Summary
Cutaneous C-unmyelinated MRGPRD+ free nerve endings and C-LTMRs innervating hair follicles convey two opposite aspects of touch sensation: a sensation of pain and a sensation of pleasant touch. The molecular mechanisms underlying these diametrically opposite functions are unknown. Here, we used a mouse model that genetically marks C-LTMRs and MRGPRD+ neurons in combination with fluorescent cell surface labeling, flow cytometry, and RNA deep-sequencing technology (RNA-seq). Cluster analysis of RNA-seq profiles of the purified neuronal subsets revealed 486 and 549 genes differentially expressed in MRGPRD-expressing neurons and C-LTMRs, respectively. We validated 48 MRGPD- and 68 C-LTMRs-enriched genes using a triple-staining approach, and the Cav3.3 channel, found to be exclusively expressed in C-LTMRs, was validated using electrophysiology. Our study greatly expands the molecular characterization of C-LTMRs and suggests that this particular population of neurons shares some molecular features with Aβ and Aδ low-threshold mechanoreceptors.
Graphical Abstract

Highlights
-
•
GINIP and IB4 staining segregate DRG neurons in four distinct subpopulations
-
•
MRGPRD+ free nerve endings and C-LTRMs display distinct transcriptional profiles
-
•
C-LTMRs share some molecular features with Aβ/Aδ low-threshold mechanoreceptors
-
•
Cav3.3 calcium channels are specific to C-LTMRs
Reynders et al. combined FACS and RNA deep sequencing to establish the transcriptional profiles of MRGPRD+ free nerve endings and C-LTMRs. They validated their data using an extensive in situ screen and demonstrated that Cav3.3 channel was exclusively expressed and functionally active in C-LTMRs.
Introduction
Skin is the largest sensory organ of the body and is densely equipped with highly specialized sensory nerve endings capable of sensing a wide range of sensory stimuli such as light touch, mechanical pressure, temperature, itch, and pain. These cutaneous sensory afferents can be distinguished by many morphological, anatomical, electrophysiological, and molecular criteria (Liu and Ma, 2011, Owens and Lumpkin, 2014). For example, based on their cell body size, degree of myelination, and axonal conduction velocity, cutaneous fibers can be split into Aβ, Aδ, or C-fibers (Abraira and Ginty, 2013, Bessou and Perl, 1969, Li et al., 2011, Smith and Lewin, 2009, Zimmermann et al., 2009). C-fibers represent the majority of cutaneous sensory afferents. They can be split into free nerve endings, found both in glabrous and hairy skin where they are embedded between the keratinocytes of the epidermis, and a particular population of C-unmyelinated fibers, found exclusively in hairy skin where they form longitudinal lanceolate endings around hair follicles (Delfini et al., 2013, Li et al., 2011) or terminate in large arborizations similar to C-fiber tactile afferent receptive fields (Liu et al., 2007, Vrontou et al., 2013). Cutaneous free nerve endings are tuned to respond to itch-inducing compounds and pain-evoking thermal, mechanical, and chemical stimuli (Zimmermann et al., 2009). MRGPRD free nerve endings belong to a subset of non-peptidergic nociceptors that convey noxious mechanical and β-alanin-induced itch stimuli (Cavanaugh et al., 2009, Liu et al., 2012), whereas C-low-threshold mechanoreceptors (C-LTMRs) have been proposed to contribute to light touch under normal conditions (Bessou et al., 1971, Douglas and Ritchie, 1957, Johansson et al., 1988, Li et al., 2011, Löken et al., 2009, Maruhashi et al., 1952, Olausson et al., 2002, Seal et al., 2009, Zotterman, 1939) and to touch hypersensitivity after injury (Delfini et al., 2013, Liljencrantz et al., 2013, Seal et al., 2009). Our knowledge of the molecular contents that contribute to the functional specialization of these diametrically different subpopulations of cutaneous afferents is still at its infancy. In a recent study (Gaillard et al., 2014), we identified a novel Gα inhibitory interacting protein (GINIP) that marks two distinct subsets of non-peptidergic neurons: the cutaneous free nerve endings MRGPRD+ neurons (Dong et al., 2001) and the V-GLUT3-, TH-, and TAFA4-expressing C-LTMRs (Delfini et al., 2013, Li et al., 2011, Seal et al., 2009). Here, we took advantage of the GINIPm-Cherry mouse model in combination with isolectin B4 (IB4) cell surface staining and fluorescence-activated cell sorting (FACS). We succeeded to purify three distinct populations of DRG neurons and subjected their respective total RNA contents to RNA deep-sequencing technology (RNA-seq). As expected, GINIP+/IB4+ neurons showed a remarkable enrichment in Mrgprd transcripts and GINIP+/IB4− neurons revealed a striking enrichment of the three known markers of C-LTMRs: Tafa4, Th, and Vglut3. Deeper comparison of the RNA-seq data revealed distinct transcriptional signatures between MRGPRD-expressing neurons and C-LTMRs that were further confirmed by in situ hybridization analysis of over 100 genes. Most importantly, in addition to providing the transcriptional signatures of two categories of primary sensory neurons, our study expands the molecular characterization of C-LTMRs and suggests that this particular subset of primary sensory neurons shares many molecular features of LTMRs. As a functional readout, we used electrophysiological recording to unravel the specific and exclusive functional expression of the low-voltage-gated Ca2+ channel Cav3.3 in C-LTMRs, where it very likely plays a key role in shaping their functional specialization.
Results
Combination of GINIP and IB4 Staining Segregates Four Non-overlapping Classes of DRG Neurons
In a recent study, we generated a mouse model that expresses the fluorescent protein m-Cherry from the Ginip locus (Gaillard et al., 2014). Double-labeling experiments using anti-GINIP antibody in combination with IB4 staining showed that DRG neurons can be split into four distinct categories: GINIP+/IB4+; GINIP+/IB4−; GINIP−/IB4+; and GINIP−/IB4− neurons. The GINIP+/IB4+ double-positive (DP) population corresponds to the cutaneous free nerve endings MRGPRD+ neurons (Figures 1A and 1B), the GINIP+/IB4− population corresponds to TAFA4-expressing C-LTMRs (Figures 1A and 1B), the GINIP−/IB4+ population contains the 20% remaining IB4+ neurons (Figure 1A), and the GINIP−/IB4− double negative (DN) neurons represents a heterogeneous population of neurons composed of peptidergic nociceptors, a subset of Ret+ neurons and TrkB+ and TrkC+ neurons (Figure 1A). As GINIPm-Cherry mouse model allows high-fidelity expression of m-Cherry in GINIP+ neurons, we sought to combine live m-Cherry fluorescence with IB4 cell surface staining and FACS to purify the four categories of DRG cells (Figures 1C and 1D). Cell suspensions were obtained from acutely dissociated DRGs, pooled from six GINIPm-Cherry mice. Neurons with a cell body size above 70 μm were eliminated by filtering the suspension, and axonal debris was removed by using a Percoll gradient. Negative gating was used to exclude dead or dying cells that incorporated Sytox Blue dye and cells that were autofluorescent in V500 channel (Figures 1C and S1). Next, we gated on m-Cherry-positive and m-Cherry-negative cells that we further subdivided into IB4+ and IB4− subsets (Figures 1C and S1). FACS-sorted IB4+ and IB4− m-Cherry-positive neurons gave rise to expected proportions of cells: about two-thirds were IB4+ (corresponding to GINIP+/IB4+) and the remaining one-third was IB4− (corresponding to GINIP+/IB4−; Figure 1D). The m-Cherry−/IB4+ subset was excluded from the transcriptional analysis, because it was highly contaminated by Cd31 (Pecam)-expressing endothelial cells, consistent with IB4 binding on this cell type (data not shown). For the remaining three sorted samples (hereafter called DP for the GINIP+/IB4+, C-LTMRs for the GINIP+/IB4−, and DN for GINIP−/IB4−), we first confirmed by RT-PCR the accurate expression of Ginip and m-Cherry transcripts (Figure S2B) and then extended the purity evaluation by analyzing the expression of Mrgprd, Tafa4, TrkA, and Trpv1 in each sorted sample (Figure S2C). As expected, Mrgprd transcripts were highly detected in the DP sample but low in the two others, whereas Tafa4 expression was high in C-LTMRs. TrkA and Trpv1 transcripts were enriched in the DN sample, consistent with the enrichment in peptidergic nociceptors in the DN fraction (Figure S2C).
Figure 1.
FACS for Transcriptomic Analysis of Three Distinct Subsets of DRG Neurons Based on GINIP Expression and IB4 Labeling
(A) Schematic proportional representation of GINIP+ subset with respect to main molecularly defined DRG subsets.
(B) ISH using antisense probes for Mrgprd (in red, top) and Tafa4 (in red, bottom), followed by immunostaining with rat anti-GINIP antibody (in blue) and labeling with Alexa-488-conjugated IB4 (in green). Filled arrowheads show GINIP+/IB4+/Mrgprd+ neurons, and empty arrowheads show GINIP+/IB4−/Tafa4+ neurons. The scale bars represent 50 μm.
(C) FACS dot plots showing the FACS strategy using GINIPm-Cherry mouse and IB4 labeling; m-Cherry/GINIP− cells (1) and m-Cherry/GINIP+ cells (2) were further separated into (1) IB4+m-Cherry− and IB4−m-Cherry− (DN) cells and (2) IB4+m-Cherry+ (DP) and IB4−m-Cherry+ (C-LTMRs) (see also Figure S1).
(D) Schematic representation of sorted samples according to m-Cherry/GINIP expression and IB4 binding; the mean numbers of sorted cells are shown underneath each subset (n = 2).
(E) Shows the reads per kilobase per million reads (RPKM) obtained for Mrgprd, P2rx3 (left), Th, Tafa4 (middle), Calca (Cgrp), and Tac1 (Sp) in DP, C-LTMRs, and DN subsets (see also Figure S2).
RNA Sequencing
High-quality RNA (Figure S2A) was extracted from the three purified populations of neurons, and their respective transcriptional profiles were generated using RNA-seq. After one round of RNA amplification, libraries were built in biological duplicate and sequenced in 100-bp paired-end cycles using the Illumina HiSeq2000 system with SBS technology. Image analysis and base calling were performed using the HiSeq Control Software and RTA component from Illumina. This approach yielded between 40 and 50 millions of reads that were further submitted to sequence mapping on mouse reference genome mm10 with CASAVA 1.8.2 software. After removal of contaminants (28S, 18S, and 5S rRNA; mitochondrial chromosomes; PhiX; Illumina controls; and Illumina adaptors), reads mapping to multiple splicing positions, and reads with no match, we obtained between 32 and 42 millions of reads that were used for the generation of the raw data file (Table S1).
Among the first-ranked genes in the raw data file, we found genes known to be related to structural organization of neurons such as Tuba1a, Prph, Sncg, and Mtap1b (Table S2). All these genes exhibited strong expression in the three sorted samples, as they generated at least 50,000 reads (Table S2). Accordingly, they were highly and broadly expressed in DRG neurons in Allen-Brain Atlas database. Interestingly, genes associated with cellular stress such as Hsp90ab1 and Fos were also highly expressed in all samples, very likely due to the cell dissociation and FACS process (Table S2). Most importantly, quick analysis of the reads per kilobase per million of reads (RPKM) generated for genes expected to be enriched in each sorted subpopulation revealed high enrichment of Mrgprd and P2rx3 in the DP subset, Th and Tafa4 in C-LTMRs, and Cgrp and Sp in the DN neurons (Figure 1E), providing the first proof of concept of the successful outcome of our experimental paradigm.
To identify genes that were preferentially enriched in one sample, we carried a pairwise comparative analysis (2-fold enrichment; p < 0.01) after libraries normalization with the trimmed mean of M values method (TMM) (provided by EdgeR statistical package). Genes that generated less than ten reads were excluded from statistical analysis. Then, for each subset, we extracted the list of genes that were upregulated in one subset as compared to the two others. We found 486 genes enriched in DP subset, 549 in C-LTMRs, and 2,916 in DN subset (Tables S3, S4, and S5). These genes encode a variety of molecules including ion channels, receptors, signaling molecules, and previously uncharacterized genes.
In Situ Hybridization Validation of RNA-Seq Data Sets
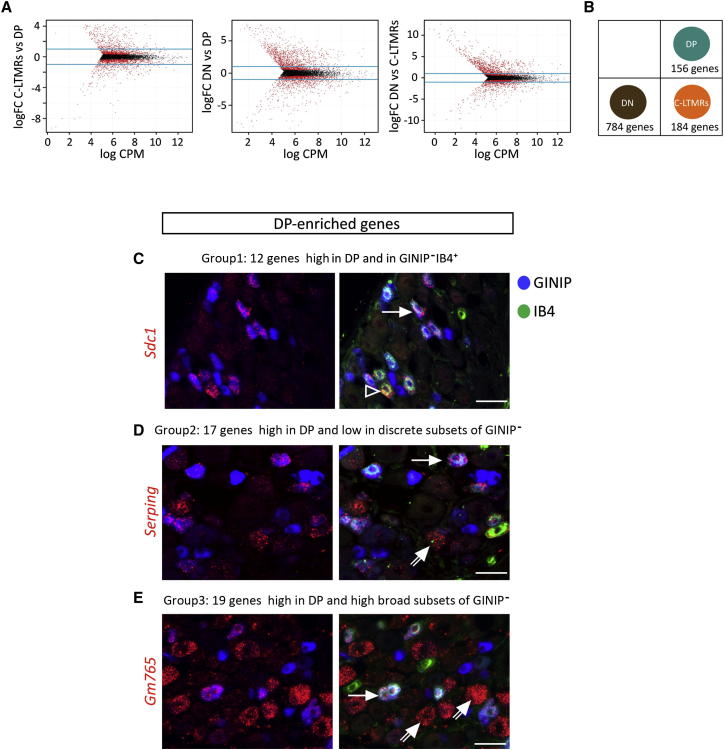
Because not all neuronal subsets were included in our FACS strategy and because FACS of acutely dissociated neurons may induce cellular stress that impacts on gene expression, the only follow-up strategy that could be employed to validate the specificity of our data sets was in situ hybridization (ISH) approach on fixed adult DRG sections. Knowing that we might miss genes that are expressed in subsets of our sorted neuronal subpopulations, we assumed that, below 1,000 number of reads, transcripts’ detection by ISH would be difficult to monitor. Therefore, we carried out a second pairwise comparative analysis by considering only the genes with values above 1,000 reads (Figure 2A). As a result, we obtained 156 transcripts preferentially expressed in the DP subset, 184 in C-LTMRs, and 784 in the DN population (Figure 2B; Tables S6, S7, and S8). We then monitored the expression of 48 DP-, 68 C-LTMRs-, and 13 DN-enriched genes using ISH followed by GINIP immunostaining and IB4 labeling (for the DP and C-LTMRs candidate genes) or GINIP and TrkA immunolabeling (for the DN candidate genes). These triple-labeling experiments provided an unambiguous and accurate expression pattern of each tested gene. The 13 DN-enriched tested genes revealed two different expression patterns: genes mainly expressed in neurons and genes exclusively expressed in non-neuronal cells that harbor satellite and/or glial cell morphology (Figure S3). In agreement with our RNA-seq data, genes expressed in neurons were excluded from DP neurons and C-LTMRs. Furthermore, we found 6/13 genes expressed in TrkA+ peptidergic neurons and others (2/13) excluded from this neuronal population, revealing the heterogeneity of the DN subset (Figure S3).
Figure 2.
Specific Transcriptional Profiles of DP, C-LTMRs, and DN Subsets and Expression Patterns of DP-Enriched Genes in DRG Neurons
(A) Shows smear plots representing pairwise comparison of normalized RNA-seq data using TMM method; fold change (FC) ≥ 2; p < 0.01. Only transcripts that generated ≥1,000 reads in the experimental replicates (n = 2) were kept for the analysis. The log FC is plotted against the log of tag concentration (counts per million [CPM]) obtained for each gene, after normalization. Genes differentially expressed are shown in red.
(B) Schematic representation of the number of genes declared to be enriched (FC ≥ 2; p < 0.01) in one subset with respect to the two others (see also Tables S6, S7, and S8).
(C–E) Representative expression patterns of DP-enriched genes assessed by ISH using antisense probes for indicated transcripts (in red), followed by immunostaining with rat anti-GINIP antibody (in blue) and Alexa-488-conjugated IB4 (in green) labeling. Plain arrows, DP neurons (GINIP+IB4+); empty arrowheads, GINIP−IB4+; double arrows, large-size neurons. Note that transcripts expressed outside DP subset (D and E) mark large-size neurons (double arrows) but are not expressed within C-LTMRs (GINIP+IB4−). The scale bars represent 50 μm (see also Figure S3 for DN-enriched genes).
Because the aim of this study consisted at deciphering the transcriptional signatures that confer the functional specializations of MRGPRD-expressing cutaneous free nerve ending and C-LTMRs, we analyzed the expression patterns of 48 DP-enriched genes and 68 C-LTMTRs-enriched genes.
In line with our RNA-seq data sets, the 48 DP-enriched probes were highly expressed in the DP neurons and virtually excluded from C-LTMRs (http://www.ibdm.univ-mrs.fr/equipes/reynders/DP-ISHlibrary.pptx). However, the vast majority of the DP-enriched genes displayed an additional expression pattern outside DP neurons. 12 genes showed some expression in few GINIP−/IB4+ neurons, 17 genes showed lower expression in discrete subsets of small (3/17) or medium-to-large size neurons (14/17), and 19 genes were expressed in broader neuronal subsets in addition to intense expression in DP neurons (Figures 2C–2E; DP-ISH library).
The same strategy applied to C-LTMRs-enriched genes showed that all 68 tested genes were expressed in C-LTMRs and massively excluded from DP neurons (http://www.ibdm.univ-mrs.fr/equipes/reynders/CLTMRs-ISHlibrary.pptx). One striking feature of C-LTMRs-enriched genes is that the vast majority was also expressed in medium-to-large size DRG neurons, whereas only two were expressed in small neurons. Indeed, out of the 68 tested genes, six were specifically expressed in C-LTMRs, as they only marked a subset of GINIP+ neurons that do not bind IB4 (Figures 3A–3F). Quantitative analyses showed that Ceacam10 is expressed in 71.2% ± 6.3% of C-LTMRs, Wfdc2 in 73.4% ± 1.7%, Cacna1i in 89.2% ± 5.9%, C1ql4 in 70% ± 1.8%, Fbp2 in 19.9% ± 4.7%, and Bok in 62.7% ± 6.4% of C-LTMRs (Figure 3G), further highlighting our previously suggested molecular heterogeneity of this particular class of neurons (Delfini et al., 2013). The remaining 60 genes could be split in three categories: 18 genes exhibited high expression in C-LTMRs but also marked a discrete subset of large-diameter neurons, 23 genes showed equal levels of expression in C-LTMRs and in large-diameter neurons, and 19 genes exhibited a very high enrichment in larger-diameter neurons and a discrete expression in C-LTMRs (Figures 4A–4C; CLTMRs-ISHlibrary). Because 88% of C-LTMRs-enriched genes displayed an additional expression preferentially in large-diameter neurons, we sought to unravel the molecular identity of these neurons. To do so, we performed double ISH using NF200 antisense probe in combination with selected C-LTMRs-enriched probes followed by TrkA immunolabeling. This triple-staining experiment showed that all the selected probes were massively excluded from TrkA+ peptidergic neurons and expressed in subsets of NF200+ neurons (Figure S4). Knowing that NF200 is a hallmark of proprioceptors and LTMRs (Bourane et al., 2009, Li et al., 2011, Luo et al., 2009, Wende et al., 2012), these data suggest that C-LTMRs share some molecular features of LTMRs.
Figure 3.
Six Novel Genes Are Exclusively Expressed in C-LTMRs Neurons
(A–F) ISH using Ceacam10, C1ql4, Wfdc2, Fbp2, Cacana1i, and Bok antisense probes (in red) followed by immunostaining with rat anti-GINIP antibody (in blue) and Alexa-488-conjugated IB4 (in green). Note that probe reactivity is detected only in C-LTMRs expressing GINIP (blue), but not IB4 (green), as indicated by the arrows. The scale bars represent 50 μm.
(G) Shows the percentage of C-LTMRs co-expressing each gene (mean ± SD; n = 3).
Figure 4.
C-LTMRs-Enriched Genes Exhibit Heterogeneous Expression Patterns in DRG Neurons
(A–C) Representative expression patterns of CLTMRs-enriched genes assessed by ISH using anti-sense probes for indicated transcripts (in red) followed by immunostaining with rat anti-GINIP antibody (in blue) and Alexa488-conjugated IB4 (in green) labelling.
(A) A representative gene highly expressed in C-LTMRs and in a discrete subset of large neurons.
(B) A gene highly expressed in C-LTMRs and in a broad subset of large neurons.
(C) A representative gene massively expressed in large diameter neurons and low expression in C-LTMRs. Filled arrowheads represent C-LTMRs neurons (GINIP+IB4−) and double arrows represent large-size neurons. Scale bars represent 50 μm (see also Figure S4).
MRGPRD-Expressing Cutaneous Free Nerve Ending Neurons and C-LTMRs Exhibit Distinct Transcriptional Profiles
MRGPRD-expressing neurons and C-LTMRs exhibit fundamental differences both at anatomical and functional levels. However, little is known about the molecular contents that instruct their respective functional specializations. In line with this, we pooled DP and C-LTMRs-enriched genes according to their biological functions by searching for statistically over-represented associations with Gene Ontology (GO) terms, using GOrillla database (Figures 5, 6, and S5). This analysis yielded, respectively, 20 and 33 functional annotations for DP and CLTMRs. DP neurons exhibited a high enrichment in G-protein-coupled receptors as well as in transcripts encoding signaling molecules, such as Gna14, Gna1, Gnaq, Plcb3, Prkca, Prkcd, and Prkcq (Figures 5A and S5A; Table S9), whereas C-LTMRs exhibited a striking enrichment in transcripts associated with voltage-gated ion channel activity (Figures 6A and S5B; Table S10).
Figure 5.
Functional Categories of DP-Enriched Genes
(A) Four out of 20 Gene Ontology functional annotations obtained upon analysis of DP-enriched data set using GOrilla database ranked from the most (top) to the least (bottom) statistically representative (p < 10−3). Part of the associated genes is listed in italics, and genes depicted in ISH experiments are shown in red (see also Figure S5 and Table S9).
(B–G) ISH using antisense probes for Agtr1a (B), Ptgdr (C), Gna14 (D), Plcb3 (E), Prkcq (F), and Prkca (G) are shown in red, immunostaining with rat anti-GINIP antibody is shown in blue, and Alexa-488-conjugated IB4 labeling is shown in green. Insets highlight (1) DP (GINIP+IB4+) and (2) C-LTMRs (GINIP+IB4−). The scale bars represent 50 μm.
Figure 6.
Functional Categories of C-LTMRs-Enriched Genes
(A) Four out of 33 Gene Ontology functional categories obtained upon analysis of C-LTMRs-enriched data set using GOrilla software ranked from the most (top) to the least (bottom) statistically representative (p < 10−3). Part of the associated genes is listed in italics; and genes depicted in ISH experiments are shown in red (see also Figure S5 and Table S10).
(B–G) ISH using antisense probes for Kcnj12 (B), Cacna1h (C), Ano6 (D), Slc12a7 (E), Cckar (F), and P2ry1 (G) are shown in red, immunostaining with rat anti-GINIP antibody is shown in blue, and Alexa-488-conjugated IB4 labeling is shown in green. Insets highlight (1) C-LTMRs (GINIP+IB4−) and (2) DP (GINIP+IB4+). The scale bars represent 50 μm (see also Figure S4).
Detailed analysis of genes associated with G protein receptor binding and signal transducer activity led to the identification of two transcripts encoding for G-protein-coupled receptors, namely the angiotensin receptor type 1a (Agtr1a) and the prostaglandin receptor D2 (Ptgdr) (Figure 5A). Our ISH experiments showed that Agtr1a is expressed in DP neurons, but not in C-LTMRs or other subsets of DRG neurons (Figure 5B), whereas Ptgdr was found at higher intensity in DP neurons compared to C-LTMRs and large-size neurons (Figure 5C). In addition to GPCRs, GO analysis revealed a striking enrichment of transcripts encoding signaling molecules in DP neurons, including Gna14, Gna1, Gnaq, Plcb3, Prkca, Prkcd, and Prkcq (Table S9). Our in situ screen showed that all these transcripts were highly expressed in DP neurons and virtually absent in C-LTMRs (Figures 5D–5G). The restricted enrichment of such number of signaling molecules likely endows DP neurons with the molecular machinery required to sense and transduce injury-induced pain hypersensitivity.
Regarding C-LTMRs, one of the most striking observations with respect to the GO analysis was the high enrichment in transcripts associated with voltage-gated ion channel activity. Other physiologically pertinent functional annotations encompass substrate-specific channel, ion transmembrane transporter, and G-protein-coupled receptor activities (Figures 6A and S5B).
Voltage-gated ion channels included potassium channels (Kcnq2, Kcnj12, Kcnv1, Kcna2, and Kcna4), sodium channels and associated beta subunits (Scn1a, Scn1b, Scn3b, Scn4b, Scn7a, and Scn8a), and low-voltage activated calcium channels (Cacna1h and Cacna1i; Figures 6A and S5B; Table S10). ISH screen showed that Cacna1i is specifically expressed in C-LTMRs (Figure 3C) and Kcnj12 and Cacna1h are expressed in C-LTMRs as well as in some large-diameter neurons but excluded from DP neurons (Figures 6B and 6C). Scn4b and Scn8a exhibited low expression in C-LTMRs, very high expression in myelinated large neurons, and no detectable signal in DP neurons (C-LTMRs-ISH library, slides 67 and 68). The combinatorial enrichment of such channels in C-LTMRs is likely to explain the tonic and repetitive firing properties of this particular class of neurons (Delfini et al., 2013, Li et al., 2011).
Interestingly, substrate-specific channels/ion transmembrane transporters encompassed anoctamin 6 (Ano6) and solute carrier 17a2 (Slc17a2). Ano6 is part of outwardly rectifying chloride channels (Martins et al., 2011), and Slc12a7 encodes for the K+/Cl− co-transporter Kcc4 (Boettger et al., 2002), suggesting that both channels could be involved in membrane excitability. In situ analysis of Ano6 and Slc12a7 showed that both transcripts were expressed in C-LTMRs, but not in DP neurons, and exhibited some expression in large DRG neurons (Figures 6D and 6E).
GPCR-activity-related transcripts were also represented in GO analysis of C-LTMRs data set, including cholecystokinin receptor A (Cckar) and purinergic receptor y1 (P2ry1). Our ISH data showed that both Cckar and P2ry1 transcripts were highly enriched in C-LTMRs, totally excluded from DP neurons, and displayed expression in some large-diameter neurons (Figures 6F, 6G, and S4B).
Together, our data provide a functional description of sets of genes differentially expressed in DP versus C-LTMRs. The differential expression of such clusters of genes likely contributes to differences in signal transduction, intrinsic membrane excitability, and the firing properties of C-LTMRs and MRGPRD+ neurons. Our data also provide a predictive basis for deciphering the mechanisms by which specific external stimuli and downstream-associated signaling pathways fine-tune the activity of these two neuronal subsets.
C-LTMRs Express Two Functionally Distinct Low-Voltage-Activated Calcium Channels
Our RNA-seq data revealed high enrichment of Cacna1h (Cav3.2) and Cacna1i (Cav3.3) in C-LTMRs. Our ISH showed that transcripts encoding both channels were expressed in C-LTMRs with Cav3.3 being exclusively expressed in this neuronal subset (Figures 3E and 7A). To explore whether both channels were functional in C-LTMRs, we used whole-cell patch-clamp recordings on primary culture of heterozygous m-Cherry-expressing neurons in the presence of Alexa 488 IB4 live staining to distinguish between C-LTMRs and DP neurons. In ionic conditions designed to isolate calcium currents, current-voltage relationships showed inward low-voltage-activated (LVA) and high-voltage-activated (HVA) currents in all recorded C-LTMRs soma, confirming the systematic presence of T-type currents in these neurons (Figures 7B and 7C). In sharp contrast, DP neurons did not express any LVA current in agreement with the absence of Cav3 transcripts in this population (Figures 7B and 7C). To test whether Cav3.2 and Cav3.3 were functional in C-LMTRs, we took advantage of Cav3.2 high sensitivity to nickel ions (Ni) (Lee et al., 1999). Blockade of T-type calcium currents in C-LTMRs by low concentration of Ni (30 μM) revealed a Ni-insensitive current with slow kinetics of activation and inactivation typical of Cav3.3 channel (Figure 7D; Chemin et al., 2002). In contrast, the Ni-sensitive fraction obtained by subtraction revealed faster kinetics with an explicit criss-cross pattern typical of Cav3.2 (Figure 7D). To date, zinc (Zn) is the only available pharmacological tool that interferes with the gating kinetics of Cav3.3 (Cataldi et al., 2007, Traboulsie et al., 2007). After blockade of Cav3.2-like fraction by Ni, subsequent application of Zn (100 μM) slowed down the remaining native Cav3.3-like current with a prominent slowing of the deactivation (Figure 7E). Analysis of several C-LTMR soma revealed that Cav3.3-like currents account for a third of the total amplitude of C-LTMRs T-type calcium currents (Figure 7F). The dramatic slowdown of the deactivation kinetics by Zn further shows that Cav3.3 is a unique feature in the portfolio of ion channels specific to C-LTMRs (Figure 7G). To test whether these two T-type calcium channel isoforms impact C-LTMR excitability, we performed current-clamp experiments. In neurons, T-type calcium channels are well known to generate rebound low-threshold spikes (LTS) following a transient hyperpolarization. In C-LTMRs, membrane depolarization triggered small LTS when the membrane is held at more and more negative values (Figure 7H). Inhibition of Cav3.2 by Ni (50 μM) did not significantly alter LTS and action potential (AP) threshold, whereas potentiation of Cav3.3 by Zn (100 μM) increased both LTS amplitude and kinetics (Figure 7H, inset). This resulted in an increased excitability with a lowering of AP activation threshold crowning the potentiated LTS (Figures 7H and 7I). In two cells out of ten, the sole application of Zn at the resting potential of C-LTMRs elicited a depolarization sufficient to trigger spontaneous AP trains (Figure 7J). Application of Ni in these conditions did not produce any change in membrane potential (not shown). To exclude TRPA1 channel contribution, also activated by Zn, these experiments were performed in presence of TRPA1 blocker, HC030031 (10 μM). Together, our electrophysiological data nicely corroborate the RNA-seq and the ISH expression data, demonstrating that both Cav3.2 and Cav3.3 channels are concomitantly expressed in C-LTMRs and that both channels are absent in the free nerve ending MRGPRD-expressing neurons.
Figure 7.
Functional Presence of Cav3.2 and Cav3.3 in C-LTMR Neurons
(A) Shows the expression patterns of Cav3.2 (Cacna1h) and Cav3.3 (Cacna1i)-encoding transcripts in DP and C-LTMRs (filled arrowheads) as assessed by ISH with corresponding antisense probes (in red) followed by double immunolabelling with anti-GINIP antibody (in blue) and Alexa-488-conjugated IB4 (in green). Cacna1h probe reactivity was also detected in larger neurons (empty arrowheads). The scale bars represent 20 μm.
(B) Typical current voltage relationships in GINIP+IB4+ and GINIP+IB4− neurons evoked by 200-ms-long ramp depolarizations from −90 to +100 mV. Note that LVA currents are only present in the GINIP+IB4− cells.
(C) Individual scatter values of LVA and HVA current densities for GINIP+IB4+ and GINIP+IB4− neurons (error bars represent the SEM calculated on the mean current).
(D) Typical example of T-type currents in C-LTMRs evoked by 100-ms-long step depolarizations from a holding potential of −90 mV to test potential of −80 to −30 mV by 5 mV increments. Left traces (in black): control condition; middle traces (in black): after 30 μM nickel (Ni) application; right traces (in gray): difference current between these two conditions.
(E) Typical example of the sequential effect of 30 μM Ni (green) and 30 μM Ni + 100 μM zinc (Zn) (red) on a C-LTMR neuron. Currents are evoked by a step depolarization from −90 mV to −30 mV.
(F) Individual scatter values showing the effect of 30 μM Ni on the peak T-type current density in C-LTMRs (error bars represent the SEM calculated on the mean current); black dots: control; green dots: +30 μM Ni.
(G) Individual scatter values showing the effect of 100 μM Zn (red dots) on the deactivation kinetics of the Ni-resistant fraction (green dots) of C-LTMRs.
(H) Typical traces of the rebound LTS evoked at the end of a 1-s-long hyperpolarizing step in control condition (black) and with 50 μM Ni + 100 μM Zn (red) in a C-LTMR cell. The hyperpolarizing steps shown are not sufficient to evoke an action potential in control whereas they evoke one in the presence of the Ni + Zn mix. The inset shows magnified traces of LTS and the effects of Ni + Zn on their kinetics.
(I) Threshold hyperpolarization for LTS-induced AP in control condition (black), in 50 μM Ni (green), and with 50 μM Ni + 100 μM Zn (red) in ten C-LTMRs. Values are presented as a “before and after” plot. The efficiency of hyperpolarization is reduced in Ni+Zn condition in comparison to control and Ni ones.
(J) Application of Ni + Zn to C-LTMRs results in membrane depolarization and spontaneous action potentials in a fraction of cells (n = 2/10). This effect is obtained in the presence of the TRPA1 channel blocker HC030001 (10 μM).
Discussion
In this study, we used our recently generated mouse model in which the cutaneous MRGPRD-expressing neurons and C-LTMRs were genetically marked by the m-Cherry protein, in combination with IB4 live staining followed by cell sorting and RNA deep sequencing to gene profile three categories of primary sensory neurons: the free nerve ending MRGPD-expressing neurons, C-LTMRs, and a heterogeneous population of DRG cells that neither bind IB4 nor are labeled by the m-Cherry protein. We provide a digital measure of the presence and prevalence of transcripts from known and previously unknown genes for each of the three neuronal subsets. Because the DN population is composed of a variety of neuronal subsets and glial cells, DN data set was used to identify genes specifically enriched in the free nerve ending MRGPRD-expressing neurons and those specifically enriched in C-LTMRs. This analysis yielded 156 genes specifically enriched in the DP neurons and 184 in C-LTMRs. These genes encode a variety of molecules including ion channels, receptors, signaling molecules, and previously uncharacterized genes.
A major concern with any gene-profiling experiment is the follow-up procedure. Which genes should one pursue, and what should be done with these lists of genes? The answer to this question was motivated by our objectives to expand the molecular characterization of C-LTMRs and to identify the combinatorial expression of the key genes that shape the functional specialization of the free nerve endings MRGPRD+ neurons and C-LTMRs. MRGPRD+ neurons are required for acute and injury-induced mechanical pain as well as for the sensation of itch (Cavanaugh et al., 2009, Liu et al., 2012, Shields et al., 2010), whereas C-LTMRs were postulated to convey light touch under physiological conditions and mechanical and chemical pain after injury (Abraira and Ginty, 2013, Delfini et al., 2013, Li et al., 2011, Löken et al., 2009, Löken et al., 2010, McGlone et al., 2014, Olausson et al., 2002). Our RNA-seq data in combination with ISH and gene ontology analysis revealed a great number of genes expressed in DP neurons, but not in C-LTMRs, and vice versa, reflecting their anatomical and functional differences. Indeed, DP neurons showed very high enrichment in genes encoding GPCRs and signaling molecules in addition to a selective expression of three isoforms of the protein kinase C family. The enrichment of such receptors and signaling molecules is in agreement with the involvement of DP neurons in acute and injury-induced mechanical pain and itch (Cavanaugh et al., 2009, Liu et al., 2012). We show that DP neurons express high levels of Agtr1a and Ptgdr, suggesting that the pro-nociceptive effect of their respective agonists, angiotensin II and prostaglandin D2, likely occurs through DP neurons activation. PGD2 has been shown to enhance neuronal excitability by increasing the amplitude of TTX-sensitive currents (Ebersberger et al., 2011, Nakae et al., 2005), and continuous perfusion of angiotensin II enhanced tactile and thermal hypersensitivity in the setting of nerve injury via the activation of Agtr1a (Pavel et al., 2013). Furthermore, DP neurons exclusively express MRGPRD, a GPCR that is activated by β-alanine, but not histamine (Liu et al., 2012), suggesting that DP neurons are part of a histamine-independent itch neural circuit and that these neurons can be targeted for treating clinical itch that is resistant to antihistamines.
C-LTMRs on the other hand expressed different GPCRs than those expressed in DP neurons such as CCKAR and the metabotropic Gαq P2Y1 receptor and showed a striking enrichment in voltage-gated channels including potassium channels (Kcnq2, Kcnj12, Kcnv1, Kcna2, and Kcna4), sodium channels and associated beta subunits (Scn1a, Scn1b, Scn3b, Scn4b, Scn7a, and Scn8a), and the low-voltage-gated calcium channels (Cacna1h and Cacna1i).
It has been shown that CCKAR was restricted to 10% of neurons that do not overlap with peptidergic CGRP+ nociceptors (Broberger et al., 2001). Our RNA-seq and ISH data demonstrate that CCKAR is expressed in C-LTMRs and excluded from DP neurons. Cholecystokinin octapeptide, which in its sulfated form binds the Gαq/11-coupled CCKAR, is involved in regulation of a large variety of physiological functions among which opioid-induced anti-nociception (Suh et al., 1995). Electrophysiological studies in rat DRG neurons have shown that activation of CCKAR evokes membrane depolarization, indicating that CCKAR mediates excitatory responses (Ma et al., 2006). Psychophysiological studies in human using cholecystokinin octapeptide could lend new insights into the role of C-LTMRs in touch sensation under normal and pathological conditions. C-LTMRs also exhibited high expression of the metabotropic Gαq P2Y1 receptor, whereas DP neurons mainly expressed the ionotropic purinergic receptor P2X3, demonstrating that, in DP neurons, ATP/ADP compounds are sensed through P2X3, whereas in C-LTMRs, these signaling intermediates are likely detected though metabotropic Gαq P2Y1 receptor.
Our data also highlighted the specific and the functional enrichment of the LVA T-type channels in C-LTMRs. Among the three T-type channels, we found Cav3.2 and Cav3.3 highly present in C-LTMRs, with Cav3.3 being exclusively expressed in this particular class of neurons. T-type currents with slow kinetics and resistance to nickel block have been described in a subset of small-sized rat DRG and trigeminal neurons expressing slowly adapting mechano-activated currents (Coste et al., 2007, Ross et al., 2008). However, the identity of such neurons has never been deciphered. Our RNA-seq data in combination with ISH and electrophysiological recordings provided a clear demonstration that Cav3.3 is a specific marker of C-LTMRs and offers a new possible selective pharmacological target to control C-LTMRs’ excitability in mice. Although zinc has plethoric effects, its use to investigate Cav3.3 contribution to C-LTMR excitability shows specificity. Particularly, the slowing of LTS kinetics is highly correlated to the massive changes in Cav3.3 currents. In line with this, the depolarizing effects observed result, at least in part, from the persistent depolarizing zinc-modulated Cav3.3 current. Whereas the contribution of other zinc targets remains possible, TRPA1 effect has been eliminated by the use of its specific blocker, and a possible inhibition of two pore potassium TASK3 channel (KCNK9) is unlikely because the transcript is not present in C-LTMRs (our study; Usoskin et al., 2015). Mechanistically, our findings suggest a substantial contribution of Cav3.3 to rebound firing reminiscent of those observed in nuclear reticularis thalamic neurons that, like C-LTMR, express both Cav3.2 and Cav3.3 and with a prominent Cav3.3 effect on burst firing (Astori et al., 2011).
Finally, a recent elegant study using an unbiased large-scale single-cell RNA sequencing revealed 11 molecularly distinct subsets of primary sensory neurons, including the TH-expressing C-LTMRs population (Usoskin et al., 2015). Very interestingly, genes enriched in C-LTMRs in our study were similar to those upregulated in the TH population described in the Usoskin and colleagues’ study. However, our ISH experiments showed that the vast majority of C-LTMRs-enriched genes were also expressed in other neurons, preferentially in the NF200+ neurons, further supporting our strategy to validate the specific expression of the identified enriched genes using ISH approach. Given that the majority of C-LTMRs-enriched genes were also expressed in NF200+ neurons and that NF200 is a hallmark of proprioceptive and Aβ and Aδ LTMRs (Abraira and Ginty, 2013), we postulate that C-LTMRs share many molecular features with some Aβ and Aδ LTMRs. In line with this hypothesis, studies of the last few years demonstrated that C-LTMRs share many functional and anatomical features of some Aβ and Aδ LTMRs (Abraira and Ginty, 2013). C-LTMRs have been described to convey low-threshold mechanical stimuli (Bessou et al., 1971, Douglas and Ritchie, 1957, Johansson et al., 1988, Li et al., 2011, Maruhashi et al., 1952, Seal et al., 2009, Zotterman, 1939). They are sensitive to skin indentation, respond to sustained mechanical stimuli with slow and intermediate adaptation rates, and form longitudinal lanceolate endings around hair follicles (Abraira and Ginty, 2013, Delfini et al., 2013, Li et al., 2011, Lou et al., 2013, Lumpkin et al., 2010, Seal et al., 2009). Cross comparison of our RNA-seq data sets with those of the five NF subsets recently published by Usoskin and colleagues (Usoskin et al., 2015) will provide a comprehensive view of the molecular signatures of C-LTMRs and Aβ and Aδ LTMRs.
In conclusion, data presented in this study provide a wealth of information about the presence and prevalence of transcripts from known and previously unknown genes in three categories of primary sensory neurons, they highlight the differential expression of a great number of genes in two diametrically opposite classes of cutaneous afferents, the ISH screen followed by immunostaining and IB4 binding led to the generation of two open-access libraries that allow monitoring the cellular distribution of 115 genes, they provide the first electrophysiological recording of a low-voltage-gated Ca2+ channel exclusively present in C-LTMRs, and they suggest that C-LTMRs share many molecular features with Aβ and Aδ LTMRs.
Experimental Procedures
The expression patterns of DN-enriched genes; the co-expression of C-LTMRs-enriched genes with NF200+ neurons; the supplemental tables enclosing DP-, C-LTMRs-, and DN-enriched genes; as well as the GO analysis are provided in the Supplemental Information.
Mice
Mice were maintained under standard housing conditions (23°C, 40% humidity, 12 hr light cycles, and free access to food and water). C57BL/6 mice were from Charles River Laboratories, and heterozygous GINIPm-Cherry knockin mice were previously generated in the laboratory (Gaillard et al., 2014). Special effort was made to minimize the number as well as the stress and suffering of mice used in this study. All protocols are in agreement with European Union recommendations for animal experimentation.
FACS
IB4+GINIP+, IB4−GINIP+, IB4+GINIP−, and IB4−GINIP− cells were sorted on FACS Aria cell sorter (BD Bioscience) by gating on Sytox Blue− m-Cherry+ for GINIP+ cells and Sytox Blue− m-Cherry− for GINIP− cells. DRGs cell suspensions from C57BL/6 mouse were used as a negative control of m-Cherry fluorescence. FACS analysis was performed with Diva software (BD Bioscience). Cells were sorted directly in ice-cold RLT lysis buffer from RNeasy Micro Kit (QIAGEN), supplemented with 10% (v/v) β-mercaptoethanol (Sigma), and snap-frozen at −80°C before RNA extraction.
RNA-Seq
RNA-seq libraries were constructed with the TruSeq RNA sample preparation (low-throughput protocol) kit from Illumina. (See the Supplemental Experimental Procedures for more details).
RNA-Seq Data Analysis
Image analysis and basecalling were performed using the HiSeq Control Software and Real-Time Analysis component provided by Illumina. The quality of the data was assessed with FastQC from the Babraham Institute and the Illumina software SAV (Sequence Analysis Viewer). Demultiplexing was performed using Illumina’s sequencing analysis software (CASAVA 1.8.2). The eland_rna module from CASAVA was used to align RNA-seq reads to the Mus musculus genome (UCSC mm10) with a set of gene-model annotations (refFlat.txt file downloaded from UCSC on October 10, 2012) in order to generate automatically the splice junctions. Reads were also aligned to a set of contaminants, including the rRNAs, the mitochondrial chromosome, the PhiX genome (Illumina control), and the Illumina adapters. Reads mapping to contaminants were discarded. The gene counting was performed using the counting module from CASAVA. This module counts the number of bases mapping to each exon and splice junctions. Then, exon counts were summed by gene and divided by the length of reads (i.e., 100) to obtain the gene counts. Only the first read in pairs has been taken into account during this step. The differential analysis was performed under the R software (version 2.15.0). Downstream statistical analysis was made on (1) genes that generated at least ten reads and (2) genes that generated at least 1,000 reads. Differentially expressed genes were identified using the Bioconductor (Robinson et al., 2010) package edgeR (Alexa et al., 2006) version 2.6.2. Data were normalized using the TMM (Robinson and Oshlack, 2010) normalization factors. Genes with adjusted p value less than 1% (according to the FDR method from Benjamini-Hochberg) were declared differentially expressed. Genes were then classified using custom rules: (1) genes declared as differentially expressed in condition A relative to condition B and to condition C and having a fold-change of at least 2 (absolute value) in both cases are considered to be specific of condition A. (2) A gene will be specific of the two conditions A and B simultaneously if it is differentially expressed in A versus C and in B versus C with an absolute fold change >2 and with an absolute fold change <2 between A and B. (3) Filtered genes are considered to be specific of none condition. (4) Genes belonging to none of the previous categories are considered to be specific of the three conditions simultaneously. Free web access GOrilla database was used to perform the GO functional annotation of the resulting list of genes as described in Eden et al. (2009).
ISH and Immunostaining
ISH and immunostaining were performed as described in Moqrich et al. (2004). (See the Supplemental Experimental Procedures for more details).
Electrophysiology
Lumbar DRGs were prepared and cultured as described previously (Francois et al., 2013) from adult male GINIPm-Cherry mice. Recordings were made within 24 hr of culture. To identify IB4+-positive neurons, we incubated cultures with Alexa-488-conjugated IB4 (1 μg/ml; 5 min at 37°C; Invitrogen) diluted in extracellular solution and washed before recordings. Conditions for recordings of voltage-gated calcium currents and of membrane potential were as described in Delfini et al. (2013) and Francois et al. (2013).
Online Information Data
Patterns of expression of over 100 genes are enclosed in DP and C-LTMRs ISH libraries. For DP, see http://www.ibdm.univ-mrs.fr/equipes/reynders/DP-ISHlibrary.pptx. For C-LTMRs, see http://www.ibdm.univ-mrs.fr/equipes/reynders/CLTMRs-ISHlibrary.pptx.
Author Contributions
A. Moqrich and A.R. designed the project, A.R. and A. Mantilleri performed the in situ analyses, P.M. performed the PCRs for probe synthesis, S.L. and E.B. performed the electrophysiological recording, C.B. contributed to unpublished data, S.N. performed the RNA-seq experiments, S.R. and A.R. analyzed the data sets, and A. Moqrich and A.R. wrote the paper.
Acknowledgments
We are grateful to the members of the laboratory for scientific discussions and to M. Barad, A. Zouine, and M. Malissen from CIML flow cytometry facility for FACS assistance. We thank S. Alonso for comments on the manuscript, M. Deage for the pilot experiment on Tafa4 DRG neurons, as well as IBDM animal facility. We thank France-BioImaging infrastructure supported by the Agence Nationale de la Recherche (ANR-10-INSB-04-01, call “Investissements d’Avenir”). Work in the E.B. laboratory was supported by grants from the AFM (AFM-PainT). This work was also funded by ERC starting grant paineurons 260435 to A. Moqrich.
Published: February 12, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, five figures, and 12 tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.01.022.
Contributor Information
Emmanuel Bourinet, Email: emmanuel.bourinet@igf.cnrs.fr.
Aziz Moqrich, Email: aziz.moqrich@univ-amu.fr.
Accession Numbers
All data, including raw reads, RPKM normalization, and TMM-normalized, have been deposited to the NCBI GEO under accession number GSE64091.
Supplemental Information
List of genes enriched in one subset versus the two others. Only genes that generated at least ten reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01). CPM, counts per million; FC, fold change; FDR, false discovery rate.
List of genes enriched in one subset versus the two others. Only genes that generated at least ten reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated at least ten reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated above 1,000 reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated above 1,000 reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated above 1,000 reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
DP-enriched clusters of genes associated with a specific molecular function GO term in GOrilla database.
C-LTMRs-enriched clusters of genes associated with a specific molecular function GO term in GOrilla database.
References
- Abraira V.E., Ginty D.D. The sensory neurons of touch. Neuron. 2013;79:618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenführer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Astori S., Wimmer R.D., Prosser H.M., Corti C., Corsi M., Liaudet N., Volterra A., Franken P., Adelman J.P., Lüthi A. The Ca(V)3.3 calcium channel is the major sleep spindle pacemaker in thalamus. Proc. Natl. Acad. Sci. USA. 2011;108:13823–13828. doi: 10.1073/pnas.1105115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessou P., Perl E.R. Response of cutaneous sensory units with unmyelinated fibers to noxious stimuli. J. Neurophysiol. 1969;32:1025–1043. doi: 10.1152/jn.1969.32.6.1025. [DOI] [PubMed] [Google Scholar]
- Bessou P., Burgess P.R., Perl E.R., Taylor C.B. Dynamic properties of mechanoreceptors with unmyelinated (C) fibers. J. Neurophysiol. 1971;34:116–131. doi: 10.1152/jn.1971.34.1.116. [DOI] [PubMed] [Google Scholar]
- Boettger T., Hübner C.A., Maier H., Rust M.B., Beck F.X., Jentsch T.J. Deafness and renal tubular acidosis in mice lacking the K-Cl co-transporter Kcc4. Nature. 2002;416:874–878. doi: 10.1038/416874a. [DOI] [PubMed] [Google Scholar]
- Bourane S., Garces A., Venteo S., Pattyn A., Hubert T., Fichard A., Puech S., Boukhaddaoui H., Baudet C., Takahashi S. Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by Ret signaling. Neuron. 2009;64:857–870. doi: 10.1016/j.neuron.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Broberger C., Holmberg K., Shi T.J., Dockray G., Hökfelt T. Expression and regulation of cholecystokinin and cholecystokinin receptors in rat nodose and dorsal root ganglia. Brain Res. 2001;903:128–140. doi: 10.1016/s0006-8993(01)02468-4. [DOI] [PubMed] [Google Scholar]
- Cataldi M., Lariccia V., Marzaioli V., Cavaccini A., Curia G., Viggiano D., Canzoniero L.M., di Renzo G., Avoli M., Annunziato L. Zn(2+) slows down Ca(V)3.3 gating kinetics: implications for thalamocortical activity. J. Neurophysiol. 2007;98:2274–2284. doi: 10.1152/jn.00889.2006. [DOI] [PubMed] [Google Scholar]
- Cavanaugh D.J., Lee H., Lo L., Shields S.D., Zylka M.J., Basbaum A.I., Anderson D.J. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc. Natl. Acad. Sci. USA. 2009;106:9075–9080. doi: 10.1073/pnas.0901507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin J., Monteil A., Perez-Reyes E., Bourinet E., Nargeot J., Lory P. Specific contribution of human T-type calcium channel isotypes (alpha(1G), alpha(1H) and alpha(1I)) to neuronal excitability. J. Physiol. 2002;540:3–14. doi: 10.1113/jphysiol.2001.013269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B., Crest M., Delmas P. Pharmacological dissection and distribution of NaN/Nav1.9, T-type Ca2+ currents, and mechanically activated cation currents in different populations of DRG neurons. J. Gen. Physiol. 2007;129:57–77. doi: 10.1085/jgp.200609665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfini M.C., Mantilleri A., Gaillard S., Hao J., Reynders A., Malapert P., Alonso S., François A., Barrere C., Seal R. TAFA4, a chemokine-like protein, modulates injury-induced mechanical and chemical pain hypersensitivity in mice. Cell Rep. 2013;5:378–388. doi: 10.1016/j.celrep.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Dong X., Han S., Zylka M.J., Simon M.I., Anderson D.J. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- Douglas W.W., Ritchie J.M. Nonmedullated fibres in the saphenous nerve which signal touch. J. Physiol. 1957;139:385–399. doi: 10.1113/jphysiol.1957.sp005899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger A., Natura G., Eitner A., Halbhuber K.J., Rost R., Schaible H.G. Effects of prostaglandin D2 on tetrodotoxin-resistant Na+ currents in DRG neurons of adult rat. Pain. 2011;152:1114–1126. doi: 10.1016/j.pain.2011.01.033. [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois A., Kerckhove N., Meleine M., Alloui A., Barrere C., Gelot A., Uebele V.N., Renger J.J., Eschalier A., Ardid D., Bourinet E. State-dependent properties of a new T-type calcium channel blocker enhance Ca(V)3.2 selectivity and support analgesic effects. Pain. 2013;154:283–293. doi: 10.1016/j.pain.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Gaillard S., Lo Re L., Mantilleri A., Hepp R., Urien L., Malapert P., Alonso S., Deage M., Kambrun C., Landry M. GINIP, a Gαi-interacting protein, functions as a key modulator of peripheral GABAB receptor-mediated analgesia. Neuron. 2014;84:123–136. doi: 10.1016/j.neuron.2014.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson R.S., Trulsson M., Olsson K.A., Westberg K.G. Mechanoreceptor activity from the human face and oral mucosa. Exp. Brain Res. 1988;72:204–208. doi: 10.1007/BF00248518. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Gomora J.C., Cribbs L.L., Perez-Reyes E. Nickel block of three cloned T-type calcium channels: low concentrations selectively block alpha1H. Biophys. J. 1999;77:3034–3042. doi: 10.1016/S0006-3495(99)77134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Rutlin M., Abraira V.E., Cassidy C., Kus L., Gong S., Jankowski M.P., Luo W., Heintz N., Koerber H.R. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljencrantz J., Björnsdotter M., Morrison I., Bergstrand S., Ceko M., Seminowicz D.A., Cole J., Bushnell M.C., Olausson H. Altered C-tactile processing in human dynamic tactile allodynia. Pain. 2013;154:227–234. doi: 10.1016/j.pain.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr. Opin. Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Vrontou S., Rice F.L., Zylka M.J., Dong X., Anderson D.J. Molecular genetic visualization of a rare subset of unmyelinated sensory neurons that may detect gentle touch. Nat. Neurosci. 2007;10:946–948. doi: 10.1038/nn1937. [DOI] [PubMed] [Google Scholar]
- Liu Q., Sikand P., Ma C., Tang Z., Han L., Li Z., Sun S., LaMotte R.H., Dong X. Mechanisms of itch evoked by β-alanine. J. Neurosci. 2012;32:14532–14537. doi: 10.1523/JNEUROSCI.3509-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löken L.S., Wessberg J., Morrison I., McGlone F., Olausson H. Coding of pleasant touch by unmyelinated afferents in humans. Nat. Neurosci. 2009;12:547–548. doi: 10.1038/nn.2312. [DOI] [PubMed] [Google Scholar]
- Löken L.S., Lundblad L.C., Elam M., Olausson H.W. Tactile direction discrimination and vibration detection in diabetic neuropathy. Acta Neurol. Scand. 2010;121:302–308. doi: 10.1111/j.1600-0404.2009.01282.x. [DOI] [PubMed] [Google Scholar]
- Lou S., Duan B., Vong L., Lowell B.B., Ma Q. Runx1 controls terminal morphology and mechanosensitivity of VGLUT3-expressing C-mechanoreceptors. J. Neurosci. 2013;33:870–882. doi: 10.1523/JNEUROSCI.3942-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin E.A., Marshall K.L., Nelson A.M. The cell biology of touch. J. Cell Biol. 2010;191:237–248. doi: 10.1083/jcb.201006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Enomoto H., Rice F.L., Milbrandt J., Ginty D.D. Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron. 2009;64:841–856. doi: 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K.T., Si J.Q., Zhang Z.Q., Zhao L., Fan P., Jin J.L., Li X.Z., Zhu L. Modulatory effect of CCK-8S on GABA-induced depolarization from rat dorsal root ganglion. Brain Res. 2006;1121:66–75. doi: 10.1016/j.brainres.2006.08.094. [DOI] [PubMed] [Google Scholar]
- Martins J.R., Faria D., Kongsuphol P., Reisch B., Schreiber R., Kunzelmann K. Anoctamin 6 is an essential component of the outwardly rectifying chloride channel. Proc. Natl. Acad. Sci. USA. 2011;108:18168–18172. doi: 10.1073/pnas.1108094108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruhashi J., Mizuguchi K., Tasaki I. Action currents in single afferent nerve fibres elicited by stimulation of the skin of the toad and the cat. J. Physiol. 1952;117:129–151. doi: 10.1113/jphysiol.1952.sp004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlone F., Wessberg J., Olausson H. Discriminative and affective touch: sensing and feeling. Neuron. 2014;82:737–755. doi: 10.1016/j.neuron.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Moqrich A., Earley T.J., Watson J., Andahazy M., Backus C., Martin-Zanca D., Wright D.E., Reichardt L.F., Patapoutian A. Expressing TrkC from the TrkA locus causes a subset of dorsal root ganglia neurons to switch fate. Nat. Neurosci. 2004;7:812–818. doi: 10.1038/nn1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae K., Hayashi F., Hayashi M., Yamamoto N., Iino T., Yoshikawa S., Gupta J. Functional role of prostacyclin receptor in rat dorsal root ganglion neurons. Neurosci. Lett. 2005;388:132–137. doi: 10.1016/j.neulet.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Olausson H., Lamarre Y., Backlund H., Morin C., Wallin B.G., Starck G., Ekholm S., Strigo I., Worsley K., Vallbo A.B., Bushnell M.C. Unmyelinated tactile afferents signal touch and project to insular cortex. Nat. Neurosci. 2002;5:900–904. doi: 10.1038/nn896. [DOI] [PubMed] [Google Scholar]
- Owens D.M., Lumpkin E.A. Diversification and specialization of touch receptors in skin. Cold Spring Harb Perspect Med. 2014;4:013656. doi: 10.1101/cshperspect.a013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavel J., Oroszova Z., Hricova L., Lukacova N. Effect of subpressor dose of angiotensin II on pain-related behavior in relation with neuronal injury and activation of satellite glial cells in the rat dorsal root ganglia. Cell. Mol. Neurobiol. 2013;33:681–688. doi: 10.1007/s10571-013-9934-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross H.R., Napier I., Connor M. Inhibition of recombinant human T-type calcium channels by Delta9-tetrahydrocannabinol and cannabidiol. J. Biol. Chem. 2008;283:16124–16134. doi: 10.1074/jbc.M707104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal R.P., Wang X., Guan Y., Raja S.N., Woodbury C.J., Basbaum A.I., Edwards R.H. Injury-induced mechanical hypersensitivity requires C-low threshold mechanoreceptors. Nature. 2009;462:651–655. doi: 10.1038/nature08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields S.D., Cavanaugh D.J., Lee H., Anderson D.J., Basbaum A.I. Pain behavior in the formalin test persists after ablation of the great majority of C-fiber nociceptors. Pain. 2010;151:422–429. doi: 10.1016/j.pain.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.S., Lewin G.R. Nociceptors: a phylogenetic view. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2009;195:1089–1106. doi: 10.1007/s00359-009-0482-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H.W., Kim Y.H., Choi Y.S., Song D.K. Involvement of different subtypes of cholecystokinin receptors in opioid antinociception in the mouse. Peptides. 1995;16:1229–1234. doi: 10.1016/0196-9781(95)02006-i. [DOI] [PubMed] [Google Scholar]
- Traboulsie A., Chemin J., Chevalier M., Quignard J.F., Nargeot J., Lory P. Subunit-specific modulation of T-type calcium channels by zinc. J. Physiol. 2007;578:159–171. doi: 10.1113/jphysiol.2006.114496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P.V. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Vrontou S., Wong A.M., Rau K.K., Koerber H.R., Anderson D.J. Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature. 2013;493:669–673. doi: 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H., Lechner S.G., Cheret C., Bourane S., Kolanczyk M.E., Pattyn A., Reuter K., Munier F.L., Carroll P., Lewin G.R., Birchmeier C. The transcription factor c-Maf controls touch receptor development and function. Science. 2012;335:1373–1376. doi: 10.1126/science.1214314. [DOI] [PubMed] [Google Scholar]
- Zimmermann K., Hein A., Hager U., Kaczmarek J.S., Turnquist B.P., Clapham D.E., Reeh P.W. Phenotyping sensory nerve endings in vitro in the mouse. Nat. Protoc. 2009;4:174–196. doi: 10.1038/nprot.2008.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotterman Y. Touch, pain and tickling: an electro-physiological investigation on cutaneous sensory nerves. J. Physiol. 1939;95:1–28. doi: 10.1113/jphysiol.1939.sp003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes enriched in one subset versus the two others. Only genes that generated at least ten reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01). CPM, counts per million; FC, fold change; FDR, false discovery rate.
List of genes enriched in one subset versus the two others. Only genes that generated at least ten reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated at least ten reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated above 1,000 reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated above 1,000 reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
List of genes enriched in one subset versus the two others. Only genes that generated above 1,000 reads were considered for the analysis. Differentially expressed (FC > 2) genes were identified using TMM method (p < 0.01).
DP-enriched clusters of genes associated with a specific molecular function GO term in GOrilla database.
C-LTMRs-enriched clusters of genes associated with a specific molecular function GO term in GOrilla database.