ABSTRACT
Tropomyosin is a coiled-coil protein that binds and regulates actin filaments. The tropomyosin gene in Schizosaccharomyces pombe, cdc8, is required for formation of actin cables, contractile rings, and polar localization of actin patches. The roles of conserved residues were investigated in gene replacement mutants. The work validates an evolution-based approach to identify tropomyosin functions in living cells and sites of potential interactions with other proteins. A cdc8 mutant with near-normal actin affinity affects patch polarization and vacuole fusion, possibly by affecting Myo52p, a class V myosin, function. The presence of labile residual cell attachments suggests a delay in completion of cell division and redistribution of cell patches following cytokinesis. Another mutant with a mild phenotype is synthetic negative with GFP-fimbrin, inferring involvement of the mutated tropomyosin sites in interaction between the two proteins. Proteins that assemble in the contractile ring region before actin do so in a mutant cdc8 strain that cannot assemble condensed actin rings, yet some cells can divide. Of general significance, LifeAct-GFP negatively affects the actin cytoskeleton, indicating caution in its use as a biomarker for actin filaments.
KEY WORDS: Actin cytoskeleton, Schizosaccharomyces pombe, Fission yeast, cdc8, Actin binding protein, LifeAct, Myosin, Fimbrin
INTRODUCTION
Actin-based cellular mechanisms in cytokinesis, intracellular transport, and establishment of cellular polarity in eukaryotes are universal. The deciphering of signaling pathways that regulate the cytoskeleton has been a focus whereas regulation at the terminal machinery has received less attention. Tropomyosin is a core actin regulatory protein, known for its role in regulating muscle contraction (Lehman and Craig, 2008), and is found in most eukaryotes. It is a two-chained α-helical coiled-coil protein that binds end-to-end along the length of both sides of the actin filament. In this position, tropomyosin regulates actin cytoskeleton dynamics. It stabilizes the actin filament and protects it against the actions of DNase I (Hitchcock et al., 1976), cofilin (Bernstein and Bamburg, 1982; DesMarais et al., 2002; Nagaoka et al., 1995; Nakano and Mabuchi, 2006; Nishida et al., 1984; Ono and Ono, 2002), and gelsolin (Pruliere et al., 1986). It inhibits Arp2/3 complex nucleation of branched actin filaments (Blanchoin et al., 2001). Tropomyosin protects the pointed, slow-growing end of the filament, alone (Broschat, 1990; Broschat et al., 1989) and with tropomodulin in vertebrate cells (Colpan et al., 2013; Kostyukova and Hitchcock-DeGregori, 2004; Kostyukova et al., 2005; Weber et al., 1994; Yamashiro et al., 2012). Tropomyosin is a positive regulator of formin function at the barbed, fast-growing end of the filament (Skau et al., 2009; Ujfalusi et al., 2009, 2012; Wawro et al., 2007) and competes with and inhibits actin crosslinking proteins, including α-actinin, filamin, and fimbrin (Abe and Maruyama, 1973; Drabikowski and Nowak, 1968; Maruyama and Ohashi, 1978; Skau et al., 2011; Skau and Kovar, 2010; Zeece et al., 1979). Tropomyosin regulates motility by making the interaction of myosin with actin positively- or negatively-cooperative, depending on the myosin and tropomyosin isoforms (Barua et al., 2014; Bremel and Weber, 1972; Clayton et al., 2010, 2012, 2015; Fanning et al., 1994; Hodges et al., 2012; Lehrer and Morris, 1982; Stark et al., 2010; Tang and Ostap, 2001). We reached this understanding of tropomyosin regulatory functions primarily based on in vitro experiments. Translation to an in vivo venue has been challenging because of the redundancy of the animal proteome. The presence of four genes encoding more than 40 isoforms in mammals (Geeves et al., 2014) makes genetic and cellular studies in vertebrates, as well as invertebrates, a challenge.
While the cytoskeletal proteins that have been investigated in yeast have homologs in most eukaryotes, tropomyosin has been identified only in animals and fungi (Ophisthokonts), but not plants, amoebae, slime molds or other protists (Barua et al., 2011; Cranz-Mileva et al., 2013). Budding yeast has two tropomyosin genes, TPM1 and TPM2 (Drees et al., 1995; Liu and Bretscher, 1989). Disruption of TPM1 results in loss of actin cables and interruptions in the secretory pathway (Liu and Bretscher, 1989, 1992). Disruption of TPM2 has no detectable phenotype but is lethal in combination with disruption of TPM1 (Drees et al., 1995).
Fission yeast has a single, essential tropomyosin gene, cdc8 (Balasubramanian et al., 1992). Disruption of the gene to create a null mutant results in the absence of actin cables, depolarization of actin patches, inability to form the actin-containing contractile ring leading to failure of cytokinesis (Balasubramanian et al., 1992), and inability to form mating tubes for fusion (no zygote formation) (Kurahashi et al., 2002). Essentially the same phenotype is observed in cdc8ts mutants at the restrictive temperature (Chang et al., 1996). Tropomyosin is required for contractile ring integrity during contraction (Mishra et al., 2013). Extensive analysis of the function of actin dynamics and assembly of actin-containing structures in fission yeast provides the context for in vivo study of structure-function relationships in tropomyosin (Kovar et al., 2011).
The fission yeast cytoskeleton uses mechanisms that are conserved in most eukaryotes for processes that include cytokinesis, intracellular transport, and establishment of cellular polarity (Mishra et al., 2014). Schizosaccharomyces pombe has emerged as a model organism for study of these processes because its simpler genome encodes a smaller and less redundant proteome than in mammals, the facility of genetic manipulation, and the amenability of the cytoskeleton to microscopic study in living cells.
For these reasons we directed our attention to development of an evolution-based molecular-genetic approach of functional analysis in fission yeast as a way to dissect the molecular basis of known and unknown functions of tropomyosin in a living cell. The overarching hypothesis is that residues required for conserved tropomyosin functions are conserved. The approach follows from our evolutionary analysis of mammalian tropomyosins using in vitro functional assays of conserved functions including actin binding and myosin regulation (Barua et al., 2011, 2012, 2013, 2014).
In earlier work (Cranz-Mileva et al., 2013) we identified the evolutionarily-conserved codons in fungal tropomyosins, and we screened a series of cdc8 Ala or Thr mutations at conserved sites on the coiled coil surface for the ability to rescue the growth and cellular phenotype of a cdc8ts mutant at the restrictive temperature. While all rescued growth, certain mutations affected actin cable organization, contractile ring formation, actin patch polarization and cellular shape. We selected sites of interest and created three gene replacement strains carrying mutations at two or three sites in the cdc8 gene. All three strains were isolated as diploids and the mutations severely reduced the in vitro affinity of recombinant tropomyosin for filamentous actin in two of the three, limiting interpretation of the results.
Here we analyze a series of gene replacement mutants that are viable as haploids, and studied the effects of the mutations on actin affinity and organization of the actin cytoskeleton and associated proteins. While all the mutants are able to divide (since they are viable) and mate, the mutations result in one or more of the following cytoskeletal phenotypes: altered actin cable morphology, abnormal or incomplete actin contractile ring assembly, depolarization of actin patches, and deficiencies in vacuole fusion. The type of cytoskeleton alteration depends on the mutation, indicating that specific functions depend on particular tropomyosin residues, some of which are in putative actin binding sites based on homology with mammalian tropomyosins. When cdc8 mutants were crossed with strains expressing cytoskeleton proteins with fluorescent protein tags, in some cases synthetic effects were observed, inferring interaction of the mutated site(s) on cdc8 with the cytoskeletal protein.
RESULTS
A cdc8 mutation that inhibits assembly of actin cytoskeletal structures enables localization of early contractile ring components
The most severe mutant in our proteomic screen of the effect of mutations at conserved sites in cdc8 on cellular morphology and function was cdc8R121A.D131A.E138A (Cranz-Mileva et al., 2013). It was the least effective mutant, among those studied, in rescuing a temperature-sensitive mutant (cdc8-27) at the restrictive temperature. The cell shape was abnormal, the actin cables, contractile ring and septum were poorly organized, and there was irregular polarization of actin patches. A gene replacement strain, cdc8R121A.D131A.E138A, isolated as a homozygous diploid, had many of the same cellular features. However, the mutant maintained some cellular function compared to a temperature-sensitive mutant that was unable to assemble any actin cytoskeletal structures or to divide at the restrictive temperature. The in vitro actin affinity of recombinant Cdc8pR121A.D131A.E138A was too weak to measure.
In order to identify the contributions of the mutations at individual residues mutated in cdc8R121A.D131A.E138A to tropomyosin function, we made gene replacement strains with single site mutations (cdc8R121A [SH39, SH40], cdc8D131A [SH43], cdc8E138A [SH34]; supplementary material Table S1) and expressed recombinant protein in E. coli with the single site mutations (Cdc8pR121A, Cdc8pD131A, Cdc8pE138A). All three mutants were isolated as haploid strains and have normal growth parameters with the exception of cdc8R121A that could not be accurately measured (supplementary material Fig. S1). The recombinant proteins were expressed with the N-acetylation mimic, AlaSer, at the N terminus (Monteiro et al., 1994) since unacetylated Cdc8p binds poorly to actin (Cranz-Mileva et al., 2013; Maytum et al., 2000). Cdc8pR121A binds actin with ∼30-fold weaker affinity than wildtype, and Cdc8pD131A and Cdc8pE138A have close to wildtype affinity (Fig. 1A). The effect of the mutations on stability is minimal (Fig. 1C).
Fig. 1.
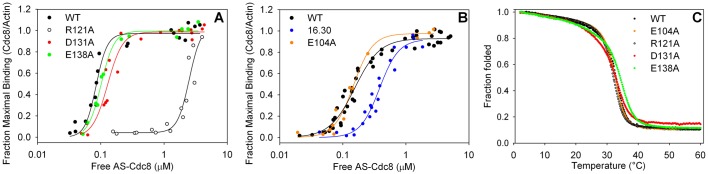
Actin affinity and thermal stability of wildtype and mutant fission yeast tropomyosins. (A,B) Actin affinity of AS-Cdc8p, wildtype and mutants, measured by cosedimentation as described in Materials and Methods (20 mM MOPS pH 7.0, 150 mM NaCl, 2 mM MgCl2, 5 µM actin). (A) AS-Cdc8pwt, Kapp=11.8×106 M−1 (n=3); AS-Cdc8pR121A, Kapp=0.40×106 M−1 (n=2); AS-Cdc8pD131A, Kapp=7.8×106 M−1 (n=3); AS-Cdc8pE138A, Kapp=9.9×106 M−1 (n=2). (B) AS-Cdc8pwt, Kapp=6.6×106 M−1 (n=2); AS-Cdc8pD16A.L30A, Kapp=2.6×106 M−1 (n=3); AS-Cdc8pE104A, Kapp=6.8×106 M−1 (n=2). The binding experiments in A and B were done at different times with different actin preparations. The data for AS-Cdc8pD16A.L30A are reproduced from (Cranz-Mileva et al., 2013) in which the wildtype Kapp=6.3×106 M−1 (n=3). (C) Thermal stability determined by measuring the ellipticity at 222 nm between 0–60°C. The ellipticity at 2°C is normalized to 1. The melting temperature (TM) is defined as the temperature where the normalized ellipticity is 0.5. The observed TM (n=1) are: AS-Cdc8pwt=33°C, AS-Cdc8pD16A.L30A=34°C (Cranz-Mileva et al., 2013); AS-Cdc8pE104A=33°C, AS-Cdc8pR121A=32°C, AS-Cdc8pD131A=33.0°, AS-Cdc8pE138A=34°.
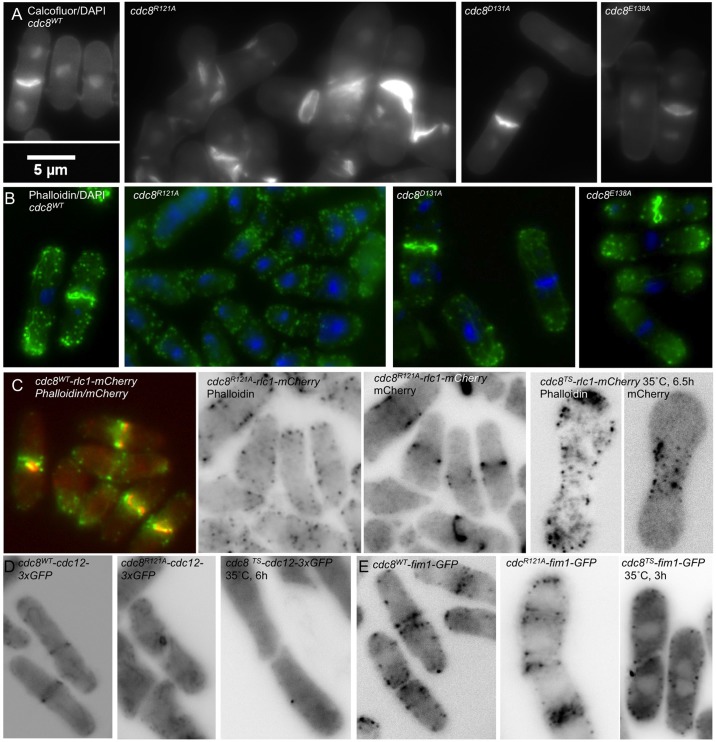
Of the three strains, the actin cytoskeleton is severely affected in cdc8R121A, while the D131A and E138A mutations have little effect. Fig. 2 shows the septum with Calcofluor, nuclei with DAPI (Fig. 2A) and actin cytoskeleton visualized with Alexa-phalloidin (Fig. 2B). While the actin cytoskeleton and septa are apparently normal in cdc8D131A and cdc8E138A, these features are disrupted in cdc8R121A resulting in the near absence of actin cables and dispersed actin patches. Alexa-phalloidin staining in the midline region indicates the presence of actin filaments, but they do not condense into contractile rings (Fig. 2B,C). Septal material accumulates but does not form a discrete septum (Fig. 2A). Tropomyosin is expressed in cdc8R121A but it is localized in patches rather than the contractile ring and cables as in wildtype or other mutants (supplementary material Fig. S2). Even though the effect of the R121A mutation on cellular morphology is severe, some cells can divide as evidenced by isolation and maintenance of a viable haploid strain.
Fig. 2.
Morphology and cytoskeleton organization of cdc8wt, cdc8R121A, cdc8D131A, cdc8E138A. (A) Fixed wildtype (SH30) and mutant cells stained with Calcofluor to show septum and septal material and DAPI to show nuclei. The cdc8R121A cells (SH39) had disorganized septa and abnormal cell shapes. Cells with up to four nuclei were observed. cdc8D131A (SH43) and cdc8E138A (SH34) appeared normal. (B) Staining of the actin cytoskeleton using Alexa-phalloidin (green), and nuclei with DAPI (blue) showed dispersed patches, the absence of actin cables and poorly condensed actin rings in cdc8R121A. The other strains appeared normal. (C) cdc8wt-rlc1-mCherry (SH49) and cdc8R121A-rlc1-mCherry (SH52) cells fixed and stained with Alexa-phalloidin (green) to visualize the actin cytoskeleton. m-Cherry (red) shows localization of myosin II in the contractile ring in cdc8wt. In cdc8R121A-rlc1-mCherry myosin II assembles into contractile rings despite the absence of organized actin in contractile rings or cables. For comparison, in cdc8-27-rlc1-mCherry (SH81), a temperature-sensitive mutant, neither the actin (Alexa phalloidin) nor myosin II (rlc1-mCherry) assembled into a contractile ring at the restrictive temperature (35°C). Other cells were grown and fixed at 30°C. (D) Formin-GFP (Cdc12p-GFP) fluoresence. Formin localized in rings in wildtype (SH57) but, like Rlc1p-mCherry, formin (Cdc12p) assembled into abnormal rings in cdc8R121A (SH60) (live cells). No midline localization was observed in a temperature sensitive strain (SH87) at the restrictive temperature (fixed cells at 35°C). (E) Fimbrin-GFP fluorescence (live cells confocal image). Fim1p-GFP localized in the midline region in cdc8wt (SH77) and cdc8R121A cells (SH75) (live cells), but not in the temperature-sensitive strain (SH83) at the restrictive temperature (cells grown and fixed at 35°C). The localization of fluorescent proteins was normal in cdc8D131A and cdc8E138A strains (supplementary material Fig. S3).
Since cdc8R121A can divide without formation of a discrete actin-containing contractile ring, we investigated the localization of proteins that are found in the area of the ring and known to be regulated by tropomyosin in fission yeast and/or mammalian cells: myosin II, fimbrin, and formin (Cdc12p) (Kovar et al., 2011). We created wildtype and mutant cdc8 strains expressing Rlc1p-mCherry (regulatory light chain of myosin II [Myo2p, Myp2p], SH49 cdc8wt, SH52 cdc8R121A), Fim1p-mEGFP (fimbrin, SH77 cdc8wt, SH75 cdc8R121A), Cdc12p-3xGFP (formin, SH57 cdc8wt, SH60 cdc8R121A) and Act1-LifeAct-GFPp, a live cell marker for filamentous actin (Riedl et al., 2008) (SH136, cdc8R121A).
We were surprised that in cdc8R121A Rlc1p-mCherry localizes to form an organized but misshapen contractile ring that closes when the cell divides (Fig. 2C). Cdc12p-3xGFP, if observed, appears to form a contracted but asymmetrically-localized ring, reminiscent of a ring that contracted without being attached to the plasma membrane (Fig. 2D). Finally, Fim1p-mEGFP localizes in the midline area in a cortical pattern that is patchy rather than ring-like (Fig. 2E). The R121A mutant retains some function compared to a cdc8-27, a temperature-sensitive mutant that is unable to initiate the first stages of contractile ring assembly at the restrictive temperature: Rlc1p-mCherry and Fim1p-mEGFP have no evident localization at the restrictive temperature (Fig. 2C,E) and Cdc12p-3xGFP is seen as a single dot (Fig. 2D). The cdc8R121A strain has an apparent mating deficiency with ain1-mEGFP (JW1144). In several attempts asci formed that were filled with a single dark mass, but discrete spores were never seen, indicative of a synthetic negative effect of the cdc8 R121A mutation and GFP modification of α-actinin.
Mirroring the wildtype-like appearance of the actin cytoskeleton in cdc8D131Aand cdc8E138A cells, strains expressing Rlc1p-mCherry (SH54 cdc8D131A, SH56 cdc8E138A), Fim1p-mEGFP (SH74 cdc8D131A, SH75 cdc8E138A) and Cdc12p-3xGFP (SH62 cdc8D131A, SH64 cdc8E138A), Ain1p-mEGFP (SH76 cdc8D131A, SH77-A cdc8E138A) all form normal contractile rings (supplementary material Fig. S3). Normal localization of tropomyosin in the contractile ring and actin cables (supplementary material Fig. S2) and of Myo52p-3xYFP at the poles in both cdc8D131A and cdc8E138A (supplementary material Fig. S3) provides further evidence of the apparent neutral effect of the cdc8 D131A and E138A mutations on these processes. The microtubule cytoskeleton, visualized in mRFP-atb2 strains is normal for all mutants in our study (supplementary material Fig. S3).
A conserved residue in fungal tropomyosins is important for polar distribution of actin patches and cell separation at the end of cytokinesis
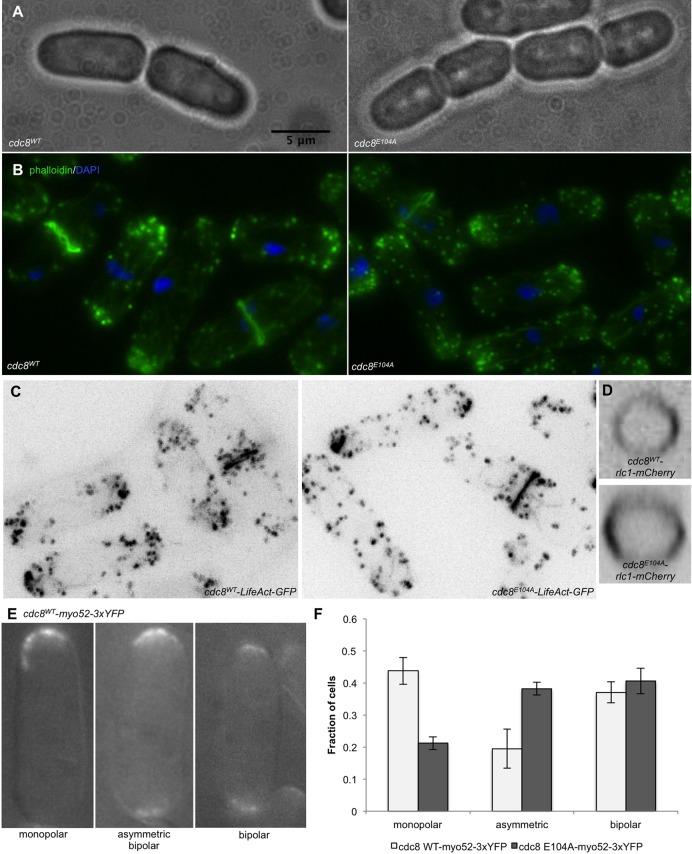
One of the most highly conserved regions in fungal tropomyosins is the third actin binding period of Cdc8p that includes residues 85–119 (Cranz-Mileva et al., 2013). A residue of particular interest to us is E104, a conserved residue in a surface c position in the coiled coil heptapeptide repeat in fungal tropomyosins. The region is less well conserved in period 3 of animal tropomyosins in which the analogous residue is Thr108 in vertebrates, but variable in other animal species (Barua et al., 2011). Therefore, we expressed Cdc8pE104A and constructed cdc8E104A strains (SH41, SH104, SH105) because of the possibility to identify a fungal-specific function for this conserved residue.
The cdc8E104A strain exhibits a prominent cellular phenotype in live cultures in which there is an increased fraction of four cells linked at the ends, as in sausage links (Fig. 3A; cdc8wt: 0.0055±0.013; cdc8E104A: 0.069±0.025; n>808, P=0.0007). The cell shape and size, nuclear number and growth rate are normal (supplementary material Fig. S1). Even though the actin affinity of recombinant Cdc8pE104A and thermal stability are normal (Fig. 1B,C), the actin cytoskeleton is not. The patches are less compact at the poles, more diffuse than in wildtype (Fig. 3B,C). In cells with a bipolar distribution of actin patches 46±0.08% of the total fluorescence is localized at the poles in wildtype, compared to 38±0.06% in cdc8E104A (n=26, P=0.0025; details in Fig. 3 legend). The contractile rings and cables appear quite normal, as visualized using Alexa-phalloidin (Fig. 3B, SH41), LifeAct-GFP (Fig. 3C, SH134), or Rlc1p-mCherry (myosin II, Fig. 3D, SH130), consistent with an apparently normal tropomyosin localization (supplementary material Fig. S2). Therefore we focused on questions related to actin patch distribution and intracellular transport. The localizations of other cytoskeletal proteins with fluorescent protein tags (tubulin (SH133), formin (Cdc12p, SH132), α-actinin (SH95), Myo1p (SH96), coronin (SH128), and Myo52p (SH98) are normal (supplementary material Fig. S3).
Fig. 3.
Cellular morphology and patch distribution in cdc8E104A. (A) Images of live cdc8wt (SH30) and cdc8E104A cells (SH41) in phase showing that cells often remain linked after division in cdc8E104A. The fraction of chains of four cells linked together was measured for wildtype (0.0055±0.013) and cdc8E104A (0.069±0.025) from 3 independent experiments of n>270 (P=0.0007). (B) Wildtype and mutant cells fixed and stained with Alexa-phalloidin (green) and DAPI (blue) to visualize F-actin and DNA. The actin patches are more dispersed in mutant than in wildtype cells, but the cables and contractile rings are similar to wildtype. (C) Confocal images of representative fields of live cdc8wt-LifeAct-GFP (KV587) and cdc8E104A-LifeAct-GFP cells (SH134) illustrate apparently normal cables and contractile rings in the mutant, but patches are less tightly polarized to the poles. The patch distribution was quantified by dividing individual cells lengthwise into three zones, two polar zones each representing 12.5% of the total length and a central zone representing 75% of the total length. The fluorescence intensity in each zone was quantified and corrected for background using ImageJ. The fraction of the total fluorescence in the polar zones was calculated: wildtype (0.46±0.08) and cdc8E104A (0.38±0.06) (n=26, P=0.00025). (D) Cross-sections of 2D reconstructions of live cdc8wt-rlc1-mCherry (SH49) and cdc8E104A-rlc1-mCherry cells (SH130) showing normal myosin II rings. (E) cdc8E104A-myo52-3xYFP cells (SH98) illustrating monopolar, asymmetric and bipolar patch distribution. (F) Fraction of cdc8wt (SH107) and cdc8E104A cells (SH98) with each distribution based on fluorescence intensity quantification from three independent measurements, each n>70 cells. The fluorescence at each cell tip was quantified and corrected for background using ImageJ. The ratio of fluorescence at one end to the fluorescence at the other end was measured. If the ratio was ≤0.33 the cell was “monopolar”, >0.33 but <0.66, the cell was “asymmetric bipolar”, or ≥0.66 the cell was “bipolar”. The fraction of bipolar cells was similar in wildtype (0.37±0.03) compared to mutant (0.41±0.04). cdc8E104A had a decreased fraction of monopolar cells (0.21±0.02) and an increased fraction of asymmetric bipolar cells (0.38±0.02) compared to wildtype cells (monopolar, 0.44±0.04; asymmetric bipolar, 0.20±0.06). In these experiments, the lengths of monopolar, asymmetric bipolar, and bipolar cells were also measured in cdc8wt-myo52-3xYFP (9.0±0.8 µm, 9.6±1.4 µm, 11.0±1.4 µm) and cdc8E104A-myo52-3xYFP (9.6±1.3 µm, 9.5±0.8 µm, 10.4±1.4 µm). Error bars show mean±standard deviation (s.d.).
To understand how the E104 mutation might affect Cdc8p function, we considered Myo52, a myosin V-class myosin that is involved in vesicular and organelle transport in cells (Hammer and Sellers, 2012). In fission yeast, deletion of myo52 results in altered polarity of actin patches (Motegi et al., 2001; Win et al., 2001) and altered vacuole fusion and distribution (Mulvihill et al., 2001). The phenotype of poor polarization of patches in cdc8E104A is shared with myo52Δ, suggesting E104 may be involved in regulating Myo52p-dependent processes. In vitro studies have shown that Cdc8p increases the affinity of Myo52p for actin, and increases the actin-activated ATPase of Myo52p (Clayton et al., 2010).
To inquire about the Myo52p function in cdc8E104A, we quantified the localization of Myo52p-3xYFP in a cdc8E104Amyo52-3xYFP strain (SH98). Myo52p-3xYFP is tightly clustered at the cell poles in interphase cells (Fig. 3E) and at the midline of dividing cells. We calculated the ratio of fluorescence intensity at the two ends and categorized the cells according to the polarity of the Myo52p fluorescence distribution. Assuming perfect bipolar distribution would have a ratio of 1.0, we created the following classifications: monopolar (≤0.33), bipolar (≥0.66), or “asymmetric bipolar” (>0.33 but <0.66). An asymmetric bipolar distribution of patches indicates a cell that is between polarities. Even though Myo52p-3xYFP localizes to the poles in cdc8E104A, the fraction of cells with monopolar localization is less than in wildtype (Fig. 3F, details in legend). Similarly, cdc8E104A has fewer cells than cdc8wt cells with a monopolar distribution of actin patches visualized using coronin (Crn1p-GFP) (supplementary material Fig. S4, details in legend). Crn1p-GFP is localized in the patches that are usually coincident with actin, as previously reported (Pelham and Chang, 2001). With both probes cdc8E104A has more cells with an asymmetric bipolar distribution than cdc8wt (Fig. 3F, supplementary material Fig. S4). The average length of the asymmetric bipolar cells is <9.5 mm, indicating they are in the pre-NETO stage of the cell cycle (Mitchison and Nurse, 1985).
In addition to vesicle and organelle movement in cells, in fission yeast Myo52 is involved in vacuole fusion (Mulvihill et al., 2001). When Myo52p-3xYFP cells are transferred from YEA medium to water, the Myo52p-3xYFP is redistributed from the cellular poles to around vacuoles within ten minutes. The effect is reversible. We observed no difference between cdc8wt and cdc8E104A cells indicating the mutation does not affect endocytosis (supplementary material Fig. S5). However, the cdc8E104A mutation alters the fusion of vacuoles.
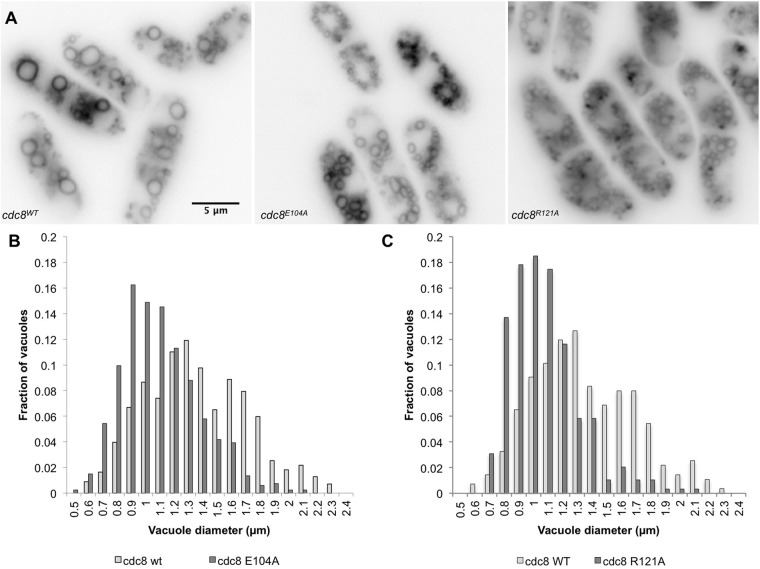
We followed the process of vacuole fusion using the lipophilic dye, FM4-64, that enters vacuoles by endocytosis (Brazer et al., 2000). Whereas the rate of labeling of vacuoles was similar in cdc8wt and cdc8E104A cells, the vacuoles are smaller but more numerous in cdc8E104A cells (cdc8wt, 6.8±1.9; cdc8E104A, 10.2±2.9 vacuoles/cell, n=48) (Fig. 4). In both cdc8wt and cdc8E104A the vacuoles increased in size with time, but they remained consistently smaller in cdc8E104 (Fig. 4B, details in legend). Also, as in myo52Δ cells (Mulvihill et al., 2001), the vacuoles tend to cluster around the nucleus (Fig. 4A). We interpret our results to indicate that in cdc8E104A cells vacuoles form normally but are delayed in fusion, a myo52-dependent process.
Fig. 4.
Analysis of vacuole fusion in cdc8E104A using the lipophilic dye, FM4-64. Cells were incubated for 45 min. in FM4-64, washed and transferred to medium as described in Materials and Methods. The images are of live cells after 60 min. (A) cdc8wt (SH30), cdc8E104A (SH41), cdc8R121A (SH39). (B) The size distribution of vacuoles in cdc8wt and cdc8E104A cells based on two independent experiments, each n>140. Mean±s.d.: cdc8wt, 1.4±0.4 µm; cdc8E104A, 1.1±0.3 µm (P=4.71×10−31). (C) The size distribution of vacuoles in cdc8wt and cdc8R121A cells in two independent experiments, each n>138. Mean±s.d.: cdc8wt, 1.3±0.3 µm; cdc8R121A, 1.1±0.2 µm (P=5.52×10−42). In both mutants the vesicles are smaller than in wildtype but greater in number. The vacuole size distributions in cdc8D16A.K30A, cdc8D131A, and cdc8E138A are indistinguishable from cdc8wt.
For comparison, we analyzed vacuole fusion in the other mutants in the present study. The D121A mutation that severely impairs actin affinity and cytoskeletal organization, as described above, results in smaller but more numerous vacuoles (Fig. 4A,C). A mutation that has a major effect on actin cable assembly would be expected to affect myo52-dependent processes that involve transport along cables. Alternatively, smaller and more numerous vacuoles may result from vacuole fragmentation in the stressed cdc8R121A mutant. Three other mutants, cdc8D16A,K30A, cdc8D131A, cdc8E138A, have no effect on vacuole fusion (supplementary material Fig. S6). The results lend credence to our postulate that the single E104A mutation affects vacuole fusion through regulation of Myo52p function, a subject for future investigation.
A synthetic effect of Cdc8p in a strain expressing fimbrin-GFP gives insights into protein function
We previously described cdc8D16A.K30A (SH22) that we isolated as a diploid strain (Cranz-Mileva et al., 2013). We noted a mild phenotype with occasional less polar distribution of patches and actin cables that tended to be more wavy and reticular than in wildtype. D16 and K30 are conserved surface residues near the N-terminal to C-terminal overlap region of tropomyosin, and D16 is part of a conserved actin binding motif in tropomyosins (Barua et al., 2013, 2011). The respective residues in human tropomyosin, E16 and K30, are among the most conserved in animal tropomyosins (Barua et al., 2011). The actin affinity of recombinant Cdc8pD16A.K30A with an AlaSer N-terminal modification is about 2-fold weaker than wildtype (Cranz-Mileva et al., 2013) (Fig. 1B), whereas a D16A mutation in an unacetylated recombinant protein increased actin affinity (East et al., 2011). A D16A mutation in a mammalian tropomyosin has little effect on the overall actin affinity (Barua et al., 2013). Therefore, we postulate D16 contributes to another conserved tropomyosin function.
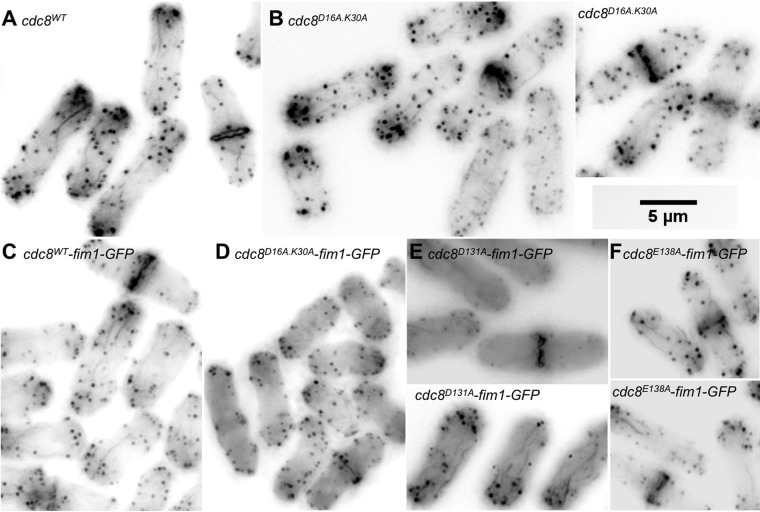
In the course of working with the cdc8D16A.K30A diploid we isolated a spontaneous haploid strain with normal growth parameters (supplementary material Fig. S1) that we analyzed in the present study since it gave us the opportunity to identify the effect of mutating these conserved sites on other cellular functions. We constructed strains of cdc8D16A.K30A expressing the following tagged cytoskeletal proteins: Rlc1p-mCherry (SH50), Fim1p-mEGFP (SH112), Cdc12p-3xGFP (SH59), Ain1-mEGFPp (SH116) and Act1-LifeActp-GFP (SH122). The distribution of the fluorescent proteins is rather normal in all (supplementary material Fig. S3), as was Cdc8p (supplementary material Fig. S2) and the actin cytoskeleton, except in cdc8D16A.K30fim1-GFP (Fig. 5). We also note that cdc8D16A.K30A had difficulty recombining with a myo1-mGFP strain (MLY422). While the asci appeared normal, in two crosses only 1/100 colonies screened in random spore analyses had a his+ura+KanR phenotype. The haploid phenotype was stable during numerous crosses, making the presence of an extragenic suppressor an unlikely explanation, but leaving open the possibility of intragenic suppression.
Fig. 5.
Actin cytoskeleton in wildtype and mutant cdc8 cells expressing GFP-fimbrin, illustrating synthetic negative effect of cdc8D16AK30A and fim1-GFP. All cells were fixed and stained with Alexa phallodin. Fim1-GFP fluorescence is weak following fixation. (A) cdc8wt cells (SH30). (B) cdc8D16AK30A cells (SH22). (C) cdc8wt fim1-GFP cells (SH77). (D) cdc8D16AK30A fim1-GFP cells (SH112). Note the absence of cables. (E) cdc8E131A fim1-GFP cells (SH74). (F) cdc8E138A fim1-GFP cells (SH75-A). The actin cytoskeleton appears normal in E and F.
Fimbrin is an actin filament crosslinking protein found in structures that have bundles of actin filaments, such as filopodia and microvilli in vertebrate cells (Bretscher, 1981; Bretscher and Weber, 1980). In fission yeast, fimbrin is localized in actin patches and in the vicinity of the actin-containing contractile ring in mitotic cells, where it bundles actin filaments and inhibits depolymerization by cofilin (Nakano et al., 2001). Fimbrin is not found in actin cables (Nakano et al., 2001; Wu et al., 2001). In contrast, Cdc8p is predominantly in actin cables and the contractile ring (Balasubramanian et al., 1992). Fimbrin-GFP localization depends on filamentous actin since it is sensitive to latrunculin and does not localize to the contractile ring area in temperature sensitive mutants of various genes, including cdc8-27, that do not form F-actin rings at the restrictive temperature (Nakano et al., 2001; Wu et al., 2001). The distribution of fimbrin-GFP in cdc8D16A.K30A (SH112) is indistinguishable from that in cdc8wt cells where it was observed, as expected, in patches and in a patchy distribution in the region of the contractile ring (supplementary material Fig. S3). However, whereas the actin cytoskeleton, visualized using Alexa-phalloidin, is apparently normal in cdc8wt-fim1-GFP cells (SH77) (compare Fig. 5A and C), the cdc8D16A.K30A-fim1-GFP cells displayed few actin cables (compare Fig. 5B and D). The actin-containing contractile rings are relatively normal, as is actin patch polarization (Fig. 5D). Expression of Fim1p-GFP did not influence the appearance of the actin cytoskeleton of three other mutants described above (cdc8D131A[SH74], cdc8E138A [SH75]; Fig. 5E,F) or cdc8E104A [SH100]. The results infer a synthetic negative effect of Cdc8pD16A.K30A and Fim1p-GFP on cable formation, even though fimbrin is not observed in cables (Nakano et al., 2001; Wu et al., 2001) and present work. These qualitative findings will need to be confirmed using more quantitative methods.
Effects of LifeAct-GFP on the actin cytoskeleton
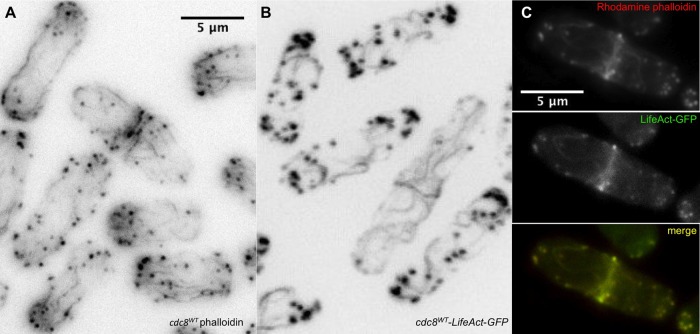
LifeAct is a 17-amino acid peptide reported to bind both G-actin and F-actin that is used as a probe for F-actin in living cells (Riedl et al., 2008). Localization of LifeAct-GFP is almost identical to that of phalloidin in fixed cells from different species, including fission yeast (Huang et al., 2012; Lemieux et al., 2014) and the present work (Fig. 6C). The synthetic peptide (no GFP) has little effect on actin polymerization in vitro (Riedl et al., 2008). For these reasons LifeAct is currently viewed as the best marker for F-actin in living cells.
Fig. 6.
Comparison of the actin cytoskeleton using Alexa-phalloidin and LifeAct. (A) cdc8wt cells (SH30) fixed and stained with Alexa-phalloidin showing long, typically straight actin cables parallel to the long axis of the cells. (B) cdc8wt-LifeAct-GFP cells (KV587, ura−) (live cells confocal image) showing wavy cables with variable orientations in relation to the long axis of the cell. The contractile rings are similar in A and B. (C) cdc8wt-LifeAct-GFP cells (KV587), fixed and stained with rhodamine-phalloidin (left, red channel), green channel showing GFP (center), and superimposed (right). The rhodamine and GFP images superimpose, showing that the phalloidin binds to the same structures that are labeled with GFP.
In our qualitative analysis, however, the actin cables in strains expressing LifeAct-GFP are altered in that they are less straight (Fig. 6A,B). When wildtype cdc8 cells expressing LifeAct-GFP (KV587) are fixed, the waviness of the cables is similar to those in living cells when visualized using the LifeAct-GFP fluorescence, or when fixed and stained with phalloidin. The LifeAct and phalloidin fluorescence are coincident (Fig. 6C). Therefore, we suggest that LifeAct affects the organization of the actin cytoskeleton in fission yeast even though it has no marked effect on the mating or growth. Similar reservations have been expressed about the use of LifeAct-mEGFP in Drosophila where strong overexpression resulted in sterility and defects in the actin cytoskeleton (Spracklen et al., 2014).
DISCUSSION
In the present work we applied an evolution-based proteomic approach towards understanding tropomyosin function in living fission yeast by creating several cdc8 mutants that have one or more Ala mutations in conserved surface residues. We found that the mutations have different effects, indicative of residue-specific functions. By combining analysis of the actin cytoskeleton with that of other proteins known to interact with Cdc8p from in vitro or cellular studies, we have inferred functional interactions that could be investigated in future studies.
Discussion of cdc8R121A
Our results show that Arg121 has a crucial role for tropomyosin function in fission yeast, primarily due to its involvement in binding actin. R121 is in a position homologous to conserved surface basic residues in mammalian tropomyosins that are part of the actin binding sites on tropomyosin in models based on experimental and molecular dynamics studies (Barua et al., 2013; Brown et al., 2005; Li et al., 2011; Zheng et al., 2013). The reduced actin affinity of Cdc8pR121A argues for a model in which the binding sites of tropomyosin for the highly-conserved protein actin are conserved in animal and fungal proteins. The finding also supports the hypothesis for universality in the functions of other cytoskeletal proteins and their interactions. Since a function shared by most tropomyosins is to stabilize actin filaments, it follows that actin cables and a condensed contractile ring do not form when tropomyosin binds poorly. It is possible that short, unstable actin filaments are present since there is sometimes diffuse or speckled phalloidin staining, versus higher-order assemblies of filaments found in cables and the contractile ring. Clearly, some tropomyosin and associated actin functions remain since cdc8R121A is viable, unlike the cdc8 disruption mutant (Balasubramanian et al., 1992) or cdc8ts mutants at the restrictive temperature (references cited in Cranz-Mileva et al., 2013).
The order of assembly of certain of the contractile ring components has been established (Wu et al., 2003). During the first stage Myo2p and its associated light chains and Cdc12p (formin) assemble in nodes in the contractile ring area (Wu et al., 2003). Actin filaments follow, forming a dense network around the equator as Myo2p and its associated light chains begin to form an equatorial ring (Vavylonis et al., 2008). Laporte et al. (2012) suggest that α-actinin and fimbrin together with Myo2p activity coalesce actin filaments that connect the nodes to form an organized contractile ring. While actin filaments are present in the area of the assembling contractile ring in cdc8R121A, as evidenced by diffuse fluorescence seen with phalloidin staining and LifeAct-GFP, coalescence of the actin into a ring is not supported. The weaker actin affinity of Cdc8pR121A may make actin filaments more susceptible to severing by cofilin, impair formin (Cdc12p) nucleation of actin filaments for formation of the ring and filaments that are crosslinked by α-actinin or fimbrin, or impair Myo2p function. Our observations support findings that the myosin molecules remain in the ring as it closes, while the actin and associated proteins do not (Mishra et al., 2013).
Discussion of cdc8E104A
Mutation of the conserved surface residue, Glu104 to Ala, has little effect on actin affinity in vitro, but it does influence the distribution of actin patches and vacuole fusion even though the cell shape, septum formation and growth are normal. The results indicate a function for cdc8 in establishing the polarity of the cellular actin cytoskeleton. We suggest this may take place in the following way.
The cdc8E104A cells remain weakly attached following cytokinesis when viewed in living cells (Fig. 3A). The handling of cells for microscopy involves centrifugation, pipetting and gentle vortexing that breaks the attachment. The asymmetric bipolar distribution of actin patch-associated proteins, including LifeAct-GFP, Myo52p-YFP and Crn1p-GFP may reflect the prior residual end-to-end attachments.
We suggest the observed phenomena of cell-cell links and altered patch distribution reflect a delay in the completion of cell division and redistribution of actin patches following cytokinesis from the new end (site of cytokinesis) to the old end. We never observed chains with more than four cells, suggesting that the cells separate when the “new end” after the first division becomes the “old end” at the next cell cycle. In cdc8E104A new end take off (NETO) appears to be normal, since the position of the end-most division scar to the new tip of the cell in relation to the cell length (Mitchison and Nurse, 1985) is the same as in cdc8wt. Also, the fraction of cells with a bipolar distribution of patches is normal. These findings, as well as the observation that cdc8E104A cells with an asymmetric bipolar patch distribution are pre-NETO (<9.5 µm) suggest that the problem with patch distribution and cell separation occurs during the restoration of monopolarity after cell division. Our results are consistent with the model that one mechanism of transport of the vesicles to the old end involves transport on actin filaments via the plus-end directed myosin, Myo52p, which is regulated by Cdc8p (Clayton et al., 2010). The altered vesicle fusion in cdc8E104A cells further supports our conclusion. We note that Glu104 is close to a region of muscle tropomyosin that is critical for regulating skeletal muscle myosin (Barua et al., 2014).
Discussion of cdc8D16A.K30A
Fimbrin and tropomyosin bind actin in different manners, yet they both stabilize filaments from severing by cofilin (Adf1p) (DesMarais et al., 2002; Nakano et al., 2001). Cdc8p and fimbrin compete with each other for binding actin in vitro (Skau and Kovar, 2010), and in vivo as inferred from cellular studies. For example, a fim1 deletion mutant is not lethal, but the actin patches contain more Cdc8p than in wildtype (Skau and Kovar, 2010). Fimbrin and tropomyosin have opposing effects on myosin 1 at actin patches (Clayton et al., 2010).
The synthetic effect of cdc8D16A.K30A and fim1-GFP in which we observe loss of cables in the cdc8D16A.K30A-fim1-GFP strain suggests a functional relationship between Cdc8p residues D16A and K30A and Fim1p with regard to cable assembly. Actin filament function in other cdc8D16A.K30A strains is not obviously affected: patch distribution and Fim1p-GFP localization appear normal. α-actinin, another actin crosslinking protein, is localized in the contractile ring, as in wildtype. Furthermore, the cells divide and mate. A straightforward explanation is that the suboptimal function of Fim1p and Cdc8p, separately, in their mutated or tagged state barely affects cable formation, but when both Fim1p and Cdc8p are compromised we observe a near total loss of cables. In this view, the functions of Cdc8pD16A.K30A and Fim1p-GFP are synergistic and may make the actin filaments more susceptible to depolymerization or severing by one or more actin binding proteins, such as cofilin-Adf1p. An interpretation more consistent with our findings is that Cdc8pD16AK30A competes less effectively for actin with Fim1p-GFP than Fim1p and more filaments are incorporated into actin patches as enabled by the Fim1p-GFP, at the expense of actin cables.
Conclusions
Our work validates an evolution-based approach as a way to identify new functions for tropomyosin and the sites of potential interactions with other cytoskeletal proteins. Analysis of the effects of site-specific mutations at conserved surface residues in fission yeast tropomyosin indicates the involvement of conserved residues in specific functions. These include actin binding, regulation of the assembly of actin-containing structures that influence the polarity of the actin cytoskeleton, contractile ring assembly, localization of other proteins in the actin cytoskeleton, and cell division.
More specifically, (1) a cdc8 mutation that has little effect on in vitro actin affinity affects patch polarization and as well as vacuole fusion, processes that depend on Myo52p, a member of the myosin V class. (2) A mutation that alone has a mild phenotype is synergistic with GFP-fimbrin, inferring involvement of the mutated tropomyosin sites in interaction between the two proteins. (3) Additional negative synthetic effects were identified by difficulties in mating certain mutants with strains expressing EGFP-α-actinin and Myo1p-mGFP. There may be additional synthetic negative effects that we did not reveal. (4) Proteins known to assemble in the region of the contractile ring before actin (Wu et al., 2003) continue to do so in a mutant cdc8 strain that fails to assemble a condensed actin ring, providing support for the contractile ring assembly pathway. (5) Of more general significance is the negative effect of the widely-used actin filament probe, LifeAct-GFP, on the actin cytoskeleton in wildtype cells, indicating caution in using this tool as a biomarker for actin filaments.
MATERIALS AND METHODS
Schizosaccharomyces pombe strains, plasmid and genetic methods
Yeast growth and general methods
Strains were grown in EMM (Edinburgh minimal medium, Sunrise Sciences, San Diego, CA) with the appropriate selective supplements (0.0225% adenine, lysine, histidine, uracil) or YEA medium (0.5% yeast extract, 3% dextrose, 0.015% adenine) following standard growth, genetic and cell biology protocols in (Forsburg, 2003a,b) and other online resources. The strains used and created in this study are listed in supplementary material Table S1. The standard growth temperature was 30°C, 35°C was the restrictive temperature for temperature-sensitive strains, and 25°C was used for permissive growth of temperature-sensitive strains and for other selected procedures. Cells were counted using a hemocytometer. Cellular transformation was carried out using the lithium acetate method (Kanter-Smoler et al., 1994) and purified plasmid or PCR-generated fragments. Genomic DNA was purified according to (Sambrook and Russell, 2001) with modifications. The breaking buffer was 2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM TrisHCl pH 8.0, 1 mM EDTA; we included an additional chloroform extraction, and RNase A digestion (10 µg/µl, 30 min at 37°C) in the final step. The cdc8 sequence was verified in the cdc8 strains and plasmids created in this study using appropriate primers (Genescript, Piscataway, NJ, USA or Genewiz, South Plainfield, NJ, USA).
Marker reconstitution mutagenesis
To make gene replacements of the cdc8+ with selected mutations, we used the method developed by Tang et al. (2011) with strains and plasmids previously described (Cranz-Mileva et al., 2013).
The plasmid pH5c-cdc8+ (Cranz-Mileva et al., 2013) is the template for introducing mutations into cdc8. The mutations were made by Mutagenex (Hillsborough, NJ, USA) and verified by DNA sequencing (Genescript or Genewiz). PCR fragments were amplified as previously described and transfected into SH13. Transformants were selected on minimal medium lacking histidine and uracil, and backcrossed twice to confirm the linkage of his+ and cdc8. The genomic sequence was confirmed. The strains created are: SH22 (cdc8D16A.K30A); SH41, SH104, SH105 (cdc8E104A); SH39, SH40 (cdc8R121A); SH43, SH44 (cdc8D131A); SH34 (cdc8E138A).
Fluorescent protein strains
Mutant cdc8 strains were crossed into strains expressing the following cytoskeletal proteins with fluorescent protein tags (supplementary material Table S1): α-actinin (ain1-mEGFP, JW1144), coronin (crn1-GFP, FC661), fimbrin (fim1-mEGFP, JW1142), formin (cdc12-3xGFP, KV344), myosin 1 (myo1-mGFP, MLY422), myosin 52 (myo52-3xYFP, MLY681), myosin regulatory light chain (rlc1-mCherry, MLY744), tubulin (atb2-mRFP, MLY1065), and LifeAct (pAct1-LifeAct-GFP, KV588). Wildtype and mutant cdc8 strains were crossed and colonies following random spore analysis were selected using nutritional, fluorescent and or kanamycin resistance, as appropriate. Except for myo1-mGFP (MLY422), crn1-GFP (FC661) and LifeAct-GFP (KV587) we constructed gene replacement cdc8+ strains expressing the fluorescent protein (grcdc8+ in supplementary material Table S1). The grcdc8+ strains are ura4+. We did not note a significant difference in the actin cytoskeleton between the grccdc8+, ura+ strains and the parental cdc8+ura− strains.
Microscopy
Staining
For microscopy cells were grown in YEA and fixed at mid-log phase. For staining with DAPI and Calcofluor cells were fixed and stored in 70% ethanol (Forsburg, 2003a; Mitchison and Nurse, 1985). To visualize the septum, ethanol-fixed cells were washed in PBS and resuspended in 5 µl 50-100 µg/ml Calcofluor (Sigma Life Science, St. Louis, MO, USA) in 50 mM sodium citrate, 100 mM sodium phosphate, pH 6.0) and incubated at ambient temperature for 5 min in the dark. For counterstaining with DAPI to visualize the nuclei, the cells were washed and resuspended in 2-5 µl PBS with the addition of 0.5 µl 50 µg/ml DAPI (Sigma Life Science). The samples were incubated for 5 min in the dark at ambient temperature. One µl of stained cells was mixed on a slide with 0.5 µl 1 mg/ml phenylenediamine in 50% glycerol, covered with a poly-l-lysine coated coverslip and sealed with clear nail polish.
Filamentous actin was visualized using Alexa Fluor 488 phalloidin or Rhodamine phalloidin (Life Technologies, Grand Island, NY, USA). Mid-log phase cells were fixed in 3.7% fresh paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) for 5 min. at the growth temperature, washed three times in PBS, and stored in PBS with 0.01% NaN3 at 4°C for 1 week or less. To stain with phalloidin, 2 µl of fixed cells were permeabilized by vortexing in 100 µl 1% Triton X-100 in PBS for 1 min, and washed 3× with PBS. Phalloidin was added to the permeabilized cells (4 µl of 0.2 U/µl phallodin) and incubated with gentle agitation for 50 min at room temperature. Counterstaining with DAPI and preparation of the slides was as described above.
The details of antibody preparation and purification, and indirect immunofluorescence are in the legend to supplementary material Fig. S2.
Fluorescence microscopy
Epifluorescence images were captured on a Nikon Optiphot 2 microscope fitted for epifluorescence using an Ushio USH-1020H mercury lamp with a Model HB-10101AF power supply and a DS epifluorescence illuminator with Chroma FITC and DAPI filters for imaging Alexa Fluor 488 phalloidin and Calcofluor/DAPI, respectively. We used a Nikon E-Plan 100×/1.25 Ph4DL oil immersion objective lens and a CoolSNAP fx camera (Photometrix, Tuscon, AZ, USA). Exposure was controlled by a Uniblitz Model VMM-D1 shutter driver (Vincent Associates, Rochester, NY, USA). IP Lab 4.0.8 (Scanalytics, Inc., Fairfax, VA USA) was used to control the microscope and its external devices. Depending on the stain intensity in the examined field, the exposure times were between 100 and 400 ms. Images of the same field were taken at different focal planes to visualize the full thickness of the cells. Binning was set to 1×1. The images were adjusted for brightness and contrast and merged using Image J 1.43u (Rasband, 1997-2012). Scale bars for all images were obtained by using an AO micrometer with 2 mm divisions subdivided into units of 10 µm.
Confocal microscopy
Confocal images were obtained at ambient temperature on an inverted Olympus IX Z spinning disk confocal microscope using an Olympus UPlan Fl 100×, 1.3NA oil immersion objective and a Hamamatsu EM-CCD C9100-02 digital camera (Hamamatsu Photonics, Hamamatsu, Shizuoka Prefecture, Japan). The binning was 1×1, 1000 ms exposure, E-gain=45, Z-slice=0.3 µm, 0.092 µm/pixel. The filters were GFP/Alx488 for GFP and Cy3 for m-Cherry.
Vacuole experiments
Cells were grown to mid-log phase and resuspended to 3×107 cells/ml. The lipophilic dye, FM4-64 (EMD Millipore, Billerica, MA, USA), was added to 50 µM from a stock of 10 mM. Cells were incubated rotating at 4°C for 30 min to allow the dye to coat the cells. The cells were then washed in YEA at 4°C and resuspended to 5×106 cells/m as described in (Brazer et al., 2000). Cells were grown shaking at room temperature and visualized at times 0–270 min to allow the cells to take up the dye by endocytosis. The vacuole sizes at each time point were analyzed using Image J 1.43u (Rasband, 1997-2012) by measuring the diameter of each vacuole (using the conversion 1 pixel=0.0535 µm). The lengths were rounded to the nearest tenth of a µm and sorted into groups to create a distribution.
Image analysis
We quantified the nuclear number and cell length in Calcofluor/DAPI stained micrographs, and other parameters using Image J 1.43u (Rasband, 1997-2012). The length of the cells was measured in pixels and converted to µm using a conversion factor of 1 pixel=0.0535 µm obtained using the AO micrometer above. The data were binned in groups to the nearest µm. Measurements of fluorescence intensity were corrected for background fluorescence. The statistics show the mean and standard deviation. Probabilities were obtained using a two-tailed t-test.
Expression, purification and analysis of recombinant Cdc8p
A plasmid encoding wildtype Cdc8p, pET3a-AS-cdc8 (gift of M. Lord, University of Vermont) (Studier et al., 1990), was used to produce Ala-Ser-Cdc8p. Mutant cdc8 variants were constructed in pET3a-AS-cdc8 by Mutagenex. The initial Met is cleaved after expression in E. coli since the second residue is Ala.
Recombinant wildtype and mutant AS-Cdc8p were expressed in E.coli BL21(DE3) using the autoinduction method (Studier, 2005) and purified using ammonium sulfate fractionation and anion ion exchange chromatography on DE52 cellulose (Whatman, GE Healthcare Life Sciences, Piscataway, NJ, USA) following established protocols (Hammell and Hitchcock-DeGregori, 1996; Hitchcock-DeGregori and Heald, 1987). The method was modified to maximize ammonium sulfate precipitation of AS-Cdc8p (45-75% or 30-75%, depending on the form). The purity was evaluated on SDS-PAGE gels and the concentration was determined using the tyrosine difference method (Edelhoch, 1967). Actin was purified from chicken pectoral muscle acetone powder using established methods (Hitchcock-De Gregori et al., 1982).
Actin binding assays
Actin 5 µM was mixed with 0.1 µM to 6 µM AS-Cdc8p in 20 mM MOPS, pH 7.0, 150 mM NaCl and 2 mM MgCl2 and cosedimented at 20°C at 60,000 rpm in a TLA100 rotor, Beckman TL-100 ultracentrifuge, for 30 min. (Hammell and Hitchcock-DeGregori, 1996). The pellets and supernatants were analyzed on 15% SDS-PAGE gels, stained with Coomassie blue, scanned and analyzed using an Image Scanner III (GE Healthcare Life Sciences) with Labscan 6.0 and Image Quant TL 7.0 image analysis software. The observed AS-Cdc8p/actin ratio was normalized to 1 by dividing the AS-Cdc8p/actin ratio obtained from densitometry by the AS-Cdc8p/actin ratio observed at saturation. The free AS-Cdc8p in the supernatant was calculated from standard curves for wildtype AS-Cdc8p. The binding constant, Kapp and Hill coefficient (αH) were determined by fitting the data to the Hill equation using SigmaPlot (Jandel Scientific, San Rafael, CA, USA):
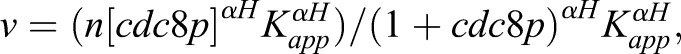 |
where α=fraction maximal AS-Cdc8p binding to actin, n=maximal AS-Cdc8p bound, and [cdc8p]=[AS-Cdc8p]free, and αH=Hill coefficient.
Circular dichroism measurements
Thermal stability was measured by following the ellipticity of AS-Cdc8p (0.2 mg/ml) at 222 nm in 0.5 M NaCl, 10 mM sodium phosphate pH 7.5, 1 mM EDTA at 0.2°C intervals from 0°C to 60°C using an Aviv model 400 CD-Spectrophotometer at the Robert Wood Johnson Medical School CD facility (Piscataway, NJ, USA). Ellipticity at 222 nm was normalized to a scale from 0 to 1. A value of 0.5 was defined as the observed melting temperature (TM) (Greenfield and Hitchcock-DeGregori, 1995).
Supplementary Material
Acknowledgements
We thank Dr Bipasha Barua (Robert Wood Johnson Medical School) for carrying out the CD measurements and analysis, Ravi Shah and Daniel Whitehead, Rutgers University students, for assistance; Dr Fred Chang (Columbia University), Dr David Kovar (University of Chicago), Dr Matthew Lord (University of Vermont) and Dr Jian-Qiu Wu (Ohio State University) for providing strains.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.C.-M., B.M. were engaged in the conception, design, execution and interpretation of the data, and preparing the article. J.R. contributed in the execution and interpretation of the data. S.E.H.-D. was involved in the conception, design and interpretation of the data, and writing the article.
Funding
This research was supported by NIH RO1 GM093065 to S.E.H.-D., the Aresty Research Center at Rutgers University (to B.M. and J.R.), the Douglass College Alumnae Association (to B.M.), and the Rodman Family Foundation (to B.M.).
Supplementary material
Supplementary material available online at http://bio.biologists.org/lookup/suppl/doi:10.1242/bio.012609/-/DC1
References
- Abe S. and Maruyama K. (1973). Effect of alpha-actinin on F-actin. A dynamic viscoelastic study. J. Biochem. 73, 1205-1210. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., Helfman D. M. and Hemmingsen S. M. (1992). A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature 360, 84-87. 10.1038/360084a0 [DOI] [PubMed] [Google Scholar]
- Barua B., Pamula M. C. and Hitchcock-DeGregori S. E. (2011). Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proc. Natl. Acad. Sci. USA 108, 10150-10155. 10.1073/pnas.1101221108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B., Winkelmann D. A., White H. D. and Hitchcock-DeGregori S. E. (2012). Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Proc. Natl. Acad. Sci. USA 109, 18425-18430. 10.1073/pnas.1212754109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B., Fagnant P. M., Winkelmann D. A., Trybus K. M. and Hitchcock-DeGregori S. E. (2013). A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin. J. Biol. Chem. 288, 9602-9609. 10.1074/jbc.M113.451161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B., Nagy A., Sellers J. R. and Hitchcock-DeGregori S. E. (2014). Regulation of nonmuscle myosin II by tropomyosin. Biochemistry 53, 4015-4024. 10.1021/bi500162z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. W. and Bamburg J. R. (1982). Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil. 2, 1-8. 10.1002/cm.970020102 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T. D. and Hitchcock-DeGregori S. E. (2001). Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 11, 1300-1304. 10.1016/S0960-9822(01)00395-5 [DOI] [PubMed] [Google Scholar]
- Brazer S.-C. W., Williams H. P., Chappell T. G. and Cande W. Z. (2000). A fission yeast kinesin affects Golgi membrane recycling. Yeast 16, 149-166. [DOI] [PubMed] [Google Scholar]
- Bremel R. D. and Weber A. (1972). Cooperation within actin filament in vertebrate skeletal muscle. Nat. New Biol. 238, 97-101. 10.1038/newbio238097a0 [DOI] [PubMed] [Google Scholar]
- Bretscher A. (1981). Fimbrin is a cytoskeletal protein that crosslinks F-actin in vitro. Proc. Natl. Acad. Sci. USA 78, 6849-6853. 10.1073/pnas.78.11.6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. and Weber K. (1980). Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J. Cell Biol. 86, 335-340. 10.1083/jcb.86.1.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broschat K. O. (1990). Tropomyosin prevents depolymerization of actin filaments from the pointed end. J. Biol. Chem. 265, 21323-21329. [PubMed] [Google Scholar]
- Broschat K. O., Weber A. and Burgess D. R. (1989). Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry 28, 8501-8506. 10.1021/bi00447a035 [DOI] [PubMed] [Google Scholar]
- Brown J. H., Zhou Z., Reshetnikova L., Robinson H., Yammani R. D., Tobacman L. S. and Cohen C. (2005). Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc. Natl. Acad. Sci. USA 102, 18878-18883. 10.1073/pnas.0509269102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Woollard A. and Nurse P. (1996). Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109, 131-142. [DOI] [PubMed] [Google Scholar]
- Clayton J. E., Sammons M. R., Stark B. C., Hodges A. R. and Lord M. (2010). Differential regulation of unconventional fission yeast myosins via the actin track. Curr. Biol. 20, 1423-1431. 10.1016/j.cub.2010.07.026 [DOI] [PubMed] [Google Scholar]
- Clayton J. E., Pollard L. W., Sckolnick M., Bookwalter C. S., Hodges A. R., Trybus K. M. and Lord M. (2014). Fission yeast tropomyosin specifies directed transport of myosin-V along actin cables. Mol. Biol. Cell 25, 66-75. 10.1091/mbc.E13-04-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J. E., Pollard L. W., Murray G. G. and Lord M. (2015). Myosin motor isoforms direct specification of actomyosin function by tropomyosins. Cytoskeleton 72, 131-145. 10.1002/cm.21213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpan M., Moroz N. A. and Kostyukova A. S. (2013). Tropomodulins and tropomyosins: working as a team. J. Muscle Res. Cell Motil. 34, 247-260. 10.1007/s10974-013-9349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranz-Mileva S., Pamula M. C., Barua B., Desai B., Hong Y. H., Russell J., Trent R., Wang J., Walworth N. C. and Hitchcock-DeGregori S. E. (2013). A molecular evolution approach to study the roles of tropomyosin in fission yeast. PLoS ONE 8, e76726 10.1371/journal.pone.0076726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Ichetovkin I., Condeelis J. and Hitchcock-DeGregori S. E. (2002). Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115, 4649-4660. 10.1242/jcs.00147 [DOI] [PubMed] [Google Scholar]
- Drabikowski W. and Nowak E. (1968). The interaction of alpha-actinin with F-actin and its abolition by tropomyosin. Eur. J. Biochem. 5, 209-214. 10.1111/j.1432-1033.1968.tb00359.x [DOI] [PubMed] [Google Scholar]
- Drees B., Brown C., Barrell B. G. and Bretscher A. (1995). Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J. Cell Biol. 128, 383-392. 10.1083/jcb.128.3.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch H. (1967). Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948-1954. 10.1021/bi00859a010 [DOI] [PubMed] [Google Scholar]
- East D.A., Sousa D., Martin S. R., Edwards T. A., Lehman W. and Mulvihill D. P. (2011). Altering the stability of the Cdc8 overlap region modulates the ability of this tropomyosin to bind cooperatively to actin and regulate myosin. Biochem. J. 438, 265-273. 10.1042/BJ20101316 [DOI] [PubMed] [Google Scholar]
- Fanning A. S., Wolenski J. S., Mooseker M. S. and Izant J. G. (1994). Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil. Cytoskelet. 29, 29-45. 10.1002/cm.970290104 [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. (2003a). Growth and manipulation of S. pombe. Curr. Protoc. Mol. Biol. 64, 16:13.16.1-13.16.17. 10.1002/0471142727.mb1316s64 [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. (2003b). S. pombe strain maintenance and media. Curr. Protoc. Mol. Biol. 64, 15:13.15.1-13.15.5. 10.1002/0471142727.mb1315s64 [DOI] [PubMed] [Google Scholar]
- Geeves M. A., Hitchcock-DeGregori S. E. and Gunning P. W. (2014). A systematic nomenclature for mammalian tropomyosin isoforms. J. Muscle Res. Cell Motil. 36, 147-53. 10.1007/s10974-014-9389-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N. J. and Hitchcock-DeGregori S. E. (1995). The stability of tropomyosin, a two-stranded coiled-coil protein, is primarily a function of the hydrophobicity of residues at the helix-helix interface. Biochemistry 34, 16797-16805. 10.1021/bi00051a030 [DOI] [PubMed] [Google Scholar]
- Hammell R. L. and Hitchcock-DeGregori S. E. (1996). Mapping the functional domains within the carboxyl terminus of alpha-tropomyosin encoded by the alternatively spliced ninth exon. J. Biol. Chem. 271, 4236-4242. 10.1074/jbc.271.8.4236 [DOI] [PubMed] [Google Scholar]
- Hammer J. A. III and Sellers J. R. (2012). Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13, 13-26. 10.1038/nrm3248 [DOI] [PubMed] [Google Scholar]
- Hitchcock S. E., Carlsson L. and Lindberg U. (1976). Depolymerization of F-actin by deoxyribonuclease I. Cell 7, 531-542. 10.1016/0092-8674(76)90203-8 [DOI] [PubMed] [Google Scholar]
- Hitchcock-De Gregori S. E., Mandala S. and Sachs G. A. (1982). Changes in actin lysine reactivities during polymerization detected using a competitive labeling method. J. Biol. Chem. 257, 8. [PubMed] [Google Scholar]
- Hitchcock-DeGregori S. E. and Heald R. W. (1987). Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. J. Biol. Chem. 262, 9730-9735. [PubMed] [Google Scholar]
- Hodges A. R., Krementsova E. B., Bookwalter C. S., Fagnant P. M., Sladewski T. E. and Trybus K. M. (2012). Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr. Biol. 22, 1410-1416. 10.1016/j.cub.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huang Y., Yu H., Subramanian D., Padmanabhan A., Thadani R., Tao Y., Tang X., Wedlich-Soldner R. and Balasubramanian M. K. (2012). Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J. Cell Biol. 199, 831-847. 10.1083/jcb.201209044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter-Smoler G., Dahlkvist A. and Sunnerhagen P. (1994). Improved method for rapid transformation of intact Schizosaccharomyces pombe cells. Biotechniques 16, 798-800. [PubMed] [Google Scholar]
- Kostyukova A. S. and Hitchcock-DeGregori S. E. (2004). Effect of the structure of the N terminus of tropomyosin on tropomodulin function. J. Biol. Chem. 279, 5066-5071. 10.1074/jbc.M311186200 [DOI] [PubMed] [Google Scholar]
- Kostyukova A. S., Rapp B. A., Choy A., Greenfield N. J. and Hitchcock-DeGregori S. E. (2005). Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry 44, 4905-4910. 10.1021/bi047468p [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Sirotkin V. and Lord M. (2011). Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 21, 177-187. 10.1016/j.tcb.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Imai Y. and Yamamoto M. (2002). Tropomyosin is required for the cell fusion process during conjugation in fission yeast. Genes Cells 7, 375-384. 10.1046/j.1365-2443.2002.00526.x [DOI] [PubMed] [Google Scholar]
- Laporte D., Ojkic N., Vavylonis D. and Wu J.-Q. (2012). alpha-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell 23, 3094-3110. 10.1091/mbc.E12-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman W. and Craig R. (2008). Tropomyosin and the steric mechanism of muscle regulation. Adv. Exp. Med. Biol. 644, 95-109. 10.1007/978-0-387-85766-4_8 [DOI] [PubMed] [Google Scholar]
- Lehrer S. S. and Morris E. P. (1982). Dual effects of tropomyosin and troponin-tropomyosin on actomyosin subfragment 1 ATPase. J. Biol. Chem. 257, 8073-8080. [PubMed] [Google Scholar]
- Lemieux M. G., Janzen D., Hwang R., Roldan J., Jarchum I. and Knecht D. A. (2014). Visualization of the actin cytoskeleton: different F-actin-binding probes tell different stories. Cytoskeleton 71, 157-169. 10.1002/cm.21160 [DOI] [PubMed] [Google Scholar]
- Li X. E., Tobacman L. S., Mun J. Y., Craig R., Fischer S. and Lehman W. (2011). Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 100, 1005-1013. 10.1016/j.bpj.2010.12.3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. and Bretscher A. (1989). Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell 57, 233-242. 10.1016/0092-8674(89)90961-6 [DOI] [PubMed] [Google Scholar]
- Liu H. and Bretscher A. (1992). Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J. Cell Biol. 118, 285-299. 10.1083/jcb.118.2.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K. and Ohashi K. (1978). Tropomyosin inhibits the interaction of F-actin and filamin. J. Biochem. 84, 1017-1019. [DOI] [PubMed] [Google Scholar]
- Maytum R., Geeves M. A. and Konrad M. (2000). Actomyosin regulatory properties of yeast tropomyosin are dependent upon N-terminal modification. Biochemistry 39, 11913-11920. 10.1021/bi000977g [DOI] [PubMed] [Google Scholar]
- Mishra M., Kashiwazaki J., Takagi T., Srinivasan R., Huang Y., Balasubramanian M. K. and Mabuchi I. (2013). In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nat. Cell Biol. 15, 853-859. 10.1038/ncb2781 [DOI] [PubMed] [Google Scholar]
- Mishra M., Huang J. and Balasubramanian M. K. (2014). The yeast actin cytoskeleton. FEMS Microbiol. Rev. 38, 213-227. 10.1111/1574-6976.12064 [DOI] [PubMed] [Google Scholar]
- Mitchison J. M. and Nurse P. (1985). Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75, 357-376. [DOI] [PubMed] [Google Scholar]
- Monteiro P. B., Lataro R. C., Ferro J. A. and Reinach F. d. C. (1994). Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J. Biol. Chem. 269, 7. [PubMed] [Google Scholar]
- Motegi F., Arai R. and Mabuchi I. (2001). Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol. Biol. Cell 12, 1367-1380. 10.1091/mbc.12.5.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill D. P., Pollard P. J., Win T. Z. and Hyams J. S. (2001). Myosin V-mediated vacuole distribution and fusion in fission yeast. Curr. Biol. 11, 1124-1127. 10.1016/S0960-9822(01)00322-0 [DOI] [PubMed] [Google Scholar]
- Nagaoka R., Kusano K., Abe H. and Obinata T. (1995). Effects of cofilin on actin filamentous structures in cultured muscle cells. Intracellular regulation of cofilin action. J. Cell Sci. 108, 581-593. [DOI] [PubMed] [Google Scholar]
- Nakano K. and Mabuchi I. (2006). Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell 17, 1933-1945. 10.1091/mbc.E05-09-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Satoh K., Morimatsu A., Ohnuma M. and Mabuchi I. (2001). Interactions among a fimbrin, a capping protein, and an actin-depolymerizing factor in organization of the fission yeast actin cytoskeleton. Mol. Biol. Cell 12, 3515-3526. 10.1091/mbc.12.11.3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E., Maekawa S. and Sakai H. (1984). Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 23, 5307-5313. 10.1021/bi00317a032 [DOI] [PubMed] [Google Scholar]
- Ono S. and Ono K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065-1076. 10.1083/jcb.200110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R. J. Jr and Chang F. (2001). Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat. Cell Biol. 3, 235-244. 10.1038/35060020 [DOI] [PubMed] [Google Scholar]
- Pruliere G., Fuller S. D., Weeds A. G., d'Albis A. and der Terrossian E. (1986). Pig platelet tropomyosin: interactions with the other thin-filament proteins. J. Mol. Biol. 192, 815-830. 10.1016/0022-2836(86)90031-8 [DOI] [PubMed] [Google Scholar]
- Rasband W. S. (1997-2012). ImageJ. US National Institutes of Health, Bethesda, MD, USA: http://imagej.nih.gov/ij/. [Google Scholar]
- Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z. et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605-607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. and Russell D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Skau C. T. and Kovar D. R. (2010). Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr. Biol. 20, 1415-1422. 10.1016/j.cub.2010.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau C. T., Neidt E. M. and Kovar D. R. (2009). Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol. Biol. Cell 20, 2160-2173. 10.1091/mbc.E08-12-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau C. T., Courson D. S., Bestul A. J., Winkelman J. D., Rock R. S., Sirotkin V. and Kovar D. R. (2011). Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26964-26977. 10.1074/jbc.M111.239004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen A. J., Fagan T. N., Lovander K. E. and Tootle T. L. (2014). The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev. Biol. 393, 209-226. 10.1016/j.ydbio.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark B. C., Sladewski T. E., Pollard L. W. and Lord M. (2010). Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol. Biol. Cell 21, 989-1000. 10.1091/mbc.E09-10-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. (2005). Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207-234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J. and Dubendorff J. W. (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60-89. [DOI] [PubMed] [Google Scholar]
- Tang N. and Ostap E. M. (2001). Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 11, 1131-1135. 10.1016/S0960-9822(01)00320-7 [DOI] [PubMed] [Google Scholar]
- Tang X., Huang J., Padmanabhan A., Bakka K., Bao Y., Tan B. Y., Cande W. Z. and Balasubramanian M. K. (2011). Marker reconstitution mutagenesis: a simple and efficient reverse genetic approach. Yeast 28, 205-212. 10.1002/yea.1831 [DOI] [PubMed] [Google Scholar]
- Ujfalusi Z., Vig A., Hild G. and Nyitrai M. (2009). Effect of tropomyosin on formin-bound actin filaments. Biophys. J. 96, 162-168. 10.1529/biophysj.108.138420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujfalusi Z., Kovacs M., Nagy N. T., Barko S., Hild G., Lukacs A., Nyitrai M. and Bugyi B. (2012). Myosin and tropomyosin stabilize the conformation of formin-nucleated actin filaments. J. Biol. Chem. 287, 31894-31904. 10.1074/jbc.M112.341230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O'Shaughnessy B. and Pollard T. D. (2008). Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 319, 97-100. 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Wawro B., Greenfield N. J., Wear M. A., Cooper J. A., Higgs H. N. and Hitchcock-DeGregori S. E. (2007). Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry 46, 8146-8155. 10.1021/bi700686p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A., Pennise C. R., Babcock G. G. and Fowler V. M. (1994). Tropomodulin caps the pointed ends of actin filaments. J. Cell Biol. 127, 1627-1635. 10.1083/jcb.127.6.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win T. Z., Gachet Y., Mulvihill D. P., May K. M. and Hyams J. S. (2001). Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114, 69-79. [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Bahler J. and Pringle J. R. (2001). Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell 12, 1061-1077. 10.1091/mbc.12.4.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., Kuhn J. R., Kovar D. R. and Pollard T. D. (2003). Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell 5, 723-734. 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Yamashiro S., Gokhin D. S., Kimura S., Nowak R. B. and Fowler V. M. (2012). Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton 69, 337-370. 10.1002/cm.21031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeece M. G., Robson R. M. and Bechtel P. J. (1979). Interaction of alpha-actinin, filamin and tropomyosin with F-actin. Biochim. Biophys. Acta 581, 365-370. 10.1016/0005-2795(79)90258-7 [DOI] [PubMed] [Google Scholar]
- Zheng W., Barua B. and Hitchcock-DeGregori S. E. (2013). Probing the flexibility of tropomyosin and its binding to filamentous actin using molecular dynamics simulations. Biophys. J. 105, 1882-1892. 10.1016/j.bpj.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.