ABSTRACT
The generation of neuronal morphology requires transport vesicles originating from the Golgi apparatus (GA) to deliver specialized components to the axon and dendrites. Drosophila Arfaptin is a membrane-binding protein localized to the GA that is required for the growth of the presynaptic nerve terminal. Here we provide biochemical, cellular and genetic evidence that the small GTPase Arl1 and the guanine-nucleotide exchange factor (GEF) Gartenzwerg are required for Arfaptin function at the Golgi during synapse growth. Our data define a new signaling pathway composed of Arfaptin, Arl1, and Garz, required for the generation of normal synapse morphology.
KEY WORDS: Arfaptin, Synapse growth, Golgi apparatus, Small GTPase
INTRODUCTION
Polarized cells, such as neurons, require highly specialized secretory pathways to grow and maintain their distinct functional domains, and failure of these processes is associated with neuronal dysfunction and pathology (Fan et al., 2008). Current data support the fundamental and conserved requirement for the Golgi apparatus (GA) during the growth and development of dendritic arbors and presynaptic nerve terminals (Ehlers, 2013; Horton et al., 2005; Mehta et al., 2005; Murthy et al., 2003; Ye et al., 2007; Zhou et al., 2014). The dysfunction of the secretory pathway and altered neuronal morphology are common cellular etiologies in motor neurons during lower motor diseases such as amyotropic lateral sclerosis (ALS). This includes lower motor diseases linked to mutations in the Dctn1 and Dhc genes, which encode components of the dynactin complex. The phenotypic analysis of disease models, as well as human patients, harboring mutations in the components of the dynactin complex suggest that dysfunction of the GA is important etiology of lower motor disease (Chang et al., 2013; Gonatas et al., 2006; Laird et al., 2008). Currently, the role of the dynactin complex at the GA in motor neurons is incompletely described and the link between dynactin complex function and motor disease remains unclear.
Drosophila Arfaptin (Arfip), the homologue of mammalian Arfaptin2, is a BAR domain protein that it required for normal growth of the Drosophila NMJ during larval development (Chang et al., 2013). Arfaptins have been implicated in vesicle formation at the GA and in the cell periphery (Mim et al., 2012; Peter et al., 2004). In Drosophila, Arfip is enriched in the nervous system where it mediates binding of the dynactin complex to the membranes of the neuronal GA consistent with the established requirement of the dynein motor for normal Golgi function and vesicle transport (Caviston and Holzbaur, 2006; Chang et al., 2013; Fath et al., 1994; Mim et al., 2012; Vaughan, 2005; Vaughan et al., 2002). Thus Arfip represents a central component of a vesicle transport system required for normal growth of the NMJ. Here we extend these results by presenting data supporting that the small GTPase Arl1 and the guanine-exchange factor (GEF) Gartenzwerg (Garz) support Arfip function at the GA during synapse growth. Our data support a model in which Arl1 and Garz work in the same genetic pathway to regulate the recruitment of Arfip to Golgi membranes in the motor neuron.
RESULTS AND DISCUSSION
The small GTPase Arl1 regulates Arfip membrane localization in Drosophila
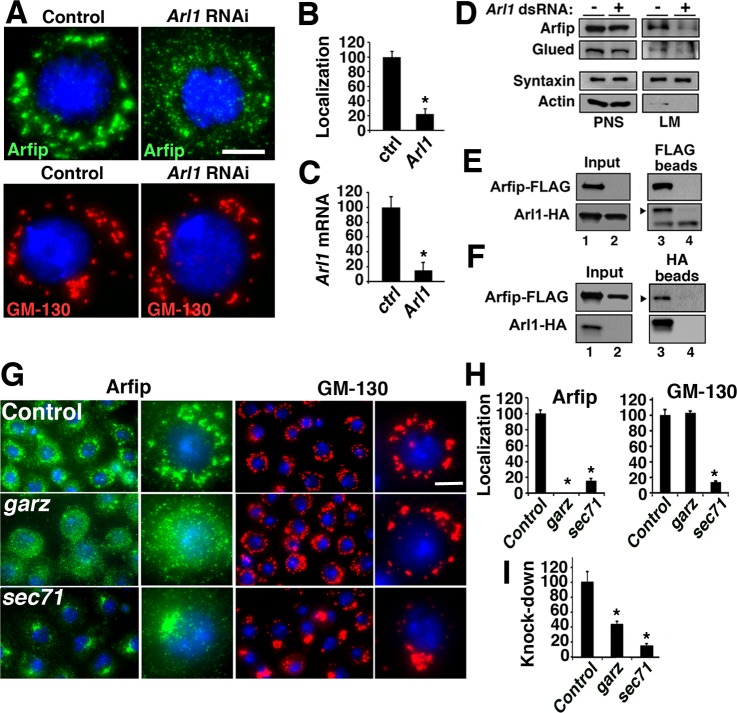
Recent results from mammalian tissue culture cells support that the Arf-like 1 (Arl1) protein regulates the interaction of mammalian Arfaptin1 and Arfaptin2 with the Golgi (Man et al., 2011). Analysis of protein sequence shows that Drosophila and human Arl1 share 78% identity in their amino acids, suggestive of well-conserved protein function between species. The Drosophila S2 cell provides a simple system to investigate the GA localization of Arfip which we will exploit to identify molecules involved in the regulation of Arfaptin function (Chang et al., 2013). In agreement with the mammalian data, perinuclear distribution of endogenous Arfip in S2 cells was abolished in cells treated with dsRNA for Arl1 (Fig. 1A,B; Chang et al., 2013). In these analyses, we observed an ∼90% reduction in Arl1 mRNA of cells treated with Arl1 dsRNA (Fig. 1C). Arl1 RNAi had no effect on the localization on GM-130 supporting that Arl1 is not required for general Golgi structure in S2 cells (Burguete et al., 2008; Lu and Hong, 2003; Panic et al., 2003; Setty et al., 2003; Wu et al., 2004). We hypothesized that the effects of Arl1 RNAi on the Arfip staining pattern was due to reduced Arfip membrane binding and not a reduction in Arfip proteins levels (Chang et al., 2013). Consistent with this hypothesis, cells treated with Arl1 dsRNA did not have a significant reduction in Arfip protein levels in post-nuclear supernatants (Fig. 1D, PNS) but did show a significant reduction of membrane-bound Arfip and Glued (Fig. 1D, LM; Chang et al., 2013).
Abbreviations
- NMJ
neuromuscular junction
- GA
Golgi apparatus
- GEF
guanine exchange factor
- MTOC
microtubule organizing center
Fig. 1.
Arl1 and Gartenzwerg are required for Arfip localization to the Golgi. (A) Control and Arl1 dsRNA-treated S2 cells stained with antibodies for either Arfip or GM-130 show that knockdown of Arl1 disrupts Arfip localization but not GM-130. (B) Graph represents the percent of treated cells with disrupted Arfip staining. (C) Graph represents the effectiveness of Arl1 RNAi determined using quantitative RT-PCR. Relative expression was determined by the ratio of message to alpha-tubulin normalized to the no dsRNA control. (D) Panel of representative immunoblots of membrane flotation analysis of S2 cells treated with Arl1 dsRNA or control dsRNA. Syntaxin and actin were markers used to monitor fractionation of light membranes (LM) and soluble proteins (PNS, post-nuclear supernatant), respectively. (E,F) Immunoblots of co-immunoprecipitation analysis from S2 cells co-expressing FLAG-Arfip and Arl1-HA fusion proteins (E and F; lane 1) incubated with either (E) anti-FLAG or (F) anti-HA antibody-coated beads. Incubation with anti-FLAG beads precipitated Arl1-HA (E; lane 3, arrowhead) and incubation with anti-HA beads precipitated Arfip-FLAG (F; lane 3, arrowhead). Lysate from S2 cells only expressing Arl1-HA (E; lane 2) when incubated with FLAG beads showed no binding of Arl1-HA to anti-FLAG beads (E; lane 4, arrowhead). Similarly, FLAG-Arfip only lysate incubated with anti-HA beads show no binding to anti-HA beads (F; lane 4, arrowhead). Cell lysate of S2 cells co-expressing FLAG-Arfip and Arl1-HA (F; lane 1) or either protein alone (F; lane 2) shows nearly equivalent amount of protein in each co-immunoprecipitation experiment. (G) Immunofluorescence images of fixed S2 cells stained with either anti-Arfip (green) or anti-GM130 (red) antibody. Nuclei are indicated by DAPI staining (blue). The cells were incubated with control dsRNA (GFP) or dsRNA targeting all Arf-like guanine-nucleotide exchange factors (GEFs) in Drosophila (see Table 1). (H) Graphs represent the percent of cells with disrupted Arfip (left graph) or GM130 (right graph) staining compared to control. (I) Graphs represent the effectiveness of garz and sec71 RNAi determined using quantitative RT-PCR. Relative expression was determined by the ratio of message to alpha-tubulin normalized to the no dsRNA control. Error bars=s.e.m. *P<0.01; Student's t-test (B,C) or ANOVA (H,I). Scale bars =5 μm.
The results above predict that Arl1 and Arfip should physically bind to each other. Co-immunoprecipitation (Co-IP) experiments using cell lysates prepared from S2 cells co-expressing HA-tagged Arl1 and FLAG-tagged Arfip incubated with anti-FLAG antibody coated beads finds that the anti-FLAG coated beads can successfully co-precipitate both the Arfip-FLAG target and Arl1-HA (arrowhead; Fig. 1E, lane 3). Cell lysate made from cells only expressing Arl1-HA (Fig. 1E, lane 2) failed to immunoprecipitate Arl1-HA when incubated with anti-FLAG antibody coated beads (Fig. 1E, lane 4) demonstrating the requirement of Arfip-FLAG for the co-immunoprecipitation of Arl1-HA. Similarly, cell lysate prepared from S2 cells co-expressing HA-tagged Arl1 and FLAG-tagged Arfip incubated with anti-HA antibody successfully co-immunoprecipitated Arl1-HA and Arfip-FLAG (arrowhead; Fig. 1F, lane 3). Control cell lysate prepared form S2 cells only expressing FLAG-tagged Arfip (Fig. 1F, lane 2) incubated with anti-HA antibody coated beads failed to immunoprecipitate the FLAG-tagged Arfip (Fig. 1F, lane 4). These results demonstrate that Arl1 and Arfip are contained within the same biochemical complex and support the assertion that Arl1 regulates the binding and localization of Arfip to Golgi membranes in Drosophila neurons (Man et al., 2011).
RNAi screen for guanine-exchange factors required for Arfip localization identifies Gartenzwerg and Sec71
The cycling between active GTP-bound forms and inactive GDP-bound forms regulates the function of small GTPases, such as Arl1. The activation of small GTPases is largely controlled by guanine-exchange factors (GEFs) that serve to exchange a GDP for a GTP. To identify the GEF responsible for the localization of Arfip to the Golgi, we performed an RNAi screen in S2 cells with dsRNAs targeting all of the identified genes encoding GEFs in Drosophila to identify dsRNAs that altered the normal localization of endogenous Arfip. We expected that RNAi knockdown of the appropriate GEF would phenocopy of the effects of Arl1 RNAi (Fig. 1A). An RNAi screen of all putative GEFs found that RNAi knockdown of gartenzwerg (garz) and Sec71 resulted in the disruption of normal Arfip localization, although the effects of Sec71 RNAi on Arfip localization were strikingly different than the effects of garz RNAi (Fig. 1G,H; Table 1). Specifically, RNAi of Sec71 resulted in the redistribution of both Arfip and GM-130 to a juxtanuclear localization within the cell (Fig. 1G-I). This is consistent with a broader role of Sec71 for the regulation of normal Golgi structure. In contrast to the effects of Sec71 knockdown, RNAi knockdown of garz phenocopied the effect of Arl1 RNAi on both Arfip and GM-130 localization (Fig. 1G-I). These data supports that Garz is required for Arfip localization to the Golgi in Drosophila.
Table 1.
RNAi analysis of Arfip localization

Co-localization of Arl1 and Arfip in cells and neurons
Our model is that Garz and Arl1 represent components of a GTPase signaling system required for normal Arfip function in the motor neuron. This model predicts that these proteins should colocalize in neurons (Chang et al., 2013). Ventral nerve cords (VNCs) from larvae expressing transgenic Arl1-HA using the panneuronal C155-Gal4 driver were fixed and processed for immunofluorescent microscopy using antibodies against endogenous Arfip and HA. Microscopic 3D imaging of motor neurons reveals that Arl1-HA co-localization with endogenous Arfip in neurons (Fig. 2A-F), similar to what we observe in S2 cells. We further confirmed these data by performing voxel by voxel co-localization analysis on 3D images to generate a Mander's Overlap Coefficient (MOC), which is a quantitative measure of co-localization (Chang et al., 2013). These analyses resulted in an average MOC value consistent with high amounts of co-localization between Arl1 and Arfip (1=perfect colocalization) (Fig. 2J). These localization data are similar to the colocalization of Arl1 and Arfaptin2 in mammalian cells (Man et al., 2011). To investigate the colocalization of Garz with Arfip we employed Garz-GFP expressed in motor neurons using the OK6-Gal4 driver and observed a similar co-localization with endogenous Arfip in the soma of the neuron (Fig. 2G-I). The Mander's Overlap Coefficients for Arfip and Garz-GFP in these analyses was consistent with significant colocalization of Arfip with Garz-GFP within the soma of the motor neurons (Fig. 2J). These data demonstrate that both Arl1 and Garz colocalize with Arfip at the GA in motor neurons.
Fig. 2.
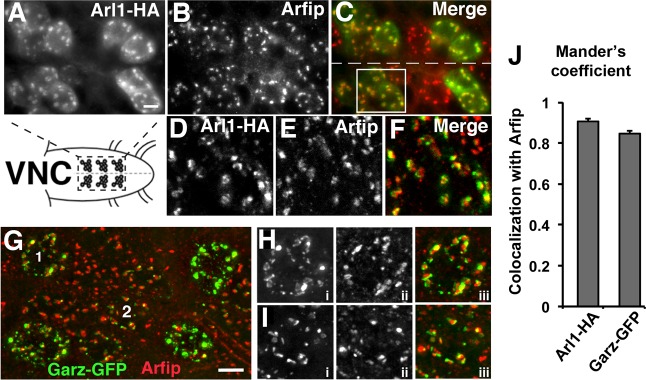
Colocalization of Arl1, Garz, and Arfip at the Golgi in motor neurons. (A-F) Images of indicated portion ventral nerve cord harboring motor neuron soma from a 3rd instar larvae expressing Arl1-HA using the C155-Gal4 line in the motor neurons co-stained for (A) Arl1-HA using the anti-HA antibody and for (B) Arfip using the anti-Arfip antibody. The merged image of panels A and B is shown in panel C. Dashed line indicates the midline. (D-F) A deconvolved image of the boxed area in C is shown demonstrating the high degree of colocalization of Arl1-HA (D; green in F) and Arfip (E; red in F) in the motor neuron soma. (G) Deconvolved immunofluorescent image of motor neuron soma in larvae expressing Garz-GFP in motor neurons using the OK6-Gal4 line co-stained with anti-GFP (green) and anti-Arfip (red). (H,I) Higher magnification images of soma 1 (H) and soma 2 (I) indicated in G. Panels i are Garz-GFP, panels ii are Arfip, and panels iii are the merged image. (J) Graphs represent the average Mander's coefficient from neuronal cell bodies comparing Arl1-HA with endogenous Arfip (left bar) or Garz-GFP with endogenous Arfip (right bar). n=12–14 soma from 3–5 VNCs. Scale bars=10 μm.
Arl1, Garz, and Arfip function in the same genetic pathway during synapse growth
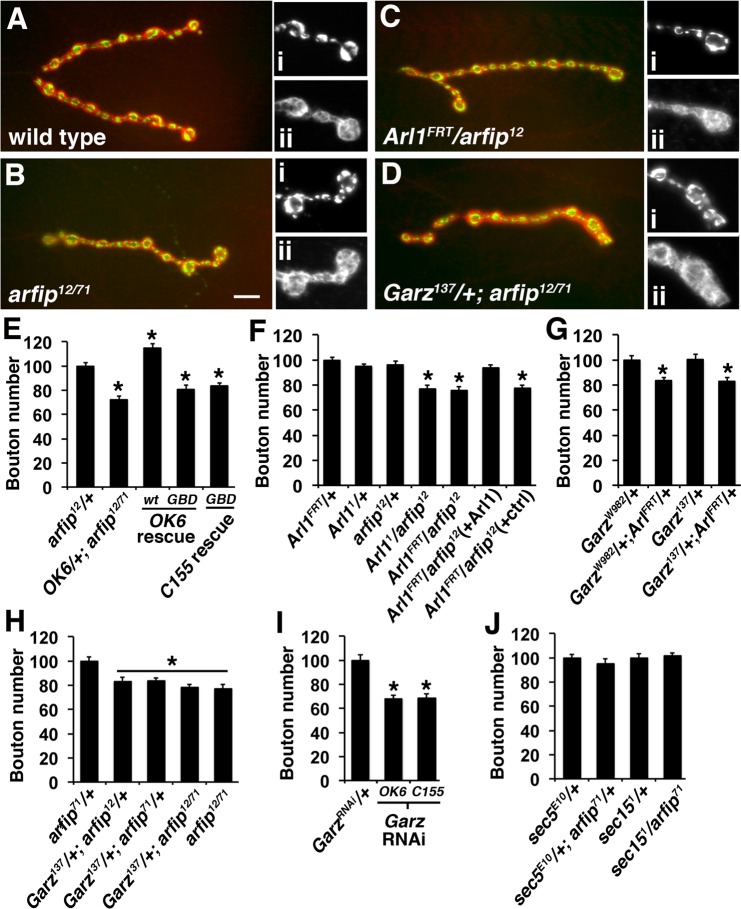
All genetic mutations used in the following studies are strong loss of function/null alleles (see Materials and Methods). We find that neuronal overexpression of an Arfip molecule with a mutated GTPase binding domain (GBD) fails to restore synapse growth in arfip mutants compared to the neuronal overexpression of wild type Arfip, which we have found results in neuronal overgrowth (Fig. 3E) (Chang et al., 2013). We confirmed this result using two independent Gal4 driver lines, the motor neuron-specific OK6-Gal4, and the panneuronal C155-Gal4 (Sanyal, 2009). These results demonstrate a role for GTPase signaling during Arfip-dependent synapse growth. These genetic analyses were extended to investigate if Garz and Arl1 regulate Arfip activity during synapse growth. Because null mutants in both Arl1 and garz are embryonic lethal (Armbruster and Luschnig, 2012; Tamkun et al., 1991; Wang et al., 2012) we have used trans-heterozygotic backgrounds to investigate whether Arl1, garz, and arfip function in the same genetic pathway during synapse growth. All alleles used in the subsequent studies have been previously characterized as representing amorphic alleles (see Methods). We observe that trans-heterozygotic combinations of Arl1FRT, Arl11, and arfip12 mutations have reduced synapse growth as determined by counting the number of boutons at the neuromuscular junction (NMJ) formed on 3rd Instar larval muscle 4 (Fig. 3A-F). Expression of Arl1-HA under control of the OK6-Gal4 in the Arl1FRT/arfip12 trans-heterozygote background restored bouton numbers to control values (Fig. 3F). These effects on synapse growth are consistent with arfip, and Arl1 functioning within the same genetic pathway during synapse growth. Reduced synapse growth phenotype is also observed in trans-heterozygotic combinations of Arl1FRT with both garz137 and garzW982 (Fig. 3G). A similar genetic interaction is observed between either arfip12or arfip71 mutations and the garz137 mutation (Fig. 3D,H). Further, removal of a single copy of garz in an arfip mutant background does not result in an enhancement of the synapse growth defect observed in arfip mutants supporting that Garz functions in the same pathway with Arfip during synapse growth (Fig. 3D,G). These results demonstrate that garz, Arl1, and arfip function in the same genetic pathway within the neuron during synapse growth. In support of this model, neuron specific RNAi of garz results in a reduction in synapse growth (Fig. 3H).
Fig. 3.
Arl1, Garz, and Arfip function in the same genetic pathway during synapse growth. (A-D) Immunofluorescent images of the neuromuscular junctions (NMJs) formed on muscle 4 from larvae of the indicated genotypes co-stained for presynaptic VGluT (green; insets i) and postsynaptic Discs-large (Dlg) (red; insets ii). Insets are higher magnification images of the terminal boutons of the synapse shown. Scale bar=10 μm. (E-I) Graphs represent the average number of boutons at the muscle 4 NMJs normalized to control values. (E) The expression of wild type Arfip (wt) but not a GTPase-binding mutant Arfip (GBD) rescued synapse growth defects in arfip12/71 mutant neurons using either the Ok6-Gal4 (Ok6 rescue) or C155-Gal4 (C155 rescue). (F) Transheterozygotic genotypes consisting of Arl11 and arfip12, Arl1FRT and arfip12 have reduced synapse growth compared to heterozygotic Arl1 controls (Arl11/+ and Arl1FRT/+). Expression of the pUAS-Arl1-HA transgene in the Arl1FRT/arfip12 heterozygotic background using the OK6-Gal4 driver [Arl1FRT/arfip12 (+Arl1)] restores synapse growth to control levels compared to the control background harboring only the OK6-Gal4 driver [Arl1FRT/arfip12 (+ctrl)]. (G) Transheterozygotic genotypes consisting of Arl1FRT and garzW982 or of Arl1FRT and garz137 have reduced synapse growth compared to heterozygotic garz controls (garzW982/+ and garz137/+). (H) Transheterozygotic genotypes containing heterozygotic mutations in arfip (arfip12 or arfip71) and garz137 have reduced synapse growth compared to arfip/+. (I) The expression of a UAS construct encoding a dsRNA to garz (UAS-garzRNAi) in motor neurons using either the Ok6-Gal4 motor neuron driver or the C155-Gal4 panneuronal driver results in reduced synapse growth. (J) Transheterozygotic genotypes between mutations in arfip71 and the exocyst genes sec5E10 or sec151 have no effect on synapse growth. Error bars=s.e.m. *P<0.05; significant difference from controls by ANOVA.
Our data here demonstrate that Arl1 is required for Arfip membrane binding, Arfip Golgi localization, and synapse growth. Because of the proposed role of BAR domain proteins during vesicle biogenesis (Campelo and Malhotra, 2012), we propose that Arfip binding to GTP-bound Arl1 facilitates the biogenesis of transport vesicles required for the growth of the nerve terminal. Consistent with this model is the observation that garz RNAi leads to both the loss of Arfip staining from the GA and the reduction in synapse growth at the NMJ. Further, we observe a reduction of synapse growth in trans-heterozygotic combinations between garz mutations and both Arl1 and arfip mutations. These data are consistent with reduction of Garz function resulting in decreased levels of Arl1-GTP and reduced Arfip function during synapse growth.
Previous data has shown that the exocyst complex, known to function during vesicle formation at the Golgi, is required for the delivery of synaptic proteins, such as synaptotagmin, to the nerve terminal (Murthy et al., 2003). The synapse growth pathway defined by Arfip, Arl1, and Garz likely functions independent of the exocyst complex as no synergistic effects were observed in transheterozygotic combinations between arfip mutations and mutations in the sec5 or sec15 genes (Fig. 3I). These data support that arfip represents a distinct pathway originating from the GA that is also required for normal synapse growth. Interestingly, there is no functional consequence of arfip mutations on synaptic vesicle release as determined quantal analysis of evoked release which is similar to the phenotype of sec5 mutants (Table 2) (Murthy et al., 2003). Our data further support that membrane trafficking required for normal neuronal growth and development are distinct from the pathways supporting neurotransmitter release, which in part has been attributed to a distinct class of Golgi-derived vesicle referred to as Piccolo-Bassoon transport vesicles (PTVs) (Shapira et al., 2003). The utilization of GTPase signaling systems could provide the biochemical diversity to generate the distinct vesicle classes required for the growth and function of the neuron.
Table 2.
Electrophysiological analysis of neurotransmission in arfaptin mutants
MATERIALS AND METHODS
Fly stocks
All fly stocks were maintained on a standard fly food (Bloomington Stock Center) at 25°C and 50% humidity. The following gartenzwerg (garz) stocks were used in this study: garz137 (small deletion generated by transposon excision), garzW982 (EMS-generated point mutation) and UAS-GFP-garz (Armbruster and Luschnig, 2012; Wang et al., 2012). Both garz mutations are strong loss of function/null alleles. The pUAS-garzRNAi line was obtained from the Bloomington Stock Center (stock# 31232). The Arl11allele (EMS-generated point mutation) is a strong loss of function/null allele (Tamkun et al., 1991). The Arl1FRT deletion was made using Flippase-mediated recombination following the protocol found at the DrosDel website (http://www.drosdel.org.uk/) using the following P-element insertions: P{XP}d07562 and PBac{WH}f02582. The resulting deletion completely removes the Arl1 gene as well as the following neighboring genes: CG10516, CG17029, CG6025 (Arl1), CG5942, CG17028, CG5949, CG17026, CG17027. The pUAS-Arl1-HA gene was generated by PCR cloning from the GM20805 cDNA (Drosophila Genomics Resource Center, Bloomington IN). To generate the pUAS-arfipGBD-HA vector, a pUASt-ArfipWT-HA vector was modified by inserting mutations H170A/Q172A/H173A into the GTPase binding pocket to abolish binding of small GTPases without altering membrane binding (Tarricone et al., 2001; TOP Gene Technologies, Montreal, Quebec, Canada). All transgenic lines were generated by microinjection into w1118 (Rainbow Transgenics, Camarillo, CA).
dsRNA interference in S2 cells
S2 cells were incubated with 10 μg/ml of dsRNA for the respective targets for five days and the media were changed daily. At the end of the RNAi treatment, cells were fixed and processed for Arfip localization using a rabbit anti-Arfip antibody. The effectiveness of the RNAi on Arfip and GM-130 localization was determined by quantifying the number of cells with correct localizaion from 10 randomly chosen fields of view (∼500 cells total). To verify knock-down, real-time PCR using Syb-R Green (Stratagene) with gene specific primers (Table 1).
Cell transfection and co-immunoprecipitations
A polyclonal S2 cell line that constitutively expresses Gal4 was used for all experiments and maintained as previously described (Chang et al., 2013). Transient transfection of S2 cells was performed using a calcium phosphate based method as described in the Drosophila Expression System Kits manual (Invitrogen). For co-immunoprecipitation (co-IP) two 80% confluent 10 cm dishes of cells co-transfected with appropriate plasmids were homogenized in cold IP buffer [IPB; 50 mM Tris (7.4), 50 mM NaCl, 1% TX-100, 1 mM MgCl2, and protease inhibitors]. Cell lysate was incubated with anti-FLAG or anti-HA coated beads (Sigma-Aldrich, St. Louis, MO USA) and samples processed as previously described (Chang et al., 2013). The following primary antibodies were used for all immunoblot analysis: rabbit anti-Arfip (1:1000; Chang et al., 2013), mouse anti-HA (1:1000; US Biologicals, Salem, MA USA), rabbit anti-FLAG (1:1000; Sigma-Aldrich). All secondary antibodies were used at a 1:10,000 dilution. Blots were exposed to autoradiographic film and visualized using the ECL Chemiluminesence kit (Amersham, Piscataway, NJ USA).
Membrane flotation
For flotation analysis, 3–4 10 cm dishes of Arfip dsRNA-treated cells and non-dsRNA-treated control cells were homogenized in homogenization buffer (8% sucrose and 3 mM imidazole, pH 7.4). A post nuclear supernatant was obtained by centrifuging at 1000×g for 7 min in standard tabletop microcentrifuge. The resulting PNS was brought to 40% sucrose, bottom loaded, and overlaid with two cushions of 35 and 8% sucrose. The gradient was centrifuged at 28,000 rpm for 1 h in a TH641 rotor. Light membranous organelles (LM) and soluble proteins were recovered and equal amounts of protein from each fraction were analyzed by SDS-PAGE and immunoblotting using the following antibodies: rabbit anti-Arfip (1:1000; Chang et al., 2013), mouse anti-syntaxin (1:500; Developmental Studies Hybridoma Bank, Iowa City, IA USA), mouse anti-actin (1:500, Sigma-Aldrich).
Microscopy
All fixed images were captured with an Orca-2 back-cooled CCD camera attached to a Zeiss Axiovert 200M. Samples were fixed for 10 min using 4% paraformaldehyde in 1× PBS followed by permeabilization with 1× PBS/0.1% TX-100. The following primary antibodies were used in these studies: rabbit anti-GM-130 (1:500; Cell Signaling Technology, Danvers, MA USA), mouse anti-Discs-large (1:400; Developmental Studies Hybridoma Bank), rabbit anti-VGlut (1:10,000; Chang et al., 2013), rabbit anti-Arfip (1:500; Chang et al., 2013), mouse anti-HA (1:500; US Biologicals), rabbit anti-FLAG (1:1000; Sigma-Aldrich), and mouse anti-FLAG (1:500; Sigma-Aldrich). All secondary antibodies were used at 1:400 dilution (Life Technologies, Grand Island, NY USA). For Mander's Overlap Coefficient (MOC) analysis z-stack images of S2 cells or larval motor neurons were generated at 160× and deconvolved using a constrained iterative algorithm that utilizes a theoretical point-spread function as previously described (Chang et al., 2013). The deconvolution and colocalization analyses are built in features of the Slidebook software controls our imaging system (Intelligent Imaging Inc., Denver, CO USA). For analysis of synaptic growth, fixed 3rd Instar larval NMJs from indicated genotypes were processed for immunofluorescent microscopy with antibodies to Drosophila VGluT and Discs-large. The number of boutons was determined at the NMJ on muscle 4 of segment A3 of the larvae. Bouton counts were normalized to muscle size and control genotype.
Electrophysiology
Recordings were performed under standard conditions as described previously (Eaton et al., 2002). Briefly, recordings were performed on muscle 6 of abdominal segment 4 in HL3 (Ca2+ – 2 mM, Mg2+ – 20 mM). Only muscles that had a resting membrane potential <−60 mV were used for data analysis. Data was acquired with a PowerLab 4/30 (ADInstruments) and LabChart7 Pro (ADInstruments). EJPs and mEJPs were measured with Mini Analysis (Synaptosoft). For each muscle 50 mEJPs and 10 compound EJPs were measured to calculate the average values. Unadjusted quantal content is reported here.
Statistical analysis
Statistical significance was determined by a Student's t-test for pair-wise comparison, and comparison of more than two conditions utilized ANOVA with a Bonferroni correction. All values are presented as an average with the error bars representing s.e.m. P values less that 0.05 were considered significant.
Acknowledgements
The authors would like to thank Yimin Wu for excellent technical support on this manuscript. An RO1 award to B.A.E. (NS062811) from the National Institute of Neurological Disorders and Stroke supported this work. The Grant T32-AG021890 from the National Institute of Aging supported L.C.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
L.C. and B.A.E. designed research. L.C., T.K.-P. and B.A.E. performed research. L.C. and B.A.E. analyzed data. L.C and B.A.E. wrote the paper.
Funding
This research was funded by NIH grants NS062811 to B.A.E. and T32-AG021890 to L.C.
References
- Armbruster K. and Luschnig S. (2012). The Drosophila Sec7 domain guanine nucleotide exchange factor protein Gartenzwerg localizes at the cis-Golgi and is essential for epithelial tube expansion. J. Cell Sci. 125, 1318-1328. 10.1242/jcs.096263 [DOI] [PubMed] [Google Scholar]
- Burguete A. S., Fenn T. D., Brunger A. T. and Pfeffer S. R. (2008). Rab and Arl GTPase family members cooperate in the localization of the Golgin GCC185. Cell 132, 286-298. 10.1016/j.cell.2007.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F. and Malhotra V. (2012). Membrane fission: the biogenesis of transport carriers. Annu. Rev. Biochem. 81, 407-427. 10.1146/annurev-biochem-051710-094912 [DOI] [PubMed] [Google Scholar]
- Caviston J. P. and Holzbaur E. L. F. (2006). Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 16, 530-537. 10.1016/j.tcb.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Chang L., Kreko T., Davison H., Cusmano T., Wu Y., Rothenfluh A. and Eaton B. A. (2013). Normal dynactin complex function during synapse growth in Drosophila requires membrane binding by Arfaptin. Mol. Biol. Cell 24, 1749-1764. S1-S5 10.1091/mbc.E12-09-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos G., Schroeder AJ., Goodman JL., Strelets VB., Crosby MA., Thurmond J., Emmert DB., Gelbart WM.; the FlyBase Consortium (2015) FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43, D690-D697. 10.1093/nar/gku1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton B. A., Fetter R. D. and Davis G. W. (2002). Dynactin is necessary for synapse stabilization. Neuron 34 (5), 729-741. 10.1016/S0896-6273(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Ehlers M. D. (2013). Dendritic trafficking for neuronal growth and plasticity. Biochem. Soc. Trans. 41, 1365-1382. 10.1042/BST20130081 [DOI] [PubMed] [Google Scholar]
- Fan J., Hu Z., Zeng L., Lu W., Tang X., Zhang J. and Li T. (2008). Golgi apparatus and neurodegenerative diseases. Int. J. Dev. Neurosci. 26, 523-534. 10.1016/j.ijdevneu.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Fath K. R., Trimbur G. M. and Burgess D. R. (1994). Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J. Cell Biol. 126, 661-675. 10.1083/jcb.126.3.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatas N. K., Stieber A. and Gonatas J. O. (2006). Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J. Neurol. Sci. 246, 21-30. 10.1016/j.jns.2006.01.019 [DOI] [PubMed] [Google Scholar]
- Horton A. C., Rácz B., Monson E. E., Lin A. L., Weinberg R. J. and Ehlers M. D. (2005). Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron 48, 757-771. 10.1016/j.neuron.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Laird F. M., Farah M. H., Ackerley S., Hoke A., Maragakis N., Rothstein J. D., Griffin J., Price D. L., Martin L. J. and Wong P. C. (2008). Motor neuron disease occurring in a mutant dynactin mouse model is characterized by defects in vesicular trafficking. J. Neurosci. 28, 1997-2005. 10.1523/JNEUROSCI.4231-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. and Hong W. (2003). Interaction of Arl1-GTP with GRIP domains recruits autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol. Biol. Cell 14, 3767-3781. 10.1091/mbc.E03-01-0864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man Z., Kondo Y., Koga H., Umino H., Nakayama K. and Shin H.-W. (2011). Arfaptins are localized to the trans-Golgi by interaction with Arl1, but not Arfs. J. Biol. Chem. 286, 11569-11578. 10.1074/jbc.M110.201442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S. Q., Hiesinger P. R., Beronja S., Zhai R. G., Schulze K. L., Verstreken P., Cao Y., Zhou Y., Tepass U., Crair M. C. et al. (2005). Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron 46, 219-232. 10.1016/j.neuron.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Mim C., Cui H., Gawronski-Salerno J. A., Frost A., Lyman E., Voth G. A. and Unger V. M. (2012). Structural basis of membrane bending by the N-BAR protein endophilin. Cell 149, 137-145. 10.1016/j.cell.2012.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy M., Garza D., Scheller R. H. and Schwarz T. L. (2003). Mutations in the exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron 37, 433-447. 10.1016/S0896-6273(03)00031-X [DOI] [PubMed] [Google Scholar]
- Panic B., Perisic O., Veprintsev D. B., Williams R. L. and Munro S. (2003). Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell 12, 863-874. 10.1016/S1097-2765(03)00356-3 [DOI] [PubMed] [Google Scholar]
- Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J. G., Evans P. R. and McMahon H. T. (2004). BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495-499. 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]
- Sanyal S. (2009). Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr. Patterns 9, 371-380. 10.1016/j.gep.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Setty S. R. G., Shin M. E., Yoshino A., Marks M. S. and Burd C. G. (2003). Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 13, 401-404. 10.1016/S0960-9822(03)00089-7 [DOI] [PubMed] [Google Scholar]
- Shapira M., Zhai R. G., Dresbach T., Bresler T., Torres V. I., Gundelfinger E. D., Ziv N. E. and Garner C. C. (2003). Unitary assembly of presynaptic active zones from Piccolo-Bassoon transport vesicles. Neuron 38, 237-252. 10.1016/S0896-6273(03)00207-1 [DOI] [PubMed] [Google Scholar]
- Tamkun J. W., Kahn R. A., Kissinger M., Brizuela B. J., Rulka C., Scott M. P. and Kennison J. A. (1991). The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc. Natl. Acad. Sci. USA 88, 3120-3124. 10.1073/pnas.88.8.3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C., Xiao B., Justin N., Walker P. A., Rittinger K., Gamblin S. J. and Smerdon S. J. (2001). The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411, 215-219. 10.1038/35075620 [DOI] [PubMed] [Google Scholar]
- Vaughan K. T. (2005). Microtubule plus ends, motors, and traffic of Golgi membranes. Biochim. Biophys. Acta 1744, 316-324. 10.1016/j.bbamcr.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Vaughan P. S., Miura P., Henderson M., Byrne B. and Vaughan K. T. (2002). A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J. Cell Biol. 158, 305-319. 10.1083/jcb.200201029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Meyer H., Ochoa-Espinosa A., Buchwald U., Onel S., Altenhein B., Heinisch J. J., Affolter M. and Paululat A. (2012). GBF1 (Gartenzwerg)-dependent secretion is required for Drosophila tubulogenesis. J. Cell Sci. 125, 461-472. 10.1242/jcs.092551 [DOI] [PubMed] [Google Scholar]
- Wu M., Lu L., Hong W. and Song H. (2004). Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 11, 86-94. 10.1038/nsmb714 [DOI] [PubMed] [Google Scholar]
- Ye B., Zhang Y., Song W., Younger S. H., Jan L. Y. and Jan Y. N. (2007). Growing dendrites and axons differ in their reliance on the secretory pathway. Cell 130, 717-729. 10.1016/j.cell.2007.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Chang J., Wang X., Savelieff M. G., Zhao Y., Ke S. and Ye B. (2014). GM130 is required for compartmental organization of dendritic Golgi outposts. Curr. Biol. 24, 1227-1233. 10.1016/j.cub.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]