) strain (C7184), a luxS quorum-sensing mutant (JDO1), the luxS mutant supplemented with the quorum-sensing molecule AI-2, and the parent strain supplemented with a quorum-sensing inhibitor are shown. Error bars represent the standard errors of the means from six replicates. Production of autoinducer-2 negatively influences chitin attachment. Different letters indicate statistically significant differences (one-way ANOVA, P < 0.0001).
Previously, we have shown that type IV pilin mutants exhibit significantly reduced chitin attachment (32). To determine if the involvement of QS in chitin attachment is an effect on type IV pili, we examined the expression of the type IV pilin genes pilA (pilin subunit A) and pilD (prepilin peptidase) in the QS mutant relative to that in the parental C-genotype strain. Figure 2 illustrates a significant upregulation of both pilA and pilD in the QS mutant relative to the level of regulation in the wild type; however, there was no change in the level of expression of mshA in the QS mutant (data not shown). These findings reveal that QS negatively regulates pilA and pilD expression in C-genotype strains. Collectively, our results suggest that the QS network either functions differently between C- and E-genotype strains or is regulated at different levels (i.e., C-genotype strains may produce more QS molecules than E-genotype strains). Further studies using an E-genotype QS mutant would help delineate how the QS system differs between these two genotypes. Such physiological differences are likely to contribute to the ecological success and pathogenic capabilities of these two genotypes.
FIG 2.
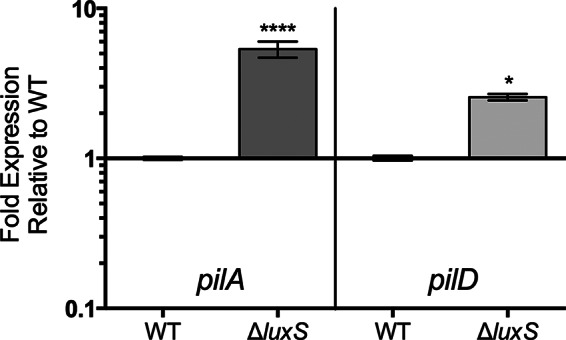
Expression of attachment genes by a quorum-sensing mutant relative to the level of expression by the wild-type strain (C7184). Error bars represent the standard errors from two biological and three technical replicates. The luxS mutant intrinsically expresses significantly higher levels of pilA and pilD than the wild-type strain. Asterisks represent statistically significant differences between the luxS mutant and the C-genotype parental strain for each gene of interest by using a one-way ANOVA. ****, P < 0.0001; *, P < 0.05.
Calcium deficiency impairs chitin attachment in E-genotype strains.
One of the environmental conditions that can vary in the natural aquatic habitats of V. vulnificus is the calcium level (47). Further, calcium levels have been shown to affect biofilm formation in V. cholerae (48). To observe the effect of calcium on chitin attachment, we allowed C- and E-genotype strains to attach to chitin in artificial seawater with or without calcium. While the absence of calcium appeared to have no impact on chitin attachment by C-genotype strains, a dramatic drop in attachment efficiency was observed in E-genotype strains (Fig. 3). Previous studies have shown that some pili possess a calcium-binding motif, and as a result, calcium can regulate type IV pilus-mediated adhesion (49, 50). To further assess this potential in V. vulnificus, we examined the effect of calcium on attachment by the QS mutant, which was previously shown to have enhanced chitin attachment and upregulation of type IV pilin genes. While the wild-type strain (C genotype) showed no change in chitin attachment in the presence or absence of calcium, we observed a significant drop in chitin attachment by the QS mutant in the absence of calcium (Fig. 4). Since calcium deficiency affected E-genotype strains and the QS mutant in a similar manner, and both produce higher levels of type IV pili than the parental C-genotype strain, this suggests that proper functioning of type IV pili is calcium dependent. Notably, calcium depletion did not completely abolish the attachment properties of C- and E-genotype strains, indicating that either (i) type IV pili are only partially impaired by calcium deficiency or (ii) additional Ca-independent surface structures also facilitate attachment to chitin, or both.
FIG 3.
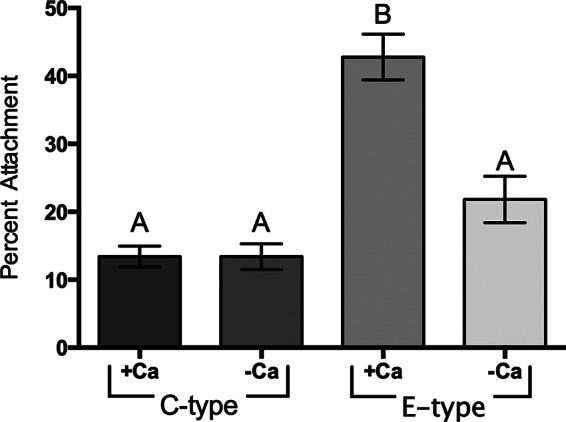
Effect of calcium on C- and E-genotype strain attachment to chitin. The chitin attachment efficiencies of two C-genotype strains (C7184 and MO6-24) and two E-genotype strains (JY1305 and JY1701) incubated in 1/2-strength ASW either with or without 0.018% (wt/vol) Ca2+ are shown. Error bars represent the standard errors of the means from three biological replicates for each strain and condition. Calcium enhances the efficiency of attachment of E-genotype strains to chitin but has no effect on the efficiency of C-genotype strain attachment. Different letters indicate statistically significant differences (one-way ANOVA, P < 0.0001).
FIG 4.
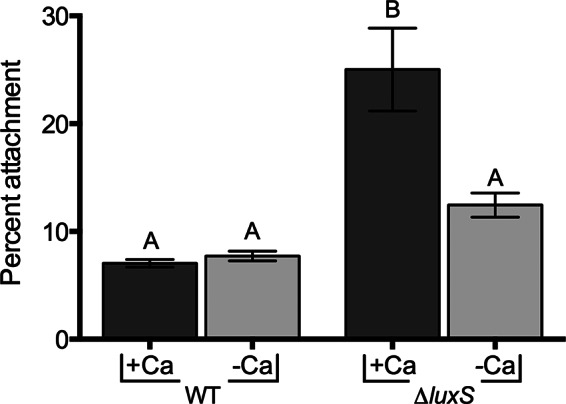
Effect of calcium on chitin attachment of the luxS mutant. The chitin attachment efficiencies of the wild-type strain (C7184) and the luxS quorum-sensing mutant incubated in 1/2-strength ASW either with or without 0.018% (wt/vol) Ca2+ are shown. Error bars represent the standard errors of the means from three biological replicates for each strain and condition. Calcium enhances the attachment of the luxS mutant to chitin but not the parental C-genotype strain. Different letters indicate statistically significant differences (one-way ANOVA, P = 0.0008).
Environmental fluctuations can alter chitin attachment efficiency.
Environmental conditions fluctuate considerably in estuarine environments; thus, bacterial inhabitants are perpetually faced with nutrient starvation, iron limitation, or changes in salinity and temperature. To cope with the feast-to-famine lifestyle, bacteria possess robust stress resistance mechanisms that help the organism adapt to these unpredictable instabilities (51). One such adaptation is the ability to attach to abiotic or biotic surfaces and subsequently build protective biofilms. In addition to the inherent stress resistance provided by biofilms, attachment to biotic substrates, such as chitin, can also provide direct access to nutrients (29, 52). Indeed, surface adhesion is often thought of as a survival strategy allowing bacteria to persist in nutrient-limited natural environments (52).
Previously, we demonstrated that environmental changes (such as a temperature upshift) can alter chitin attachment efficiency in C- and E-genotype strains of V. vulnificus (32). By increasing the attachment incubation temperature from 20°C to 37°C, we demonstrated a significant increase in chitin attachment by C-genotype strains. This enhanced ability to attach to chitin prompted us to further investigate how common environmental fluctuations, such as nutrient starvation and cold temperatures, can influence the attachment efficiency of V. vulnificus.
To investigate the role of nutrient starvation on V. vulnificus attachment to chitin, we incubated cells in artificial seawater (ASW) at room temperature (ca. 22°C) to induce starvation. At 2 and 18 days of starvation, cells were tested for their ability to attach to chitin particles. Figure 5 shows that cells were significantly more capable of attaching to chitin after 2 and 18 days of starvation than they were under nonstarvation conditions, although the attachment efficiency dropped at 18 days relative to that at 2 days.
FIG 5.
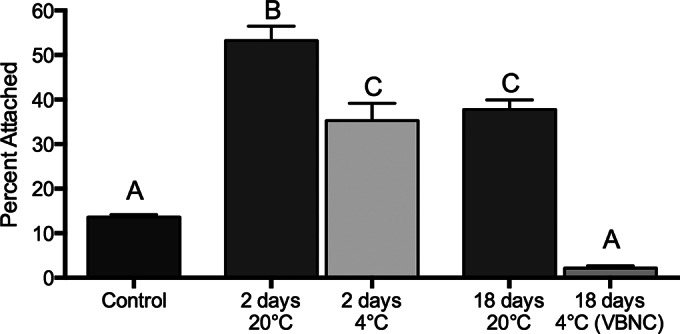
Effect of starvation and cold temperature (4°C) on chitin attachment. The chitin attachment efficiency of a C-genotype strain (C7184) during prolonged starvation in 1/2-strength ASW at 20°C or 4°C is shown. Cells were VBNC at day 18 at 4°C. Error bars represent the standard errors of the means from three biological replicates. Starvation increases the ability of V. vulnificus to attach to chitin. However, longer-term starvation (18 days) leads to decreased attachment to chitin relative to that after short-term starvation (2 days). There was significantly less chitin attachment by cells incubated at 4°C for 2 and 18 days than cells incubated at 20°C. Different letters indicate statistically significant differences (one-way ANOVA, P < 0.0001).
Environmental stress, such as nutrient deprivation, salinity fluctuations, and water temperature, can also induce a state of dormancy referred to as the viable but nonculturable (VBNC) state (53, 54). Cells that enter the VBNC state are still viable but lose culturability due to a reduction in metabolic activity. In this state of dormancy, cells exhibit notable changes in cellular properties and gene expression patterns. Our results show that, during exposure in ASW for 2 and 18 days at 4°C, cells retained the ability to attach to chitin, though the cells attached significantly less efficiently when they were in the VBNC state (Fig. 5). Reduced attachment by VBNC cells may be attributed to cell rounding, cell surface modifications, and reduced metabolism, which have been documented to take place in VBNC cells (see references 53 and 54 and references therein).
The VBNC state is recognized to be an important bacterial strategy for survival under adverse conditions, and here we show that despite a significant reduction in attachment ability, these cells still maintain the capability to attach to chitin. Both starvation-induced attachment and the VBNC state are ecologically relevant phenomena that likely promote the persistence of V. vulnificus in the estuarine environment. Furthermore, some chitin-binding ligands in V. cholerae have been shown to act as dual-role colonization factors, facilitating human intestinal colonization in addition to chitin attachment (55). Thus, the environmental cues that modulate chitin attachment can potentially have significant implications for the pathogenic potential of V. vulnificus.
Clinical E-genotype strains attach to chitin in a manner similar to that for C-genotype strains.
Recently, it has become increasingly apparent that a subset of E-genotype strains may have enhanced pathogenic potential relative to other environmentally isolated E-genotype strains. These E-genotype wound isolates are of particular interest, given the recent rise in wound isolates within the United States as well as in other countries. The genetic factors that distinguish this subset of E-genotype strains remain unknown; however, we recently demonstrated that these strains are capable of surviving in human serum in a manner nearly identical to that for C-genotype strains (13). In the current study, we assessed the ability of these wound isolates to attach to chitin and found that, again, these isolates behave more like C-genotype strains (Fig. 6). Furthermore, studies examining biofilm formation found that E-genotype wound isolates behave similarly to C-genotype strains, forming very poor biofilms, whereas E-genotype environmental isolates formed much more robust biofilms under the same conditions (data not shown). Considering the various lines of study, evidence suggests that E-genotype wound isolates phenotypically behave much like C-genotype strains, indicating that these strains may share some genetic features which can be attributed to their success in colonizing the human host. Previous studies have already identified some of these commonalities (16), and genome sequencing of multiple E-genotype wound isolates is under way in order to gain further insight into the relatedness of these strains.
FIG 6.
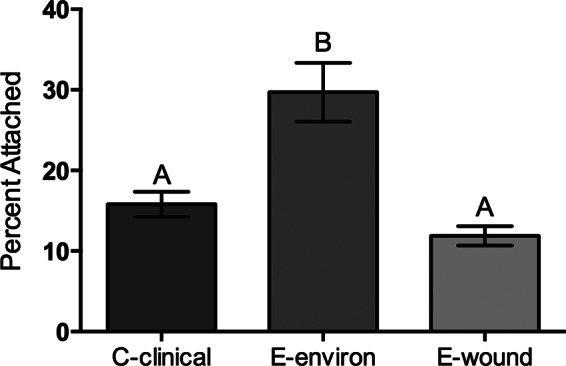
Effect of isolation source on V. vulnificus attachment to chitin. The attachment efficiencies of two clinically isolated C-genotype strains (MO6-24 and CMCP6), two environmentally isolated E-genotype strains (JY1701 and JY1305), and two E-genotype strains isolated from wound infections (E64MW and LSU1657) are shown. Error bars represent the standard errors of the means from three replicates per strain. Clinical isolates (C-clinical and E-wound) attach to chitin significantly less than environmentally isolated E-genotype (E-environ) strains. Different letters indicate statistically significant differences (one-way ANOVA, P = 0.0002).
Conclusion.
The primary goal of this study was to elaborate on previous studies investigating the interactions of C- and E-genotype strains with chitinous substrates, with the intent of obtaining a more thorough understanding of the molecular foundations. Previously, we revealed the involvement of type IV pili in facilitating chitin attachment in V. vulnificus, particularly in E-genotype strains. In this study, we identified quorum sensing to be a negative regulator of type IV pilus expression, as a C-genotype QS mutant displayed enhanced chitin attachment in a manner similar to that for E-genotype strains. These results suggest that the observed difference in chitin attachment properties between C- and E-genotype strains is a result of differential regulation somewhere within the QS pathway, which ultimately affects the level of type IV pilus expression. Further investigation of the molecular components involved in this pathway is needed to identify the root of this difference. We also found that calcium deficiency impairs chitin attachment in E-genotype strains and suggest that this reduction in chitin attachment is likely due to a defect in the function of type IV pili when calcium is not available.
Environmental parameters, such as increasing temperatures, have previously been shown to alter chitin attachment efficiency in V. vulnificus and V. cholerae (32, 56); thus, we further investigated how additional environmental fluctuations, such as nutrient starvation and cold temperatures, can influence chitin attachment. Both short-term (2-day) starvation and cold temperature incubation significantly enhanced V. vulnificus attachment to chitin, indicating that this biotic substrate may serve as a critical reservoir for V. vulnificus under suboptimal conditions. In the natural environment, planktonic cells must successfully navigate through the dynamic estuary toward a suitable niche. In the face of starvation or death, locating a chitinous oasis within a vast aquatic expanse can turn the tides for the organism by providing access to nutrients, protecting the organism from predators, and setting the stage for growth and biofilm formation. Even dormant cells of V. vulnificus retained their attachment properties (albeit to a lesser extent), despite the significant reduction in cell size and changes in cell wall structure. Overall, these results reveal that physical parameters can alter the attachment efficiency of V. vulnificus and these factors likely contribute to the environmental persistence of this organism.
Geographical regions which have experienced warming patterns have been linked with the emergence of Vibrio outbreaks (6, 57). Indeed, V. vulnificus infections have been increasing in the United States and worldwide, with wound infections becoming the more prevalent disease manifestation (6, 57). Despite genotypic schemes which correlate with pathogenicity, a subset of E-genotype strains has been isolated from wound infections. Previous studies demonstrated the ability of these strains to survive in human serum (13), and here we show again that these strains behave more like C-genotype strains than archetypal E-genotype strains. These results indicate that E-genotype wound isolates share some virulence characteristics with C-genotype strains, and identification of these features would likely aid our understanding of the pathogenic capabilities of V. vulnificus.
ACKNOWLEDGMENTS
We thank Matthew Parrow of the University of North Carolina at Charlotte for providing synthetic autoinducer-2.
This study is based on work supported by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, award no. 2009-03571.
Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Agriculture.
REFERENCES
- 1.Baker-Austin C, Lemm E, Hartnell R, Lowther J, Onley R, Amaro C, Oliver JD, Lees D. 2012. pilF polymorphism-based real-time PCR to distinguish Vibrio vulnificus strains of human health relevance. Food Microbiol 30:17–23. doi: 10.1016/j.fm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Oliver JD. 2006. Vibrio vulnificus, p 253–276. In Belkin S, Colwell RR (ed), Oceans and health: pathogens in the marine environment. Springer Science, New York, NY. [Google Scholar]
- 3.Oliver JD. 2006. Vibrio vulnificus, p 349–366. In Thompson FL, Austin B, Swings J (ed), The biology of vibrios. American Society for Microbiology, Washington, DC. [Google Scholar]
- 4.Oliver JD. 2012. Vibrio vulnificus: death on the half shell. A personal journey with the pathogen and its ecology. Microb Ecol 65:793–799. doi: 10.1007/s00248-012-0140-9. [DOI] [PubMed] [Google Scholar]
- 5.Oliver JD. 2005. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol Infect 133:383–391. doi: 10.1017/S0950268805003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, Martinez-Urtaza J. 2013. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat Clim Change 3:73–77. [Google Scholar]
- 7.Warner E, Oliver JD. 2008. Multiplex PCR assay for detection and simultaneous differentiation of genotypes of Vibrio vulnificus biotype 1. Foodborne Pathog Dis 5:691–693. doi: 10.1089/fpd.2008.0120. [DOI] [PubMed] [Google Scholar]
- 8.Warner JM, Oliver JD. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other vibrio species. Appl Environ Microbiol 65:1141–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol Immunol 49:381–389. doi: 10.1111/j.1348-0421.2005.tb03731.x. [DOI] [PubMed] [Google Scholar]
- 10.Senoh M, Miyoshi S, Okamoto K, Fouz B, Amaro C, Shinoda S. 2005. The cytotoxin-hemolysin genes of human and eel pathogenic Vibrio vulnificus strains: comparison of nucleotide sequences and application to the genetic grouping. Microbiol Immunol 49:513–519. doi: 10.1111/j.1348-0421.2005.tb03756.x. [DOI] [PubMed] [Google Scholar]
- 11.Aznar R, Ludwig W, Amann RI, Schleifer KH. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotide probes. Int J Syst Bacteriol 44:330–337. doi: 10.1099/00207713-44-2-330. [DOI] [PubMed] [Google Scholar]
- 12.Bogard RW, Oliver JD. 2007. Role of iron in human serum resistance of the clinical and environmental Vibrio vulnificus genotypes. Appl Environ Microbiol 73:7501–7505. doi: 10.1128/AEM.01551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams T, Ayrapetyan M, Ryan H, Oliver J. 2014. Serum survival of Vibrio vulnificus: role of genotype, capsule, complement, clinical origin, and in situ incubation. Pathogens 3:822–832. doi: 10.3390/pathogens3040822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HY, Ayrapetyan M, Oliver JD. 2014. Survival of Vibrio vulnificus genotypes in male and female serum, and production of siderophores in human serum and seawater. Foodborne Pathog Dis 11:119–125. doi: 10.1089/fpd.2013.1581. [DOI] [PubMed] [Google Scholar]
- 15.Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. 2012. Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553. doi: 10.1371/journal.pone.0037553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gulig PA, de Crecy-Lagard V, Wright AC, Walts B, Telonis-Scott M, McIntyre LM. 2010. SOLiD sequencing of four Vibrio vulnificus genomes enables comparative genomic analysis and identification of candidate clade-specific virulence genes. BMC Genomics 11:512. doi: 10.1186/1471-2164-11-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiaville PC, Bourdage KL, Wright AC, Farrell-Evans M, Garvan CW, Gulig PA. 2011. Genotype is correlated with but does not predict virulence of Vibrio vulnificus biotype 1 in subcutaneously inoculated, iron dextran-treated mice. Infect Immun 79:1194–1207. doi: 10.1128/IAI.01031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motes M, DePaola A, Cook D, Veazey J, Hunsucker J, Garthright W, Blodgett R, Chirtel S. 1998. Influence of water temperature and salinity on Vibrio vulnificus in northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl Environ Microbiol 64:1459–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaspar CW, Tamplin ML. 1993. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl Environ Microbiol 59:2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froelich BA, Williams TC, Noble RT, Oliver JD. 2012. Apparent loss of Vibrio vulnificus from North Carolina oysters coincides with a drought-induced increase in salinity. Appl Environ Microbiol 78:3885–3889. doi: 10.1128/AEM.07855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith B, Oliver JD. 2006. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol 72:1445–1451. doi: 10.1128/AEM.72.2.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randa MA, Polz MF, Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol 70:5469–5476. doi: 10.1128/AEM.70.9.5469-5476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson FL, Iida T, Swings J. 2004. Biodiversity of vibrios. Microbiol Mol Biol Rev 68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner E, Oliver JD. 2008. Population structures of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl Environ Microbiol 74:80–85. doi: 10.1128/AEM.01434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Froelich B, Ayrapetyan M, Oliver JD. 2013. Integration of Vibrio vulnificus into marine aggregates and its subsequent uptake by Crassostrea virginica oysters. Appl Environ Microbiol 79:1454–1458. doi: 10.1128/AEM.03095-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vezzulli L, Pezzati E, Repetto B, Stauder M, Giusto G, Pruzzo C. 2008. A general role for surface membrane proteins in attachment to chitin particles and copepods of environmental and clinical vibrios. Lett Appl Microbiol 46:119–125. [DOI] [PubMed] [Google Scholar]
- 27.Pruzzo C, Crippa A, Bertone S, Pane L, Carli A. 1996. Attachment of Vibrio alginolyticus to chitin mediated by chitin-binding proteins. Microbiology 142(Pt 8):2181–2186. doi: 10.1099/13500872-142-8-2181. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko T, Colwell RR. 1975. Adsorption of Vibrio parahaemolyticus onto chitin and copepods. Appl Microbiol 29:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalin DR, Daya V, Reid A, Levine MM, Cisneros L. 1979. Adsorption and growth of Vibrio cholerae on chitin. Infect Immun 25:768–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 31.Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. 2011. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front Microbiol 2:260. doi: 10.3389/fmicb.2011.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams TC, Ayrapetyan M, Oliver JD. 2014. Implications of chitin attachment for the environmental persistence and clinical nature of the human pathogen Vibrio vulnificus. Appl Environ Microbiol 80:1580–1587. doi: 10.1128/AEM.03811-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beam DM. 2004. The role of AI-2 in Vibrio vulnificus. M.S. thesis University of North Carolina at Charlotte, Charlotte, NC. [Google Scholar]
- 34.Ayrapetyan M, Williams TC, Oliver JD. 2014. Interspecific quorum sensing mediates the resuscitation of viable but nonculturable vibrios. Appl Environ Microbiol 80:2478–2483. doi: 10.1128/AEM.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitesides MD, Oliver JD. 1997. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl Environ Microbiol 63:1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Srinivas S, He H, Gong G, Dai C, Feng Y, Chen X, Wang S. 2013. Quorum sensing in Vibrio and its relevance to bacterial virulence. J Bacteriol Parasitol 4:172. [Google Scholar]
- 38.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Stirling FR, Zhu J. 2007. Temporal quorum-sensing induction regulates Vibrio cholerae biofilm architecture. Infect Immun 75:122–126. doi: 10.1128/IAI.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDougald D, Lin WH, Rice SA, Kjelleberg S. 2006. The role of quorum sensing and the effect of environmental conditions on biofilm formation by strains of Vibrio vulnificus. Biofouling 22:133–144. doi: 10.1080/08927010600691879. [DOI] [PubMed] [Google Scholar]
- 41.Paranjpye RN, Johnson AB, Baxter AE, Strom MS. 2007. Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Appl Environ Microbiol 73:5041–5044. doi: 10.1128/AEM.00641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paranjpye RN, Strom MS. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect Immun 73:1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiavelli DA, Marsh JW, Taylor RK. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol 67:3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frischkorn KR, Stojanovski A, Paranjpye R. 2013. Vibrio parahaemolyticus type IV pili mediate interactions with diatom-derived chitin and point to an unexplored mechanism of environmental persistence. Environ Microbiol 15:1416–1427. doi: 10.1111/1462-2920.12093. [DOI] [PubMed] [Google Scholar]
- 45.Shime-Hattori A, Iida T, Arita M, Park KS, Kodama T, Honda T. 2006. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol Lett 264:89–97. doi: 10.1111/j.1574-6968.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- 46.Watnick PI, Fullner KJ, Kolter R. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol 181:3606–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riley JP, Tongudai M. 1967. The major cation/chlorinity ratios in sea water. Chem Geol 2:263–269. doi: 10.1016/0009-2541(67)90026-5. [DOI] [Google Scholar]
- 48.Bilecen K, Yildiz FH. 2009. Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11:2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng Y, Johnson MD, Burillo-Kirch C, Mocny JC, Anderson JE, Garrett CK, Redinbo MR, Thomas CE. 2013. Mutation of the conserved calcium-binding motif in Neisseria gonorrhoeae PilC1 impacts adhesion but not piliation. Infect Immun 81:4280–4289. doi: 10.1128/IAI.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutz C, Erken M, Noorian P, Sun S, McDougald D. 2013. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol 4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dawson MP, Humphrey B, Marshall K. 1981. Adhesion: a tactic in the survival strategy of a marine vibrio during starvation. Curr Microbiol 6:195–199. doi: 10.1007/BF01566971. [DOI] [Google Scholar]
- 53.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 54.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol. 43(Spec No):93–100. [PubMed] [Google Scholar]
- 55.Vezzulli L, Guzman CA, Colwell RR, Pruzzo C. 2008. Dual role colonization factors connecting Vibrio cholerae's lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr Opin Biotechnol 19:254–259. doi: 10.1016/j.copbio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Stauder M, Vezzulli L, Pezzati E, Repetto B, Pruzzo C. 2010. Temperature affects Vibrio cholerae O1 El Tor persistence in the aquatic environment via an enhanced expression of GbpA and MSHA adhesins. Environ Microbiol Rep 2:140–144. doi: 10.1111/j.1758-2229.2009.00121.x. [DOI] [PubMed] [Google Scholar]
- 57.Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J. 2010. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ Microbiol Rep 2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 58.Nilsson WB, Paranjype RN, DePaola A, Strom MS. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J Clin Microbiol 41:442–446. doi: 10.1128/JCM.41.1.442-446.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen ALV, Oliver JD, DePaola A, Feil EJ, Fidelma Boyd E. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl Environ Microbiol 73:5553–5565. doi: 10.1128/AEM.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


