Summary
The ADAR RNA-editing enzymes deaminate adenosine bases to inosines in cellular RNAs. Aberrant interferon expression occurs in patients in whom ADAR1 mutations cause Aicardi-Goutières syndrome (AGS) or dystonia arising from striatal neurodegeneration. Adar1 mutant mouse embryos show aberrant interferon induction and die by embryonic day E12.5. We demonstrate that Adar1 embryonic lethality is rescued to live birth in Adar1; Mavs double mutants in which the antiviral interferon induction response to cytoplasmic double-stranded RNA (dsRNA) is prevented. Aberrant immune responses in Adar1 mutant mouse embryo fibroblasts are dramatically reduced by restoring the expression of editing-active cytoplasmic ADARs. We propose that inosine in cellular RNA inhibits antiviral inflammatory and interferon responses by altering RLR interactions. Transfecting dsRNA oligonucleotides containing inosine-uracil base pairs into Adar1 mutant mouse embryo fibroblasts reduces the aberrant innate immune response. ADAR1 mutations causing AGS affect the activity of the interferon-inducible cytoplasmic isoform more severely than the nuclear isoform.
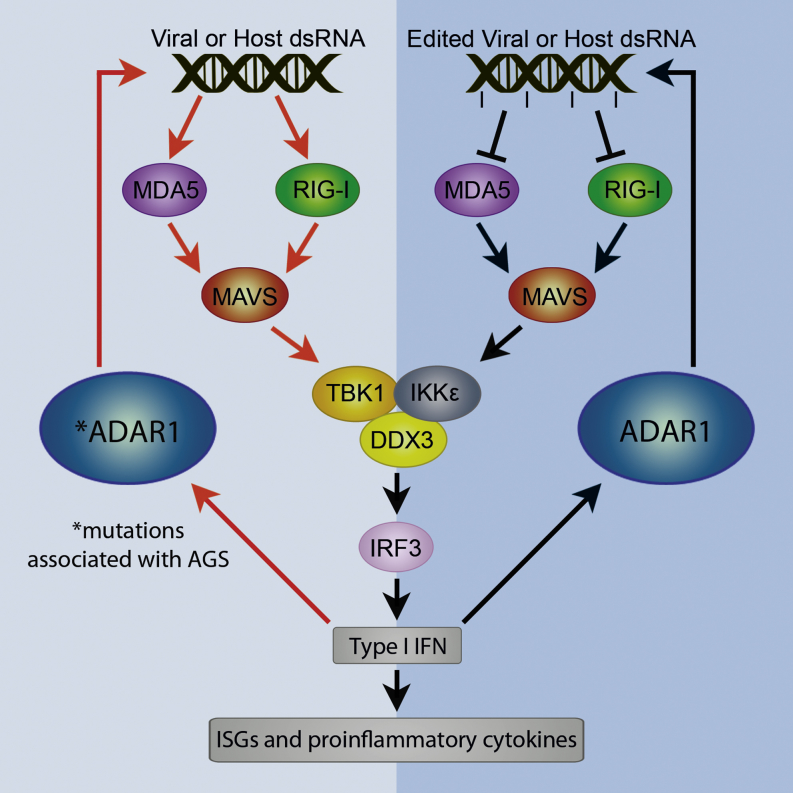
Graphical Abstract
Highlights
-
•
Adar1 mutant mouse embryonic lethality is rescued in Adar1; Mavs double mutants
-
•
Aberrant antiviral responses in the Adar1 mutant are due to loss of RNA editing
-
•
Human ADAR1 mutations causing AGS affect primarily the interferon-inducible isoform
-
•
We propose that inosine helps innate immunity to distinguish cellular from viral RNA
Mice lacking Adar1 have a heightened immune response and stress-related apoptosis. Mannion et al. demonstrate that this mutation can be rescued to birth by generating a double mutant with Mavs, an innate immune gene, indicating the central role ADAR1 plays in innate immunity.
Introduction
In vertebrates, viral double-stranded RNAs (dsRNAs) in the cytoplasm bind and activate RIG-I (retinoic acid-inducible gene I)-like cytoplasmic viral sensor proteins (RLRs) (for review, see Takeuchi and Akira, 2010). The known features that these sensors use to discriminate virus and pathogen RNAs from host cytoplasmic RNAs include the presence of dsRNA ends and 5′ triphosphates. RLRs translocate on dsRNA, and some RLRs may scan dsRNA to help distinguish between host and virus molecules. Cytoplasmic viral RNAs usually lack modifications, because most viruses do not encode modifying enzymes. It has been proposed that nucleic acid modifications of cellular RNAs help innate immune sensors to avoid aberrant activation by host nucleic acids (Gehrig et al., 2012; Karikó et al., 2005; Vitali and Scadden, 2010). Consistent with this idea, transfection of in-vitro-transcribed RNA into cultured cells generates an innate immune response. However, if the RNA is synthesized to contain naturally occurring modified bases, then the modified RNA does not cause innate immune induction (Karikó et al., 2005; Warren et al., 2010).
Suppression of responses of innate immune RNA sensors by modifications normally present in host RNAs could act as thresholding mechanisms to help prevent aberrant responses. Response thresholding mechanisms may be required because some cellular RNAs do contain RNA duplexes or have 5′ triphosphates. RNA duplexes in host RNAs are particularly hazardous; some Alu hairpins are still present in 3′ UTRs of mature mRNAs in the cytoplasm (Capshew et al., 2012). Transcription also occurs over most of the human genome, which inevitably generates further dsRNA that may reach the cytoplasm (Kapranov et al., 2007). The RIG-I and MDA5 (melanoma differentiation associated gene 5, IFIH1; interferon induced with helicase C domain 1) sensors are activated by binding RNA duplexes and signal through the MAVS (mitochondrial antiviral signaling) adaptor protein to activate NFκB, IRF3/7, and AP1. This activates transcription of genes encoding type I interferon (IFN) and proinflammatory cytokines. Secreted type I IFN binds to cell-surface type I IFN receptors to amplify and spread the antiviral response, inducing transcription of a large set of antiviral, IFN-stimulated genes (ISGs). Aberrant or chronic activation of IFN-stimulated defense processes is very damaging to the host.
Experiments in cultured cells do not test the overall importance of RNA modification for restraining innate immune responses in whole-model organisms or in human diseases. However, recent findings on human gene mutations causing autoimmune diseases are consistent with possible significant roles for RNA modifications in autoimmune diseases. For instance, mutations in the ADAR1 gene encoding one RNA-editing adenosine deaminase acting on RNA cause Aicardi-Goutières syndrome (AGS) (Rice et al., 2012). AGS is a fatal childhood encephalopathy with aberrant IFN expression and symptoms resembling those caused by congenital virus infection. Similar rare mutations in ADAR1 also cause dystonia due to bilateral striatal neurodegeneration associated with IFN overproduction (Livingston et al., 2014). Over 100 mutations in ADAR1 have been identified in East-Asian patients with dyschromatosis symmetrica hereditaria (DSH), a mild genodermatosis with mostly unknown IFN status, in which the dominant phenotype appears to be often due to ADAR1 haploinsufficiency (Liu et al., 2006).
ADARs catalyze the deamination of adenosine to inosine, which is the most common base modification known to occur in mammalian RNA (Gerber and Keller, 2001). Inosine is readily detected in transcriptome sequence data because inosine prefers to base pair with cytosine, leading to replacement of genome-encoded adenosine (A) by guanosine (G) at edited positions in cDNA sequences. ADARs edit particular adenosine residues at specific positions in short RNA duplexes in protein-coding transcripts, and they also edit numerous adenosines promiscuously in longer dsRNAs (for review, see Heale and O’Connell, 2009). Site-specific RNA-editing events in transcript open reading frames that generate new isoforms of encoded proteins represent the best-understood mechanism of ADAR action. Vertebrates have two enzymatically active ADAR proteins: ADAR1 (ADAR) and ADAR2 (ADARB1). Editing of the critical Q/R site in the transcript encoding the key AMPA receptor subunit is performed by ADAR2. Mutant Adar2 mice die from seizures within 3 weeks of birth, and seizures and death are prevented by knocking in an editing-equivalent A to G mutation in the AMPA receptor subunit gene (Higuchi et al., 2000).
Adar1 mutant mice die by embryonic day E12.5 with massive overproduction of IFN, loss of embryonic liver hematopoietic cells, liver disintegration, and widespread apoptosis (Hartner et al., 2004; Wang et al., 2004). Cultured Adar1 mutant embryonic fibroblasts are also highly sensitive to stress-induced apoptosis (Hartner et al., 2004; Wang et al., 2004). For many years, research has focused on identifying a key ADAR1 target transcript analogous to the AMPA receptor transcript. However, among the approximately 60 known editing events that recode open reading frames, many of the key events are catalyzed primarily by ADAR2, and none appears likely to account for the Adar1 mutant embryonic lethal phenotype. However, only 0.4% of human A-to-I editing occurs within protein-coding sequences (Peng et al., 2012); the vast majority of known A-to-I-editing sites, now estimated at over 100 million in the human genome (Bazak et al., 2013), have been found in RNA duplex-forming pairs of Alu elements embedded in inverted orientations near each other in introns and 3′ UTR regions of transcripts.
Editing of cellular dsRNA leads to formation of I-U wobble base pairs that cause bending and alter the properties of the dsRNA helix; multiple sequential I-U base pairs and high levels of promiscuous editing destabilize dsRNA. Editing of endogenous RNA duplexes may lower the risk that they aberrantly induce innate immune responses. The constitutive ADARp110 isoform is a shuttling protein that accumulates mainly in the nucleus and edits dsRNA before nuclear export (Desterro et al., 2003). The N-terminally extended ADARp150 isoform, expressed from a late IFN-inducible promoter, is predominantly cytoplasmic and has been shown to edit viral RNAs (Samuel, 2011). Transfected dsRNA oligonucleotides containing inosine-uracil (I:U) base pairs have been shown to bind to RLRs competitively with poly I:C and to suppress activation of innate immune responses (Vitali and Scadden, 2010).
We reveal that loss of ADAR1 RNA-editing activity and the resulting loss of inosine bases in RNA are critical in producing aberrant RLR-mediated innate immune responses in Adar1 mutant mice and cultured mouse cells. We characterize the immune response-blocking actions of human ADAR1 protein in Adar1 mutant mouse cells and show that most AGS-associated ADAR1 mutant proteins have impaired RNA-editing activity.
Results
IFN Receptor Mutation Partially Rescues Mouse Adar1 Mutant Embryonic Lethality, whereas Mavs Mutation Rescues to Live Births
We wished to investigate whether aberrant immune responses are critical to the Adar1 mutant embryonic lethal phenotype in mice. Preventing type I IFN signaling is sufficient to fully rescue embryonic lethality in embryos mutant for DNaseII (Yoshida et al., 2005) that also die with type I IFN overproduction. We constructed mouse strains that generate Adar1; Ifnar1 (IFN-α and -β receptor 1) double-mutant embryos in crosses. No live Adar1−/−; Ifnar1−/− progeny were obtained (Table S1). Staged embryo collections from crosses of Adar1+/−; Ifnar1−/− parents suggest that the embryonic lifespan of the double homozygous Adar1−/−; Ifnar1−/− embryos was extended due to Ifnar1−/− (Table 1; Figure 1); the mixed C57Bl6/129 strain background in these crosses had little effect (Table S1). Responses to secreted type I IFN are not the main reason for Adar1 mutant embryonic lethality.
Table 1.
Survival of Mouse Embryos with Different Adar1 Genotypes from Crosses between Adar1+/−; Ifnar1−/− (C57Bl6N/129) or Adar1+/−; Mavs−/− (C57Bl6N/J) Parents
| Day | Litters | Embryos | Adar1+/+ | Adar1+/− | Adar1−/− | Exp. Adar1−/− |
|---|---|---|---|---|---|---|
| Parents crossed; Adar1+/−; Ifnar1−/− | ||||||
| E 11.5 | 2 | 14 | 4 | 7 | 3 | 3.50 |
| E 12.5 | 2 | 13 | 6 | 7 | 0 | 3.25 |
| E 13.5 | 4 | 22 | 4 | 14 | 4 | 5.50 |
| E 14.5 | 4 | 28 | 8 | 15 | 5 | 7.00 |
| E 15.5 | 5 | 33 | 16 | 16 | 1 | 8.25 |
| E 16.5 | 2 | 9 | 0 | 6 | 3 | 2.25 |
| E 17.5 | 2 | 10 | 5 | 5 | 0 | 2.50 |
| Total | 129 | 43 | 70 | 16 | ||
| Parents crossed; Adar1+/−; Mavs−/− | ||||||
| E 11.5 | 7 | 59 | 17 | 32 | 10 | 14.75 |
| E 12.5 | 2 | 15 | 4 | 7 | 4 | 3.75 |
| E 13.5 | 2 | 19 | 5 | 10 | 4 | 4.75 |
| E 14.5 | 2 | 11 | 3 | 7 | 1 | 2.75 |
| E 15.5 | 4 | 29 | 8 | 15 | 6 | 7.25 |
| P 0.5 | 7 | 60 | 17 | 35 | 8 | 15.00 |
| Total | 193 | 54 | 106 | 33 | ||
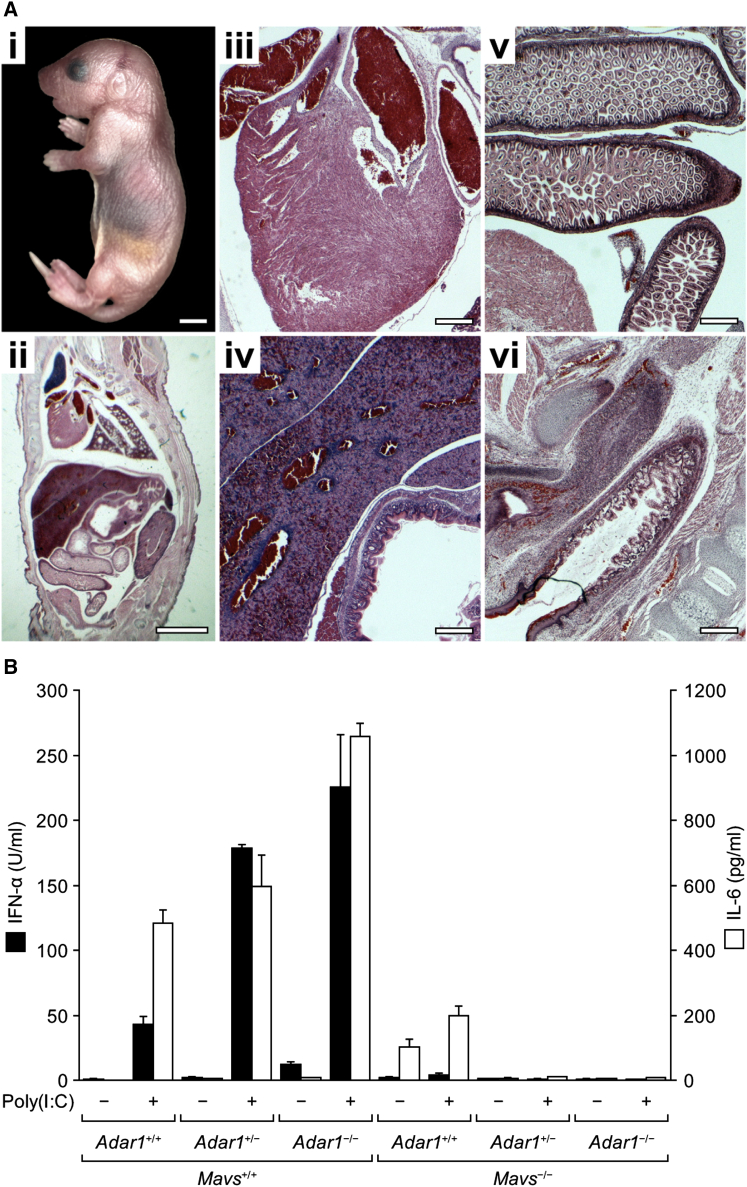
Figure 1.
Partial Rescue of Adar1 Mutant Embryo Viability and Liver Integrity in Adar1; Ifnar1 Double Mutant
(A and B) Whole embryos and corresponding liver sections of wild-type (A) and Adar1 mutant (B) mice at E11.5.
(C–F) Whole embryos and corresponding liver sections of Adar1; Ifnar1 (C and E) and Adar1; Ifnar1 (D and F) mice at E14.5 (C and D) and E15.5 (E and F).
(G and H) Sectioned whole embryos of Adar1+/+; Ifnar1−/− (G) and Adar1−/−; Ifnar1−/− (H) mice at E15.5. Fetal livers outlined by red boxes.
Scale bars, liver sections (A–F), 25 μm; all others, 1 mm.
Although embryonic death is delayed, histological analysis of the Adar1; Ifnar1 embryos revealed that the rescued embryos die with defects similar to those seen in the Adar1 mutant (Figure S1A). Rescue is incomplete and variable at E14.5–E15.5 (Figures 1A–1F), and some embryos still have major defects in liver structure with apoptotic nuclei both in hepatocytes and hematopoietic cells (Figures 1E–1H). Liver, heart, and lungs are underdeveloped in Adar1; Ifnar1 embryos compared to wild-type; there are fewer densely staining hematopoietic cells in the Adar1; Ifnar1 liver, and there is an apparent lack of peripheral blood in or around any of the organs (Figure 1). In the E15.5 Adar1; Ifnar1 liver, 58% of the erythrocytes are still nucleated, versus ∼5% in the wild-type (Figure S1B). A highly similar phenotype was observed in Adar1−/−; Stat1−/− embryos, with hematopoietic defects and subsequent lethality occurring around E15.5 (Figure S1C; Table S1). Stat1 is a key mediator of systemic IFN responses (Stark and Darnell, 2012). Embryonic lethality occurs earlier in Adar1 mutant than in the DNaseII mutant and does not depend on the amplifying effect of high type 1 IFN secretion.
Because Adar1; Ifnar1 double-mutant embryos do survive for 3–4 additional days, this suggested that rescue of Adar1 mutant embryonic lethality might be achieved with a mutation giving a more potent, cell autonomous block to aberrant immune antiviral activation, IFN production, and apoptosis. If cytoplasmic dsRNA in the Adar1 mutant triggers an aberrant antiviral response by activating RLRs, then the key adaptor protein in that signaling pathway is the mitochondrial antiviral signaling protein MAVS. To investigate the effect of blocking signaling from RLRs, we generated strains that produce Adar1−/−; Mavs−/− double-mutant embryos in crosses. The Adar1; Mavs double-mutant embryos have a significantly extended survival up to live birth of pups (Table 1; Figure 2A). Histological sections through a newborn Adar1; Mavs pup show apparently normal liver, heart, and other internal organs (Figure 2A). The Adar1; Mavs double-mutant pups are able to feed, but they die within a day of birth. Preliminary data suggest that the newborn pups still show heightened IFN and interleukin 1 (IL-1) expression in blood (data not shown). This rescue shows that Adar1 mutant embryonic lethality is largely due to aberrant RLR/MAVS pathway signaling triggering apoptosis.
Figure 2.
Rescue of Adar1 Mutant Phenotypes in Adar1; Mavs Double-Mutant Mice and MEF Cultures
(A) Gross visceral anatomy of Adar1−/−; Mavs−/− newborn mice. (i) Appearance of pups at P0.5. (ii–vi) Sections showing general visceral anatomy (ii) with further detail of heart (iii), liver (iv), intestines (v), and rectum (vi). Scale bars in (i) and (ii), 3 mm and (iii)–(vi), 400 μm.
(B) ELISA showing mean levels of IFN-α and IL-6 in cell-culture supernatants of Adar1+/+, Adar1+/−, and Adar1−/− MEFs with Mavs+/+ or Mavs−/− backgrounds following transfection with poly(I:C) (1 μg/ml; +) or water (−). The units on the y axis are expressed per 10,000 cells. Error bars, SD.
To investigate innate immune responses in more detail, six primary mouse embryonic fibroblast (MEF) cultures were established representing all possible Adar1 genotypes in Mavs mutant or wild-type genetic backgrounds. Constitutive and polyriboinosinic:polyribocytidylic acid (poly I:C)-induced levels of IFN-α and interleukin-6 (IL-6) proteins were measured in early-passage MEF culture supernatants by ELISA. Adar1−/−; Mavs+/+ MEFs show a detectably elevated basal level of IFN-α and respond to poly I:C by producing higher levels of both IFN-α and IL-6 than the wild-type (Adar1+/+; Mavs+/+) MEFs. Surprisingly, heterozygous Adar1-/+ MEFs behave somewhat like homozygous Adar1−/− mutant MEFs and respond vigorously to poly I:C treatment with heightened expression of both IFN-α and IL-6. This suggests that prevention of aberrant immune responses in Adar1-/+ MEFs is very sensitive to levels of ADAR1, consistent with evidence of ADAR1 haploinsufficiency in DSH patients (Liu et al., 2006). Confirming that Mavs mutation blocks the aberrant innate immune responses caused by the Adar1 mutation, aberrant expression of type I IFN and proinflammatory cytokines observed in Adar1−/−;Mavs+/+ MEFs is prevented in the Adar1−/−; Mavs−/− double-mutant MEFs; these MEFs also do not induce type I IFN or IL-6 in response to poly I:C treatment (Figure 2B).
Transcriptional Alterations in Adar1 Mutant Embryos Are Primarily Consequences of Aberrant Immune Induction
To investigate whether there is an altered transcriptional profile in Adar1 mutant embryos quantitative RT-PCR (qRT-PCR) analyses were performed to determine the expression levels of 12 interferon-stimulated gene (ISG), transcripts in total RNA from E11.5 whole embryos. Some ISG transcripts are highly elevated in Adar1 mutant whole embryos compared to wild-type embryos and levels return to normal in Adar1; Mavs and Adar1; Ifnar embryos (Figure 3A), though the Adar1; Ifnar1 double-mutant embryo does not normalize ISG transcript levels as fully as Adar1; Mavs. The levels of these ISG transcripts in the Mavs and Ifnar1 mutant embryos are also somewhat different from wild-type levels (Figure S2), and therefore the relevant background has been subtracted from the double-mutant levels (Figure 3A). The qRT-PCR data confirm the ELISA data on cultured fibroblasts showing that the abnormal IFN responses in Adar1 mutant embryos are mediated by aberrant immune signaling through the MAVS adaptor protein.
Figure 3.
Rescue of Adar1 Mutant Embryo Aberrant Proinflammatory Cytokine Expression in Adar1; Ifnar1 and Adar1; Mavs Double-Mutant Embryos
(A) Expression levels for an array of 12 ISGs measured in E11.5 whole embryos. For each gene, Adar1−/−, Adar1−/−; Ifnar1−/−, and Adar1−/−; Mavs values are expressed relative to wild-type, Ifnar1, or Mavs background levels, respectively (one on y axis, dashed lines). Error bars, SEM. ∗p ≤ 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus wild-type; ††p < 0.01; †††p < 0.001 versus Adar1; ‡‡‡p < 0.001 versus Adar1−/−; Ifnar1−/−.
(B) Heatmap showing expression levels for an array of 96 proinflammatory cytokine and control transcripts measured in E11.5 whole embryos. For each gene, Adar1−/−, Adar1−/−; Ifnar1−/−, and Adar1−/−; Mavs−/− values are expressed relative to wild-type, Ifnar1−/−, or Mavs−/− background levels, respectively, which are all zero following log10-transformation and hence are not shown (white in color).
To characterize alterations in gene expression across the whole transcriptome in Adar1 mutant Adar1; Mavs double-mutant embryos, we sequenced ribosomal RNA-depleted total RNA from whole E11.5 embryos. Overall, more transcripts decrease in whole Adar1 embryos than increase (Figure 3B); this may be related to losses of hematopoietic cells or to more widespread defects consistent with apoptosis observed in Adar1 mutant embryos. We identified a set of 61 protein-coding transcripts that are upregulated at least 3-fold in Adar1 mutant embryos and restored to near-wild-type levels in Adar1; Mavs embryos (Table S2). Gene Ontology term analysis on the upregulated transcripts confirms enrichment of immune and antiviral response transcripts (Table S2). To confirm the increased expression of immune gene transcripts in the Adar1 mutant, qRT-PCR expression analyses were performed with total RNA from E11.5 whole embryos. Changes in proinflammatory cytokine transcript expression in Adar1; Ifnar1 and Adar1; Mavs embryos were calculated relative those in Ifnar1 and Mavs mutants, which were again slightly different from wild-type levels (Figure S2). Consistent with the sequencing data, levels of certain ISGs and proinflammatory cytokines were highly increased in the Adar1 mutant compared with wild-type (Figures 3A and 3B).
Altered expression levels of immune gene transcripts in the Adar1 mutant embryo are restored close to wild-type levels in both Adar1; Ifnar1 and Adar1; Mavs double-mutant embryos. The increase in the expression of the transcription factor Stat1 is very striking in the Adar1 embryo at E11.5 and the Stat1 transcript is reduced in both Adar1; Ifnar1 and Adar1; Mavs double-mutant embryos. STAT1 protein is critical to the cellular response to extracellular IFN (for review, see Stark and Darnell, 2012); however, consistent with the similarly limited effect of the Ifnar1 mutation, generating an Adar1; Stat1 double mutant also does not rescue Adar1 mutant embryonic lethality fully (Figure S1C). The stronger rescue of embryonic lethality by Mavs mutation may not be due mainly to stronger effects on transcription, but rather the Mavs mutation may also block apoptosis or other nontranscriptional effects of aberrant RLR signaling. Together both the qRT-PCR and ELISA data confirm that the abnormal responses in Adar1 mutant embryos are mediated by immune signaling through MAVS.
To examine transcription in more detail in embryonic hematopoietic cells, we sequenced ribosomal RNA-depleted total RNA from E15.5 embryonic livers of Adar1; Mavs and Mavs sibling embryos; liver is the main site of hematopoiesis at this stage. Adar1 mutant embryos die by E12.5 when embryonic liver is still small. Adar1; Mavs E15.5 fetal livers appear normal and comparisons of RNA sequencing (RNA-seq) data from Adar1; Mavs and Mavs livers could not identify any protein-coding transcripts expressed at significantly differing levels. The lack of aberrant transcription in Adar1; Mavs livers shows that all observed alterations in levels of protein-coding transcripts in the Adar1 mutant are effects of aberrant RLR signaling and interferon production. Transcriptional alterations are not caused directly by loss of ADAR1 protein per se.
It has been proposed that RNA editing could increase turnover of edited dsRNA due to cleavage by an inosine-specific ribonuclease, but the physiological effectiveness of this turnover process is not known (Scadden, 2005). If increased levels of repetitive transcripts with dsRNA-forming potential do arise in the Adar1 mutant, these might cause the aberrant antiviral response. If repetitive transcripts increase as a direct result of the absence of Adar1, the aberrant repetitive transcript levels should be detected also in the Adar1; Mavs double mutant. To assess expression of transcripts likely to form dsRNA, a detailed analysis of repetitive sequence expression in Adar1; Mavs and Mavs E15.5 liver total RNA-seq data was performed (Figure 4). Increased expression of individual members of ERV and IAP subfamilies was observed (Table S3), and qRT-PCR analyses confirmed increased expression of MMERVK10C transcripts (Figure 4E). Overall, however, changes in repetitive transcripts parallel those seen for protein-coding transcript; i.e., levels of most repetitive elements did not change between Adar1; Mavs and Mavs. This suggests that a general dramatic increase in levels of repetitive transcripts due to slowed turnover is not the initiating event in the aberrant immune response in Adar1 mutants. Changes in some individual repetitive transcripts could contribute to initiating aberrant immune responses in the Adar1 mutant, but the reduction in inosine levels within cellular dsRNA is perhaps more critical.
Figure 4.
Repetitive Element Expression in Adar1; Mavs Embryonic Mouse Liver
(A–C) Expression profiles of repetitive element classes (A), retrotransposon subfamilies (B), and specific LTR retrotransposon populations (C) in Adar1−/−; Mavs−/− E15.5 liver total RNA relative to Mavs−/− (zero on x axis).
(D) Schematic of the prototypical mouse RLTR10C-flanked MMERVK10C retrotransposon (generated using Jurka et al., 2005). Positions and lengths of the gag, pro, pol, and env genes are shown. Black bars indicate qRT-PCR products (i–xi) generated using specific primer pairs (Table S4).
(E) Expression levels of RLTR10C-flanked MMERVK10C retrotransposon regions shown in (D) in Adar1−/−; Mavs−/−E15.5 livers relative to Mavs−/− (one on y axis, dashed line).
Error bars, SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001 versus Mavs for each region.
Restoring Expression of Editing-Competent ADARs Prevents Aberrant Innate Immune Responses in Adar1 Mutant Mouse Fibroblasts
To determine whether the aberrant antiviral response in Adar1 mutant cells is due to reduced inosine modification in intracellular RNAs, we established MEF cell cultures from Adar1; p53 double-mutant embryos. Adar1 MEFs could not be cultured long term due to cell death (Figure S3), and Adar1; Mavs double-mutant MEFs are not useful because the aberrant innate immune response is blocked. When compared with p53 mutant MEFs, elevated expression of ISGs is observed in Adar1; p53 MEFs when cultures are stressed by nutrient starvation (Figure 5A); without stress, ISG expression is very low in Adar1; p53 MEFs. We chose the p53 mutant background because this prevents cell death associated with a range or other mutations (Dittmer et al., 1993). The reduction in aberrant ISG expression in Adar1; p53 versus Adar1 MEFs is consistent with the idea that preventing cell death is also an important feature of the Mavs mutant rescue.
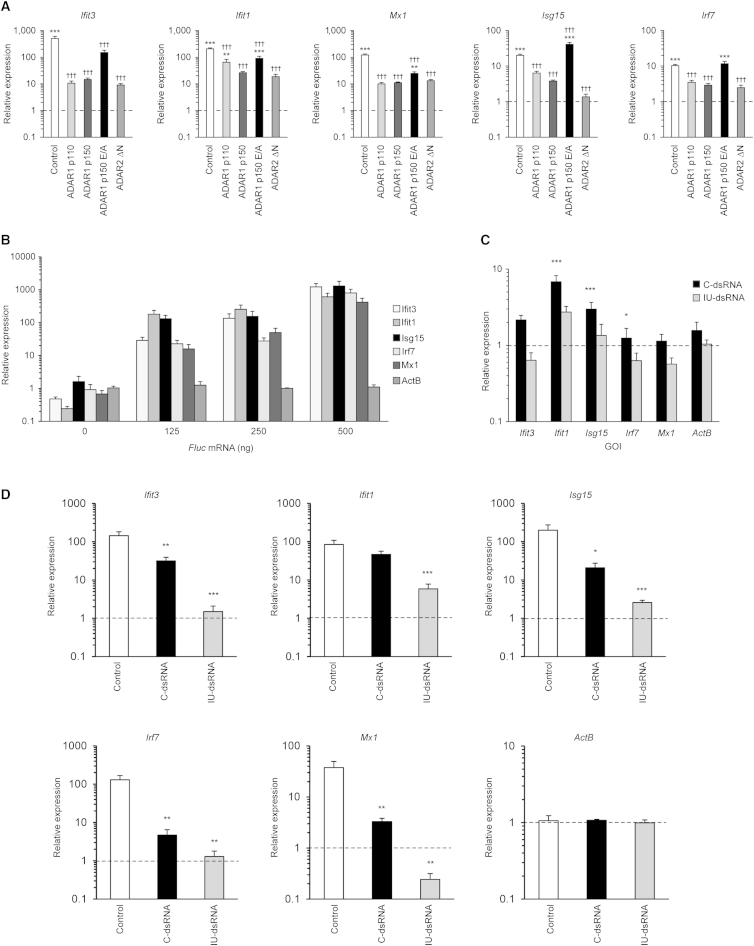
Figure 5.
Aberrant Innate Immune Responses in Adar1−/−; p53−/− Double-Mutant MEFs Are Suppressed by Expression of ADAR Proteins or IU-dsRNA
(A) Expression levels for an array of five ISGs measured in nutrient-starved (72 hr) Adar1−/−; p53−/− MEF cultures following stable knockin of GFP (control) or ADAR proteins. For each gene, values are expressed relative to p53−/− (one on y axis, dashed lines) and normalized to the housekeeping gene ActB. Error bars, SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus p53−/−; ††p < 0.01; †††p < 0.001 versus Adar1−/−; p53−/−.
(B) Expression levels for an array of 5 ISGs following transfection of Adar1−/−; p53−/− MEF cultures with 0–500 ng Fluc mRNA after 12 hr. For each gene, values are expressed relative to mock transfection (0 ng) at 12 hr and normalized to the housekeeping gene ActB. Error bars, SEM.
(C) Expression levels for an array of five ISGs following transfection of Adar1−/−; p53−/− MEF cultures with control dsRNA (C-dsRNA) or IU-dsRNA after 12 hr. For each gene, values are expressed relative to that seen after 6 hr in cells transfected with control dsRNA (C-dsRNA) (one on y axis, dashed lines) and normalized to the housekeeping gene ActB. Error bars, SEM. ∗p < 0.05; ∗∗∗p < 0.001.
(D) Expression levels for an array of five ISGs and one housekeeping gene following transfection of Adar1−/−; p53−/− MEF cultures with 500 ng Fluc mRNA (control), with either control dsRNA (C-dsRNA) or IU-dsRNA. For each gene, values are expressed relative to expression after 6 hr in cells transfected with control dsRNA (C-dsRNA) (one on y axis, dashed lines) and normalized to the housekeeping gene ActB.
Error bars, SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 versus control.
To test for rescue of Adar1 mutant phenotypes by ADAR protein variants, we next established stable Adar1; p53 MEF lines expressing wild-type or mutant human ADAR1 proteins from stably integrated PiggyBac constructs that we previously characterized (Heale et al., 2009). The Adar1; p53 PiggyBac MEFs were starved for 72 hr to stress the cells, total RNA was isolated and qRT-PCR was performed to measure ISG transcript levels. Expression levels of ISG transcripts were normal in p53 mutant cells and highly elevated in Adar1; p53 cells (Figure 5A). When either the predominantly nuclear ADAR1 p110 isoform or the IFN-inducible mainly cytoplasmic ADAR1p150 isoform were expressed, ISG transcripts were significantly reduced (Figure 5A). Expressing the ADAR1p150 (E912A) mutant that inactivates the deaminase catalytic site gave the least effective reduction in ISG levels. These data indicate that robust suppression of ISG transcription requires a catalytically active ADAR capable of deaminating adenosine to inosine into RNA.
Expressing human ADAR2ΔN, a nuclear localization mutant of ADAR2 that aberrantly accumulates in the cytoplasm (Wong et al., 2003), in Adar1; p53 MEFs also significantly reduced expression of the ISGs (Figure 5A). This suggests that restoring RNA-editing activity in the cytoplasm is particularly important. MEF cells already express endogenous, active, ADAR2 in the nucleus, and site-specific editing of protein-coding transcripts usually involves transient exon-intron RNA duplexes formed before splicing. ADAR2 proteins have different specific site preferences from ADAR1 proteins (Keegan et al., 2011). The ADAR2ΔN is more likely to rescue nonspecific, promiscuous editing events lost in the Adar1 mutant than it is to edit some transcript containing a highly ADAR1-specific site. The sufficiency of either catalytically active ADAR for strong rescue is consistent with the idea that loss of promiscuous editing in intracellular dsRNA is critical to the Adar1 mutant phenotype.
Transfecting dsRNA Containing I-U Base Pairs Prevents Aberrant Antiviral Responses in Adar1 Mutant Mouse Fibroblasts
If cellular dsRNA lacking inosine in Adar1 mutant cells is sufficient to induce an aberrant innate immune response, then isolating RNA from Adar1 mutant cells and transfecting it into wild-type cells might replicate this effect. Therefore, we extracted total RNA from livers of Adar1; Mavs E15.5 embryos and transfected this into RIG-I reporter cells but could not detect any difference between the responses induced by Adar1; Mavs and wild-type RNA samples. Because the majority of a total RNA sample is rRNA or tRNAs, this experiment may not mimic in vivo conditions. We decided to perform instead the converse experiment, which was to determine if inosine-containing RNAs interfere with immune induction by transfected RNA.
To elucidate if transfecting dsRNA oligonucleotides containing inosine-uracil base pairs suppress aberrant ISG expression in Adar1 mutant phenotype, we established a test system using Adar1; p53 MEFs. To partially induce innate immune responses in Adar1; p53 MEFs, we transfected these cells with levels of an in-vitro-transcribed Fluc mRNA that has been shown previously to induce a modest innate immune response in HeLa cells (Vitali and Scadden, 2010). Transfecting similar concentrations of Fluc RNA into the hypersensitive Adar1; p53 MEF cells caused a dramatic increase in expression of ISG transcripts (Figure 5B), as in the case of serum starvation (Figure 5A). This strong response of Adar1; p53 MEFs to transfected RNA is likely to reflect the aberrant immune induction due to the Adar1 mutation.
Transfecting dsRNA oligos alone into Adar1; p53 MEFs has very little effect on immune induction at the concentrations used (Figure 5C). dsRNA oligonucleotide (C-dsRNA) or IU-dsRNA that contained four inosine-uracil base pairs were then cotransfected with 500 ng Fluc RNA to look for specific inhibitory effects of IU-dsRNA oligonucleotides on ISG transcript induction (Figure 5C). IU-dsRNAs further reduce ISG transcript expression below reductions caused by control C-dsRNAs (Figure 5D). These reductions in ISG transcript levels were maintained for 24 hr with the IU-dsRNA, whereas the C-dsRNA was unable to reduce the ISG response at this time point (data not shown). The background inhibitory effect of the control C-dsRNA oligonucleotide in these experiments could arise if the 20-mer dsRNA oligonucleotides bind only one RLR molecule each, thus impeding oligomerization of RLR CARD domains (Peisley et al., 2014). It is clear that transfection of I-U dsRNA specifically inhibits aberrant immune induction further. Both this specific effect of I-U base pairs and the general suppressive effect of 20-mer dsRNA oligonucleotides indicate that aberrant RNA complexes formed by RLRs are central to the Adar1 mutant phenotype.
Mutations in Human ADAR1 Associated with AGS Have More Severe Effects on the IFN-Inducible Isoform of ADAR1
Most of the mutations in ADAR1 occurring in patients with AGS change residues on the surface of the catalytic domain at the C terminus of the protein (Figure 6), close to where dsRNA is predicted to bind. Previously, we tested the effects of these AGS mutations on enzymatic activity in ADAR1p110 (Rice et al., 2012). Constructs expressing ADAR1p110 were transiently cotransfected into HEK293 cells with a construct expressing a known editing substrate for ADAR1 and editing activity was measured. Surprisingly, most of the AGS mutant proteins, with the exception of the ADAR1 G1007R mutation, still exhibited robust editing activity, though with statistically significant reductions.
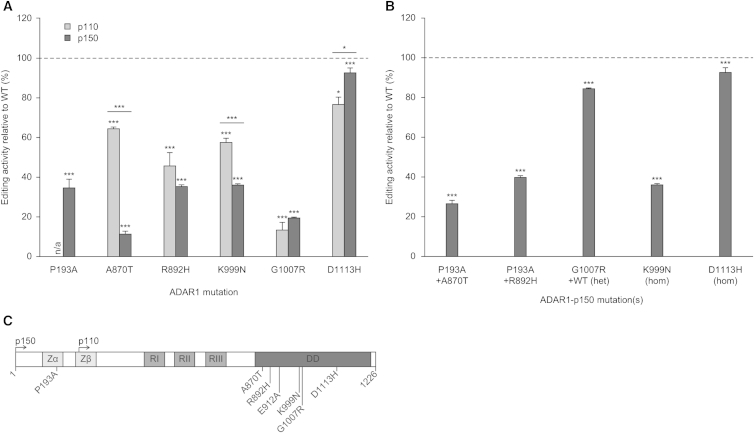
Figure 6.
Mutations in ADAR1 that Cause Aicardi-Goutières Syndrome Affect RNA-Editing Activity
The first letter denotes the original amino acid and the second letter the mutation; the number is the position of the amino acid in the p150 isoform.
(A) Editing activity of each ADAR1 AGS mutant in either the p110 or p150 isoform expressed in HEK293T cells relative to wild-type p110 or p150, respectively (100% on y axis, dashed line). Error bars, SEM. ∗p ≤ 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(B) Editing activity of the ADAR1 mutant combinations found in an AGS patient cohort in the p150 isoform expressed in HEK293T cells relative to wild-type p150 (100% on y axis, dashed line). Error bars, SEM. ∗∗∗p < 0.001 versus wild-type p150.
(C) A diagram of ADAR1 illustrating where the AGS mutations occur in the protein.
We now tested the effects of the same AGS mutations in ADAR1p150 in a similar manner (Figure 6A). Each of the AGS mutations causes a significant decrease in editing activity of ADAR1p150 that is greater than the effect of the same mutation in the ADAR1p110 isoform. This large difference between mutant effects in the two isoforms was not anticipated because the AGS mutations are predominantly in the C-terminal domains shared by the two isoforms. We provide a report of mutations in ADAR1 that have different effects on the two isoforms, revealing a significant separation of roles for the two isoforms. These findings also imply that the effects of AGS mutations in ADAR1 are mediated particularly through ADAR1p150 and primarily involve reductions in RNA-editing activity. Loss of the IFN-inducible cytoplasmic ADAR1 isoform may be important in AGS and dystonia patients. Clinical observations have suggested that symptoms may develop following an infection when type I IFN expression should have led to later expression of active ADAR1p150 (Livingston et al., 2014)
ADAR1 mutant proteins found in AGS patients were also tested for effects on RNA-editing activity in HEK293 cells when expressed in combinations matching those observed in the AGS patient cohort (Figure 6B). Evidence for interactions between different AGS mutations in the same ADAR1 protein and for interactions between different ADAR1 mutant proteins in heterozygotes was obtained. For instance, we found that the ADAR1p150 P193A mutant exhibits a significant decrease in editing activity (Figure 6A). The P193A mutation is expressed only in the N-terminally extended ADAR1p150 isoform and increases the effect of other AGS mutations in the same protein (Figure 6B). The crystal structure of the ADAR1 N-terminal Z-alpha domain complex with Z-RNA has been solved, and the P193A mutation is predicted to change a highly conserved amino acid involved in nucleic acid contact (Schade et al., 1999). This variant is enriched in the AGS cohort but is also present in 41 of 6,553 control patients in the Exome Variant Server database. An instance of strong interaction between different ADAR1 protein variants in AGS heterozygote patients is seen with the ADAR1p110 G1007R mutant that binds dsRNA but is catalytically inactive and which exerts a dominant-negative effect on editing by ADAR1p110 wild-type protein (Rice et al., 2012) (Figure 6B).
Discussion
This study elucidates a key conserved role for ADARs in preventing aberrant IFN responses by cytoplasmic antiviral RLR innate immune RNA sensors. Mouse Adar1 mutant embryos die by embryonic day E12.5 with aberrant IFN expression, failure of hematopoiesis, and degeneration of the embryonic liver (Hartner et al., 2004, 2009; Wang et al., 2004). We obtained partial rescues of Adar1 mutant embryonic lethality by combination with Ifnar1 or Stat1 mutations and a much more substantial rescue to live birth by combination with a Mavs mutation. The Mavs rescue, in particular, indicates that the Adar1 embryonic lethal mutant phenotype is largely due to an aberrant antiviral response. Some mutations affecting RLRs have phenotypes similar to Adar1 mutants, for example, a mutation in the mouse Ifih1 gene encoding MDA5 that causes constitutive IFN signaling (Funabiki et al., 2014), giving rise to a Lupus-like autoimmune disorder. Mutations in Mavs but not Ifnar1 also rescued the aberrant immune response in those Ifih1 mice. In addition, with striking similarity to ADAR1, mutations in human IFIH1 also cause AGS (Rice et al., 2014). Our preliminary data indicate, however, that combining Ifih1 with Adar1 does not rescue the lethality of the Adar1 mutant mouse embryo (data not shown), suggesting that MDA5 is not the sole mediator of the Adar1 mutant phenotype.
Loss of the late IFN-inducible cytoplasmic ADAR1 p150 isoform, in particular, may be critical to the Adar1 mutant phenotype. This isoform interacts in a complex way with viruses and may participate in the resolution phase of the IFN response by editing residual viral RNAs (Ward et al., 2011). ADARs have been reported as being “proviral” because ADARs facilitate replication of viruses in cultured cells (Samuel, 2011). However, proviral effects are unusual among proteins induced by IFN, and these experiments in cell cultures may be somewhat misleading. Studies on Adar1 mutant mice lacking only the ADAR1 p150 isoform instead suggest an overall negative effect of ADAR1 p150 on measles virus propagation, illustrating the importance of whole-animal studies (Ward et al., 2011). Hepatitis C virus encodes a protease that cleaves MAVS, and it would be interesting to determine whether this virus or protease rescue aspects of Adar1 mutant phenotypes.
We show that signaling from antiviral innate immune sensors through MAVS is inhibited by dsRNA oligonucleotides containing inosine-uracil base pairs. Transfection of I-U dsRNA corresponding to an ADAR1-edited section of an artificial dsRNA substrate into Adar1 mutant MEFs suppresses the aberrant antiviral response. The data suggest that the aberrant antiviral response in the Adar1 mutant results from loss of inosine in cytoplasmic dsRNA that normally inhibits antiviral sensor activation. Other examples of how changes in modification states of RNAs determine innate immune responses come from viruses encoding ribose 2′-O-methylation enzymes that methylate viral mRNA cap structures to match host mRNA caps so that they avoid activating the MDA5 sensor (Züst et al., 2011). Vertebrate tRNAs also have specific modifications at certain positions in their structures that allow innate immune sensors to discriminate between them and bacterial tRNAs (Gehrig et al., 2012). Modifications that occur naturally in host RNAs also prevent activation of other innate immune sensors such as PKR and Toll sensors (Nallagatla et al., 2008). Because transfection of I-U dsRNA blocks the aberrant immune activation in Adar1; p53 MEFs, it is most likely that the aberrant immune activation is due to an endogenous dsRNA that activates RLRs. We cannot exclude the possibility that a site-specific editing event contributes, but, if the Adar1 mutant aberrant immune induction was due to loss of editing in a transcript encoding an innate immune protein or any other indirect effect, then inhibiting RLRs at the level of RNA interaction would be unlikely to rescue.
In the case of I-U dsRNA inhibition of RLRs, the significant effect of sequential I-U wobble base pairs bending and destabilizing dsRNA (McLaughlin and Keegan, 2014) makes it possible to envisage why the sensors are able to detect edited dsRNA (Figure S4). I-U dsRNA binds the RLRs competitively with activating dsRNA (Vitali and Scadden, 2010), probably making grossly similar contacts to the helicase domains and carboxy-terminal domains. RLRs are suitable to assess dsRNA structure because they surround the dsRNA using multiple protein domains to make many contacts to both strands (Kowalinski et al., 2011; Luo et al., 2011). The Hel2i domain, in particular, could have a role in scanning the dsRNA minor groove to detect I-U base pairs and other imperfections (Figures S4C and S4D), possibly leading to alterations in dynamic RLR domain rearrangements required for signaling. Natural RNA duplexes formed by repetitive elements in transcripts will usually be imperfectly paired, and RNA editing will further reduce helical regularity. Formation of regular filaments by RLRs on dsRNA facilitates formation of CARD domain complexes activating MAVS signaling (Kowalinski et al., 2011; Peisley et al., 2014). RLR signaling may be exquisitely sensitive to mismatches and RNA-editing events present in RNA duplexes that indicate the dsRNA is not a direct product of virus replication.
In Adar1; p53 mutant MEFs, the catalytically inactive ADAR1 protein is the least effective at reducing elevated ISG transcripts though it is still RNA binding competent, demonstrating that RNA-editing activity is required. Catalytically active ADAR1 proteins and even a cytoplasmically mislocalized ADAR2 mutant protein (Wong et al., 2003) reduce the aberrantly elevated ISG transcript levels. This emphasizes the role of RNA-editing activity, argues against a dominant role for ADAR1-specific protein-protein or protein-RNA interactions, and suggests that promiscuous editing by ADARs is involved rather than a highly ADAR1-specific editing event. No ADAR1 protein fully suppressed ISG transcripts to levels observed in the p53−/− mutant, but the reintroduced ADAR1 proteins are not expressed under the control of the IFN-inducible Adar1 promoter itself. Because transfection of general dsRNA oligonucleotides containing inosine-uracil base pairs significantly reduces elevated ISG levels in Adar1; p53 MEFs, the importance of inosine in RNA in preventing the innate immune response is supported in two distinct ways.
Several of the AGS mutations alter residues on the surface of the deaminase domain close to where RNA is predicted to bind (Rice et al., 2012), and it is possible that the D1113H mutation, which affects editing activity the least, alters ADAR1 function in other ways such as by perturbing protein and/or RNA interactions required for suppression of innate immunity. In addition to the catalytically active ADAR1 and ADAR2 proteins, two other vertebrate ADARs lack catalytic deaminase activity (for review, see Heale and O’Connell, 2009). Therefore, editing-independent ADAR roles are maintained and could involve protein interactions on dsRNA or sequestration of dsRNA.
While this manuscript was under review, Wang and colleagues showed a substantially stronger editing-independent role of ADAR1 in decreasing innate immune responses in HEK293 (Yang et al., 2014). We also observe some editing-independent rescue, and we suspect that the difference is due to our use of Adar1; p53 MEFs completely lacking ADAR1 and their use of HEK293 cells.
We do not know why Adar1; Mavs newborn pups die within a day of birth, but preliminary data on cytokine expression in blood from these pups implicate some residual inflammatory effects. Activated RLRs can bypass MAVS and induce inflammasome responses via an alternative pathway (Lucioli et al., 2013). Cell death is a very prominent effect in the Adar1 mutant, and possible MAVS-bypass signaling to cell death pathways in some cells in Adar1; Mavs newborn pups also remains to be investigated. Many important autoimmune diseases show transcriptional signatures of elevated ISG expression. Our findings suggest, however, that preventing the aberrant systemic IFN response may not always be sufficient to treat these diseases. When aberrant RLR signaling is activated, the key defects may be cell autonomous. Correcting them will require a much more detailed understanding of intracellular nucleic acid sensing. When the important cellular modified nucleic acids that modulate innate immune responses have been defined, they may point the way to new therapeutic approaches to autoimmune and neurodegenerative diseases.
Experimental Procedures
Additional methods are online in the Supplemental Information.
Mouse Genetics
Adar1Δ2-13 mice in the C57Bl6N background were obtained from M. Higuchi and P. Seeburg (strain C192) (Hartner et al., 2004). These mice were crossed to the IFN receptor subunit mutant (Ifnar1) in the 129 strain background (B&H Animal Suppliers). Two sublines breed Adar1−/+ in the Ifnar1−/− background, and crosses of these mice generate Adar1−/−; Ifnar1−/− embryos. Two further sublines generate heterozygous Adar1−/+ animals with the mixed B6/129 strain background, and crosses of these generate control Adar1−/− embryos without the Ifnar1 mutation. The CARDIF−/− (MAVS, IPS-1, or VISA) mutant line generated by the Tschopp group in Lausanne was obtained from Caetano Reis e Sousa in London. It is homozygous-viable null mutant in the C57B6J background. All experiments on mice were performed in accordance with Edinburgh University rules and National Guidelines on animal experimentation.
Knockin of ADAR cDNA in MEFs
cDNAs encoding ADAR proteins were subcloned into the PiggyBac expression system (System Biosciences) and transfected into Adar1−/−; p53−/− MEFs using LyoVec (InvivoGen). Cells were grown for 2 weeks, and those with successful integration were isolated by fluorescence-activated cell sorting (GFP positive).
Mouse Immune Array
The TaqMan Array Mouse Immune Panel (CN: 4367786; Life Technologies) was used for quantitative gene expression analysis of the immune response signatures in E11.5 whole embryos. Total RNA was extracted and purified, and cDNA was generated as described above. The Taqman array card was loaded with the first strand cDNA sample and Taqman Universal PCR Master Mix II UNG (Life Technologies) according to the manufacturer’s protocol. The amplification was performed on an upgraded version of the Applied Biosystems 7900HT Fast Real-Time PCR System, and the analysis of the data was performed as described above.
Author Contributions
N.M.M. performed experiments and assisted with writing the manuscript; S.M.G. performed experiments and assisted with writing the manuscript; R.Y. performed bioinformatic analyses of mouse sequences; S.C. performed experiments; J.B. performed genotyping of mouse strains; D.R. performed genotyping of mouse strains; C.N. performed bioinformatic analyses of mouse sequences; C.V. performed experiments; C.P.P. assisted with bioinformatic analyses of mouse sequences; P.J.M. performed RNA-protein structural analyses, M.F.J. designed experiments; J.D. advised on innate immunity and mouse genetics; I.R.A. performed bioinformatic analyses of repeats; A.D.J.S. designed experiments and provided reagents; M.Ö. assisted with writing the manuscript; L.P.K. performed experiments, designed experiments, and wrote the manuscript; and M.A.O. designed experiments and wrote the manuscript.
Acknowledgments
We would like to thank Prof. P. Seeburg and Dr. M. Higuchi for the Adar1 mutant mice, Prof. Caetano Reis e Sousa for the Mavs mice, and Prof. N. Hastie for his support. This work was funded by the Medical Research Council.
Published: November 13, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes four figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.10.041.
Contributor Information
Liam P. Keegan, Email: liam.keegan@igmm.ed.ac.uk.
Mary A. O’Connell, Email: mary.oconnell@igmm.ed.ac.uk.
Accession Numbers
The GEO Accession number for the mouse sequence data from this project is GSE62917.
Supplemental Information
References
- Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2013;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capshew C.R., Dusenbury K.L., Hundley H.A. Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res. 2012;40:8637–8645. doi: 10.1093/nar/gks590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro J.M., Keegan L.P., Lafarga M., Berciano M.T., O’Connell M., Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Dittmer D., Pati S., Zambetti G., Chu S., Teresky A.K., Moore M., Finlay C., Levine A.J. Gain of function mutations in p53. Nat. Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- Funabiki M., Kato H., Miyachi Y., Toki H., Motegi H., Inoue M., Minowa O., Yoshida A., Deguchi K., Sato H. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Gehrig S., Eberle M.E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W.M., Erdmann V.A., Sprinzl M. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 2012;209:225–233. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P., Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- Hartner J.C., Schmittwolf C., Kispert A., Müller A.M., Higuchi M., Seeburg P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Hartner J.C., Walkley C.R., Lu J., Orkin S.H. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale B.S.E., O’Connell M.A. Biological Roles of ADARs. In: Grosjean H., editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience; Austin: 2009. pp. 243–258. [Google Scholar]
- Heale B.S., Keegan L.P., McGurk L., Michlewski G., Brindle J., Stanton C.M., Caceres J.F., O’Connell M.A. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Keegan L.P., McGurk L., Palavicini J.P., Brindle J., Paro S., Li X., Rosenthal J.J., O’Connell M.A. Functional conservation in human and Drosophila of Metazoan ADAR2 involved in RNA editing: loss of ADAR1 in insects. Nucleic Acids Res. 2011;39:7249–7262. doi: 10.1093/nar/gkr423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Liu Q., Jiang L., Liu W.L., Kang X.J., Ao Y., Sun M., Luo Y., Song Y., Lo W.H., Zhang X. Two novel mutations and evidence for haploinsufficiency of the ADAR gene in dyschromatosis symmetrica hereditaria. Br. J. Dermatol. 2006;154:636–642. doi: 10.1111/j.1365-2133.2006.07133.x. [DOI] [PubMed] [Google Scholar]
- Livingston J.H., Lin J.P., Dale R.C., Gill D., Brogan P., Munnich A., Kurian M.A., Gonzalez-Martinez V., De Goede C.G., Falconer A. A type I interferon signature identifies bilateral striatal necrosis due to mutations in ADAR1. J. Med. Genet. 2014;51:76–82. doi: 10.1136/jmedgenet-2013-102038. [DOI] [PubMed] [Google Scholar]
- Lucioli J., Pinton P., Callu P., Laffitte J., Grosjean F., Kolf-Clauw M., Oswald I.P., Bracarense A.P. The food contaminant deoxynivalenol activates the mitogen activated protein kinases in the intestine: interest of ex vivo models as an alternative to in vivo experiments. Toxicon. 2013;66:31–36. doi: 10.1016/j.toxicon.2013.01.024. [DOI] [PubMed] [Google Scholar]
- Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin P.J., Keegan L.P. Conflict RNA modification, host-parasite co-evolution, and the origins of DNA and DNA-binding proteins1. Biochem. Soc. Trans. 2014;42:1159–1167. doi: 10.1042/BST20140147. [DOI] [PubMed] [Google Scholar]
- Nallagatla S.R., Toroney R., Bevilacqua P.C. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Cheng Y., Tan B.C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., del Toro Duany Y., Jenkinson E.M., Forte G.M., Anderson B.H., Ariaudo G., Bader-Meunier B., Baildam E.M., Battini R., Beresford M.W. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden A.D. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Schade M., Turner C.J., Lowenhaupt K., Rich A., Herbert A. Structure-function analysis of the Z-DNA-binding domain Zalpha of dsRNA adenosine deaminase type I reveals similarity to the (alpha + beta) family of helix-turn-helix proteins. EMBO J. 1999;18:470–479. doi: 10.1093/emboj/18.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Darnell J.E., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Vitali P., Scadden A.D. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Miyakoda M., Yang W., Khillan J., Stachura D.L., Weiss M.J., Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Ward S.V., George C.X., Welch M.J., Liou L.Y., Hahm B., Lewicki H., de la Torre J.C., Samuel C.E., Oldstone M.B. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:331–336. doi: 10.1073/pnas.1017241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Sato S., Lazinski D.W. Elevated activity of the large form of ADAR1 in vivo: very efficient RNA editing occurs in the cytoplasm. RNA. 2003;9:586–598. doi: 10.1261/rna.5160403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Deng P., Zhu Z., Zhu J., Wang G., Zhang L., Chen A.F., Wang T., Sarkar S.N., Billiar T.R., Wang Q. Adenosine Deaminase Acting on RNA 1 Limits RIG-I RNA Detection and Suppresses IFN Production Responding to Viral and Endogenous RNAs. J. Immunol. 2014;193:3436–3445. doi: 10.4049/jimmunol.1401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Okabe Y., Kawane K., Fukuyama H., Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.