Abstract
Gene regulatory circuits are to the cell what arithmetic logic units are to the chip: fundamental components of information processing that map an input onto an output. Gene regulatory circuits come in many different forms, distinct structural configurations that determine who regulates whom. Studies that have focused on the gene expression patterns (functions) of circuits with a given structure (form) have examined just a few structures or gene expression patterns. Here, we use a computational model to exhaustively characterize the gene expression patterns of nearly 17 million three-gene circuits in order to systematically explore the relationship between circuit form and function. Three main conclusions emerge. First, function does not follow form. A circuit of any one structure can have between twelve and nearly thirty thousand distinct gene expression patterns. Second, and conversely, form does not follow function. Most gene expression patterns can be realized by more than one circuit structure. And third, multifunctionality severely constrains circuit form. The number of circuit structures able to drive multiple gene expression patterns decreases rapidly with the number of these patterns. These results indicate that it is generally not possible to infer circuit function from circuit form, or vice versa.
Gene expression is tightly regulated in both space and time1,2. Much of this regulation is carried out by sequence-specific DNA-binding proteins known as transcription factors (TFs), which activate or repress the expression of genes by promoting or preventing the recruitment of RNA polymerase. TFs often regulate the expression of other TFs, resulting in the formation of small regulatory circuits that are nested within a cell’s larger regulatory network3. The gene expression patterns of such circuits embody crucial biological functions, ranging from chemotaxis in bacteria4 to limb development in vertebrates5.
In many biological systems, form or structure can hint at function. Examples range from the fusiform shapes of fast-swimming animals like sharks and squid to the three-dimensional shapes of proteins. The possibility that this may also hold for gene regulatory circuits has motivated an intense research effort to understand how the form or structure of such circuits (i.e., the wiring diagram of who regulates whom) governs their function (i.e., gene expression pattern)6.
Certain circuit structures — commonly referred to as motifs7 — are statistically enriched in the gene regulatory networks of organisms as diverse as bacteria8 and humans9. These structures include three-gene motifs such as the feedforward loop (TF a regulates TF b, and both regulate TF c)10 and four-gene motifs such as the bi-fan (TFs a and b both regulate TFs c and d)11. Such motifs are involved in important physiological and developmental processes, including response acceleration in the galactose utilization system of bacteria12 and the interpretation of morphogen gradients during embryogenesis in fruit flies13. The functions of these and other circuit motifs have therefore been the topic of several theoretical10,13,14,15,16,17 and experimental studies12,18,19.
A common interpretation of the statistical enrichment of a circuit motif is that it is a signature of adaptation, and that it reflects a function that the circuit performs. This view is contentious20,21,22,23 and several studies suggest that circuit function cannot always be inferred from circuit structure11,24,25,26, as structure, by itself, does not provide sufficient information to uniquely infer function. For example, the motif that drives circadian rhythms in Drosophila melanogaster27 can function either as a resonator or as an integrator, depending on various circuit parameters, such as protein degradation rates25. Despite such anecdotal examples, we still know very little about the extent to which a circuit’s function can be inferred from its form. This is because earlier studies of circuit function usually focused on just a few motifs and studied them only under a limited subset of all possible initial gene expression states. In addition, they only considered a few of the possible regulatory programs — the rules governing the dynamics of gene expression28,29,30 — that each motif implements. And furthermore, they did not consider the fact that regulatory circuits can be multifunctional31, forming distinct metastable gene expression patterns in different tissues32 and developmental stages1, or in response to different physiological conditions33. A systematic analysis of the relationship between form and function in gene regulatory circuits is therefore lacking.
To help fill this knowledge gap, we build upon our earlier work31,34 with Boolean circuits35 — a prominent model of genetic regulation — to systematically investigate the relationship between circuit form and function. We choose to study the Boolean model for three reasons. First, despite its many simplifying assumptions, the model and its variants have successfully recapitulated the gene expression patterns of regulatory circuits that drive processes as different as the embryonic specification of endomesoderm in the sea urchin36 and circadian oscillations in fungi and plants37. Moreover, the model has been widely adopted. Its application domains range from cell and developmental biology to community ecology38 and evolutionary robotics39, making the study of form and function in Boolean circuits broadly relevant. Second, the model allows one to study not only the structural variants of a circuit (i.e., motifs), but also the signal-integration logic of a circuit’s cis-regulatory regions. These regions specify a circuit’s regulatory program, encoding the input-output mapping of regulatory signals (e.g., the presence or absence of TFs) to gene expression patterns28,29,30. Third, small Boolean circuits are amenable to exhaustive enumeration; the maximum number of encodings of a circuit with N genes is 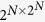 , which remains a manageable number for small N. Importantly, this facilitates a comprehensive and systematic characterization of every regulatory program in every circuit motif under all possible initial gene expression states.
, which remains a manageable number for small N. Importantly, this facilitates a comprehensive and systematic characterization of every regulatory program in every circuit motif under all possible initial gene expression states.
Methods
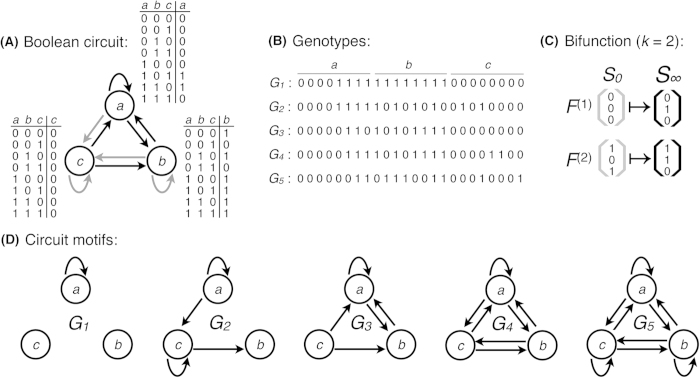
We consider Boolean circuits with N = 3 genes (Fig. 1A). Not only are circuits of this size the typical focus of circuit motif analyses7,8,9, they are also involved in important physiological and developmental processes. Examples include the kaiABC gene cluster in Cyanobacteria, which drives circadian oscillations40, and the Oct4-Sall4-Nanog circuit in mice, which controls pre-implantation development41. We compactly represent each circuit as a binary vector of length L = N × 2N = 24, which we refer to as the circuit’s genotype G (Fig. 1B). The genotype specifies the signal-integration logic of the circuit’s constituent genes. In biological circuits, this logic is encoded by the number and affinity of TF binding sites in a gene’s cis-regulatory region42, in addition to the spacing between such sites42,43, their distance to the transcription start site44, and their genomic context45. The genotype G also specifies the circuit’s structure, since a circuit’s signal-integration logic may render some regulatory interactions inactive (gray arrows in Fig. 1A). Because different genotypes may encode the same regulatory interactions in different ways, a single motif may be represented by several genotypes. For example, in Fig. 1A, the autoregulatory interaction b → b is not present because the signal-integration logic of gene b encodes the statement “a or not c”. Different signal-integration logic may encode statements such as “a and c” or “not a and not c”, which are also independent of gene b, and would therefore render the autoregulatory interaction b → b inactive.
Figure 1. Schematic illustration of Boolean circuits.
(A) A Boolean circuit with N = 3 genes, which are labeled as open circles a, b, c. Gene expression states are binary, such that genes are either on (1) or off (0). A directed edge a → b connects two genes if the expression state of b is dependent upon that of a (black arrows). Such dependencies are captured in the lookup table associated with each gene, which deterministically maps the 2N possible expression states of N genes to an output expression state. These mappings may render some edges inactive (gray arrows). For example, the lookup table associated with gene b encodes the logical statement “a or not c”, which is independent of gene b. The autoregulatory interaction b → b is therefore inactive, as indicated by the gray arrow. (B) Five circuit genotypes Gi, each represented as a vector of length L = N × 2N = 24. Each genotype is constructed by concatenating the rightmost columns of the lookup tables of the circuit’s constituent genes. For example, the circuit shown in (A) is represented by the genotype G3, which — along with the circuits represented by the other four genotypes G1, G2, G4, G5 — has the bifunction shown in (C) i.e., 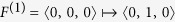 ,
, 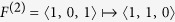 . These circuits have other k-functions as well. For example, the circuit encoded by genotype G3 has a total of 12 distinct bifunctions, because six initial states result in the equilibrium state 〈0, 1, 0〉 and two initial states result in the equilibrium state 〈1, 1, 0〉. (D) The circuit motifs that correspond to the five genotypes in (B) illustrating how a circuit’s genotype determines its structure. Note how the inactive edges (gray arrows) in the circuit shown in (A) are not present in the motif that corresponds to genotype G3. The five motifs increase in complexity — measured as their number of edges13 — from left to right.
. These circuits have other k-functions as well. For example, the circuit encoded by genotype G3 has a total of 12 distinct bifunctions, because six initial states result in the equilibrium state 〈0, 1, 0〉 and two initial states result in the equilibrium state 〈1, 1, 0〉. (D) The circuit motifs that correspond to the five genotypes in (B) illustrating how a circuit’s genotype determines its structure. Note how the inactive edges (gray arrows) in the circuit shown in (A) are not present in the motif that corresponds to genotype G3. The five motifs increase in complexity — measured as their number of edges13 — from left to right.
In the Boolean model, genes can be in one of two states, on (1) or off (0). Gene states are updated according to the states of the other genes in the circuit, as prescribed by the signal-integration logic encoded in the genotype. Specifically, for each gene i, the genotype encodes a mapping fi that updates the gene’s state σi(t) at time t according to
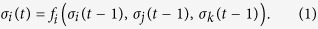 |
An example of such a mapping is shown as a look-up table in Fig. 1A, and all such mappings can be represented as a look-up table.
We refer to a circuit’s set of gene states at time t as a gene expression state St. Each circuit is initialized with a gene expression state S0, which represents physiological conditions, such as the presence or absence of a sugar12 or hormone46, or exogenous regulatory influences, such as those from a higher level in a cell’s regulatory hierarchy3. From S0, the circuit’s gene expression state is updated deterministically and synchronously until it reaches an equilibrium expression state S∞ with period p, which can be a fixed-point (p = 1) or periodic (p > 1). Circuits with fixed-point expression patterns are exemplified by the gap gene circuit in Drosophila melanogaster, which interprets a maternally-deposited morphogen gradient to produce specific concentrations of protein along the anterior-posterior axis of the developing embryo. Examples of periodic expression patterns include circadian rhythms47 and the cell cycle48.
Each function that a circuit could potentially perform can be represented as a pairing of initial and equilibrium expression states, F = (S0, S∞)31,34. (Although a computational model can only be used to study potential circuit functions, we refer to them for brevity simply as functions.) This definition is motivated by circuits in development and physiology that produce specific, temporally invariant gene expression patterns in response to specific combinations of physiological or regulatory signals1. A circuit can have up to k ≤ 2N functions. We refer to the set of these functions as a multifunction or as a k-function31,34: 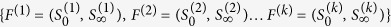 . Multifunctional circuits are not unusual32,33,49,50,51 and are typified by the hedgehog circuit in butterflies, which both patterns the wing blade and helps to form the wing’s eyespots52. Our only requirements of a k-function are that the equilibrium expression states of its k constituent functions are all (i) fixed-points and (ii) unique (i.e.,
. Multifunctional circuits are not unusual32,33,49,50,51 and are typified by the hedgehog circuit in butterflies, which both patterns the wing blade and helps to form the wing’s eyespots52. Our only requirements of a k-function are that the equilibrium expression states of its k constituent functions are all (i) fixed-points and (ii) unique (i.e., 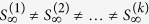 ). We use the terms monofunction, bifunction, trifunction, etc. to indicate the number of functions k in a multifunction. An example bifunction is shown in Fig. 1C.
). We use the terms monofunction, bifunction, trifunction, etc. to indicate the number of functions k in a multifunction. An example bifunction is shown in Fig. 1C.
We emphasize that a circuit can realize its k functions individually, or in various combinations, meaning that while a circuit genotype can be said to have between one and k functions, it can realize these functions in many different ways31. As an extreme example, a circuit with k = 8 functions (i.e., {F(1) = (〈0, 0, 0〉, 〈0, 0, 0〉), F(2) = (〈0, 0, 1〉, 〈0, 0, 1〉)…F(8) = (〈1, 1, 1〉, 〈1, 1, 1〉)} can realize these functions in a total of 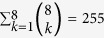 distinct combinations. This is because a circuit’s functions vary depending on the initial expression states that the circuit experiences. Continuing with the example above, this circuit may either realize the bifunction {F(1) = (〈0, 0, 0〉, 〈0, 0, 0〉), F(2) = (〈0, 0, 1〉, 〈0, 0, 1〉)} or {F(1) = (〈0, 0, 0〉, 〈0, 0, 0〉), F(3) = (〈0, 1, 1〉, 〈0, 1, 1〉)}, and this distinction only depends upon the initial expression states experienced by the circuit.
distinct combinations. This is because a circuit’s functions vary depending on the initial expression states that the circuit experiences. Continuing with the example above, this circuit may either realize the bifunction {F(1) = (〈0, 0, 0〉, 〈0, 0, 0〉), F(2) = (〈0, 0, 1〉, 〈0, 0, 1〉)} or {F(1) = (〈0, 0, 0〉, 〈0, 0, 0〉), F(3) = (〈0, 1, 1〉, 〈0, 1, 1〉)}, and this distinction only depends upon the initial expression states experienced by the circuit.
The space of circuits we consider here comprises 2L = 16, 777, 216 genotypes. For each of these genotypes, we determine the circuit’s motif (Fig. 1D) and all of its functions. This is accomplished by applying Eq. 1 to every genotype under all 2N = 8 possible initial expression states S0 (i.e., physiological conditions or exogenous regulatory influences). Since there are only 32, 399 possible k-functions31,34 and 104 possible circuit motifs (after accounting for graph isomorphisms), both the genotype-to-function and the genotype-to-motif mappings are many-to-one. (We note that by including multiple arrow types — e.g., by explicitly distinguishing inhibitory interactions using blunted arrows6 — the number of circuit motifs would increase.) We explore these mappings to ask a series of questions about the form and function of Boolean circuits. These include (i) How many circuit motifs have more than one function? (ii) Are some of a motif’s functions realized by more circuit genotypes than others? (iii) How many distinct circuit motifs have the same function? (iv) How does multifunctionality constrain circuit structure?
Results
Most circuit genotypes have more than one function
To provide a baseline for comparison to the number of functions per circuit motif, we first determine the set Kg of functions per circuit genotype, which we refer to as the genotype’s functional repertoire. We denote the size of this set as |Kg| and refer to a circuit as viable if it has at least one function (i.e., |Kg| > 0). We reiterate that the maximum size of this set is 255, not 8, because a circuit can realize its functions individually or in various combinations31. Figure 2A shows the probability that a circuit genotype has a functional repertoire size of at least |Kg|. Of the 16, 777, 216 possible genotypes, 11, 012, 415 (66%) have at least one function31. This implies that 34% of circuit genotypes are incapable of producing a fixed-point equilibrium expression pattern. Among the viable circuit genotypes, the median number of functions per circuit is seven; 74% of viable circuit genotypes have fewer than 10 functions and 92% have fewer than 25 functions. Only one circuit genotype realizes the maximum functional repertoire size of 255. It is the sole circuit genotype capable of an 8-function, a circuit with three edges: one autoregulatory interaction per gene. Finally, we observe no meaningful correlation between circuit complexity — measured as the number of active edges in the circuit — and the size of the circuit’s functional repertoire (Spearman’s rho = −0.008, p < 1 × 10−50; Fig. 2A, inset).
Figure 2. All circuit motifs have more than one k-function.
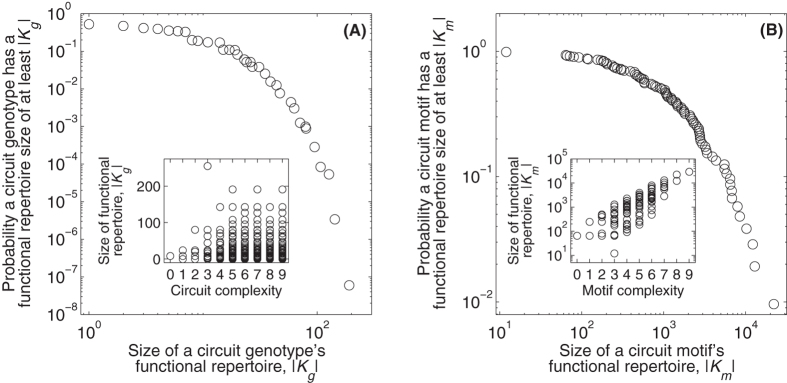
Each data point shows the probability that a circuit (A) genotype or (B) motif has a functional repertoire size of at least |Kg| or |Km|, respectively. The insets show the functional repertoire size in relation to the motif complexity, measured as the number of edges in the motif13, for (A) individual circuits and (B) all circuits with a given motif. Note the logarithmic scale of the x- and y-axes in the main panels and of the y-axis in the inset of (B).
All circuit motifs have more than one function
Previous studies have found that some circuit motifs have more than one function11,24,25,26. We explore the generality of this result by exhaustively analyzing the circuit functions of all 104 three-gene motifs, including all possible regulatory programs (i.e., signal-integration logic) and initial gene expression states. We refer to the set Km of k-functions realized by a motif as the motif’s functional repertoire. We denote the size of this set as |Km|. Figure 2B shows the probability that a circuit motif has a functional repertoire size of at least |Km|. Every circuit motif has at least 12 k-functions and most circuit motifs realize many more. The motif with the fewest k-functions is the feedback loop; its functional repertoire of 12 k-functions includes 8 monofunctions and 4 bifunctions. The feedforward loop can realize 64 distinct k-functions, as can the motif with no regulatory interactions. In both cases, the functional repertoire comprises all of the 64 possible monofunctions. Remarkably, the median number of k-functions per circuit motif is 994 and the maximum number is 28, 990, which corresponds to the fully connected circuit. These observations suggest that motifs with more than one function are the rule, rather than the exception, in gene regulatory circuits.
We also find that the size of a motif’s functional repertoire is positively correlated with motif complexity (Spearman’s r = 0.78, p < 1.43 × 10−22; Fig. 2B, inset), despite the lack of correlation between the size of a circuit’s functional repertoire and its complexity (Fig. 2A, inset). Complex motifs, being rich in potential functionality, are therefore highly versatile.
No one function dominates a motif’s functional repertoire
We have shown that each of the 104 three-gene motifs has more than one function. However, it may be the case that most of these functions are realized by just a small proportion of a motif’s constituent genotypes, while one or a few functions are realized by a large majority of these genotypes. This would indicate that a motif is much more likely to have some functions than others. To determine if this is the case, we calculate the entropy of a motif’s functional repertoire as 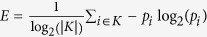 , where pi is the number ni of a motif’s constituent genotypes that have function i divided by
, where pi is the number ni of a motif’s constituent genotypes that have function i divided by 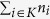 . This measure takes on its maximum value of 1 when each of a motif’s functions are realized by an equal number of genotypes. It approaches its minimum value of 0 when a single function is realized by the vast majority of a motif’s genotypes. Figure 3 shows the distribution of this measure for all 104 circuit motifs. The distribution is skewed toward high entropy values, indicating that no one function dominates a motif’s functional repertoire.
. This measure takes on its maximum value of 1 when each of a motif’s functions are realized by an equal number of genotypes. It approaches its minimum value of 0 when a single function is realized by the vast majority of a motif’s genotypes. Figure 3 shows the distribution of this measure for all 104 circuit motifs. The distribution is skewed toward high entropy values, indicating that no one function dominates a motif’s functional repertoire.
Figure 3. No one function dominates a motif’s functional repertoire.
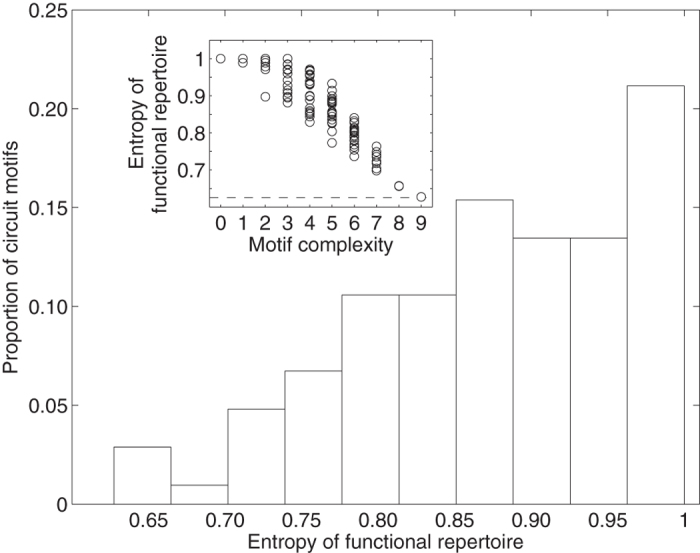
Entropy distribution of the functional repertoires of all 104 circuit motifs. The inset shows the entropy of a motif’s functional repertoire in relation to motif complexity. The dashed vertical line indicates the entropy of the functional repertoire of all 2L circuits, regardless of their motif. This was calculated as 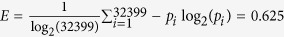 , where pi is the number of circuit genotypes with function i divided by 2L.
, where pi is the number of circuit genotypes with function i divided by 2L.
We also find that the complexity of a motif and the entropy of its functional repertoire are inversely correlated (r = −0.89, p = 1.89 × 10−36; Fig. 3, inset). This indicates that while complex motifs are more functionally versatile than simple motifs (Fig. 2, inset), their constituent genotypes are not as uniformly distributed amongst these functions. We note that this observation is largely driven by a strong correlation between motif complexity and the number of circuit genotypes per motif (r = 0.90, p = 1.61 × 10−38). This is confirmed by a randomization procedure in which circuit genotypes are randomly assigned to circuit motifs while holding constant the number of genotypes per motif and the functional repertoire per genotype (see Supplementary Fig. S1 online). As the number of circuit genotypes per motif increases, the entropy of the functional repertoire approaches that of the entire space of 2L circuit genotypes (Fig. 3, inset, dashed horizontal line), indicating that the low entropy of the functional repertoires of complex motifs is largely the result of a more complete sampling of the genotype space. However, even for the fully connected circuit, which has the lowest entropy (E = 0.627), the maximum proportion of circuit genotypes with any one function is only 0.02, and there are eight functions with this proportion of genotypes — all monofunctions in which the initial and equilibrium expression states are the same (e.g., F = (〈0, 0, 0〉, 〈0, 0, 0〉)). Thus, even for complex circuit motifs, no one function dominates the functional repertoire.
Most functions are realized by more than one circuit motif
Earlier theoretical13,53, empirical54,55, and experimental19 work has shown that certain circuit functions can be realized by more than one circuit motif. We explore the generality of this result by determining which of the 104 three-gene circuit motifs realize each k-function. Figure 4 shows the proportion of k-functions that are realized by a given proportion of the 104 circuit motifs, for all multifunctions with k ≤ 3. This figure indicates that circuit functions are generally realized by multiple circuit motifs. For example, of the 64 possible monofunctions31, all are realized by at least 60% of the 104 circuit motifs and 8 are realized by all 104 circuit motifs (Fig. 4A).
Figure 4. The same function can be realized by many distinct circuit motifs.

The distribution of the proportion of all possible circuit motifs per k-function is shown for all (A) monofunctions, (B) bifunctions, and (C) trifunctions. Bar heights are therefore normalized by (A) 64, (B) 1204, and (C) 7616, the number of possible monofunctions, bifunctions, and trifunctions, respectively31.
Multifunctionality constrains circuit structure
In our previous work31,56, we found that the number of circuit genotypes that can have k functions decreases exponentially as the number of functions k increases, indicating that multifunctionality constrains the number of viable circuit genotypes. Figure 4 hints that multifunctionality may also constrain circuit structure, because the distributions shift toward the left as k increases. To assess the extent of this constraint, we consider all 32, 399 k-functions and quantify the proportion of the 104 circuit motifs that realize each of these k-functions. Figure 5 shows that this proportion decreases rapidly as the number of functions k increases, implying that multifunctionality severely constrains circuit structure. Said differently, the more functions a circuit needs to perform, the fewer circuit structures that can perform all of them. For example, the average monofunction is realized by 89% of circuit motifs, whereas the average bifunction is realized by only 42% of circuit motifs. This constraint is also evident in the proportion of k-functions that are realized by only a single circuit motif, which quickly approaches 1 as k exceeds 3 (Fig. 5, inset). In total, 23% of all k-functions (7353 of 32,399) are realized by only one circuit motif. While this clearly demonstrates that multifunctionality constrains circuit structure, the fact remains that 77% of all k-functions are realized by more than one circuit motif, indicating that it is generally not possible to directly infer circuit form from circuit function.
Figure 5. Multifunctionality constrains circuit structure.
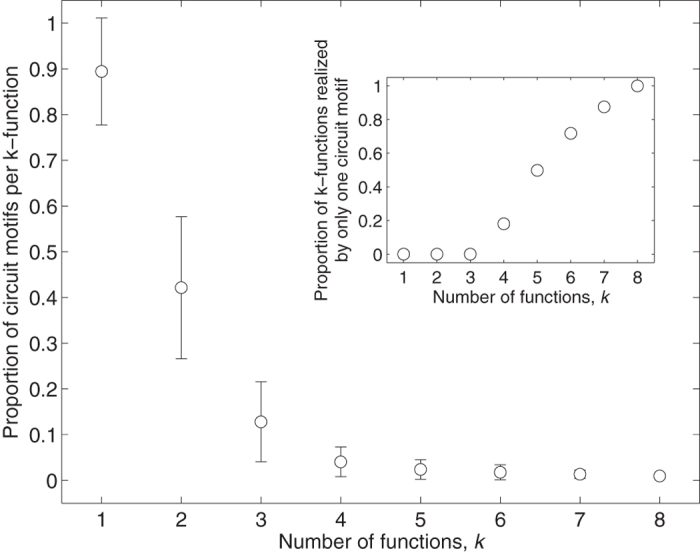
Data points show the average proportion of the 104 circuit motifs per k-function, in relation to the number of functions k. Error bars correspond to a single standard deviation. The inset shows the proportion of k-functions that are realized by just one circuit motif, in relation to the number of functions k.
Discussion
We have used a Boolean model of gene regulatory circuits35 to exhaustively characterize the gene expression patterns (functions) of nearly 17 million three-gene circuits and all possible structures (motifs) that these circuits can form, together with the functions they can perform, as embodied in the equilibrium gene expression patterns they reach from every possible initial gene expression state. Three main conclusions emerge. First, function does not follow form, that is, circuits with a given structure usually have multiple functions. Specifically, we found that every single one of the 104 possible three-gene motifs can perform at least 12 distinct functions and that nearly 90% of these motifs (93 of 104) can have more than one hundred functions. Moreover, each of a motif’s functions is realized by a roughly equal number of circuit genotypes, indicating that no one function dominates a motif’s functional repertoire. It is therefore not possible to directly infer circuit function from circuit structure without detailed information of the circuit’s signal-integration logic. These results complement and extend earlier observations that were based on the analysis of a small number of circuit motifs and on a limited subset of regulatory programs and initial gene expression states11,24,25,26. Further, they caution against the inference of circuit function from motif enrichment statistics20. However, it should be noted that the distribution of functions per motif depends crucially upon how the motifs are defined. By including inhibitory interactions, for example, the number of possible motifs would increase and some of these motifs may have fewer than the minimum number of 12 functions reported here (Fig. 2B).
Our second main conclusion is that form usually does not follow function, that is, circuits that can perform the same function can have very different structure. Over three-quarters of the 32,399 possible circuit functions are realized by more than one circuit motif. Remarkably, some functions are realized by all 104 motifs. These observations are in line with those from earlier computational models of large regulatory networks57,58, which found that many structurally distinct networks are capable of producing the same gene expression pattern. They are also in line with findings from comparative genomics, which have shown that the inter-species conservation of gene expression patterns does not imply conservation in the DNA sequences involved in regulating these expression patterns59,60,61,62. Such divergence in regulatory sequence has also lead to circuit rewiring55,63,64,65, as exemplified by the circuits controlling mating behavior in yeast54 and eye development in Drosophila66.
The third main conclusion is that multifunctionality constrains circuit structure. We found that the number of distinct circuit motifs with a given number of functions decreases rapidly as the number of functions increases. This observation may be relevant to synthetic biologists, who aim to understand the design constraints of circuits with specific biological functions. For example, a recent study designed several distinct three-gene circuit structures that are capable of interpreting a morphogen gradient to form a single spatial stripe19, inspired by the gene expression stripes formed during Drosophila embryogenesis. While such studies have understandably not yet considered multifunctional circuits, our results suggest that when they do, they will find far fewer circuit structures yielding multifunctions than monofunctions.
Our results also suggest that complex circuit motifs are more functionally versatile than simple motifs, as they have large functional repertoires that are not dominated by any one function. A recent analysis of the transcriptional regulatory networks of mouse and human revealed that complex motifs are statistically enriched, while simple motifs are statistically depleted, relative to randomized networks61. This suggests that in these organisms, highly versatile circuits are especially abundant. Combining our results with observations concerning the dynamical stability of circuit motifs17 may further our understanding of motif enrichment patterns in transcriptional regulatory networks.
Several studies have asked how functional constraints on transcriptional regulatory networks might favor some circuit motifs over others58,67,68,69. On the one hand, some studies have found that functional constraints lead to motif enrichment67,69. For instance, selection for oscillating gene expression patterns leads to the enrichment of certain four-gene motifs, such as the bifan69. On the other hand, a recent study focused on model regulatory networks capable of reproducing the gene expression patterns that embody flower organ specification in Arabidopsis showed that this functional constraint shapes the network’s pattern of edge usage (i.e., the presence or absence of specific regulatory interactions), but has almost no impact on motif enrichment58. Similar results were obtained in an earlier analysis of the set of regulatory networks capable of reproducing the gene expression pattern of the yeast cell cycle68.
Other studies have asked how the global topological properties of a network, such as its degree distribution, may influence local topological properties, such as the enrichment of motifs, even in the absence of functional constraints. For example, transcriptional regulatory networks are hierarchical and possess a heavy-tailed degree distribution, two global topological properties that have been shown to bias motif enrichment toward less complex motifs70. However, computational models have demonstrated that motif enrichment is easily fine-tuned via edge rewiring, even while global topological properties are held constant71, suggesting that it is possible, in principle, to select for particular motifs without changing the network’s global topological properties. Nevertheless, while selection for one of a motif’s several functions may lead to the enrichment of that motif, it remains impossible to use enrichment statistics to directly infer which of the motif’s several functions was selected. The reason is that, as we have shown, function does not follow form.
These findings highlight the limitations of diagrammatic representations of gene regulatory circuits, and underscore the importance of collecting detailed information about a circuit’s signal-integration logic28,29,72. Such information includes the location, number, orientation, order, and affinity of the regulating factors’ binding sites, whether the regulating factors interact cooperatively with themselves or with cofactors, and how target gene expression levels vary with the abundance and activity of the regulating factors73,74,75. We are only beginning to understand how these various facets of promoter architecture affect gene expression42,43,44,76, rendering the prediction of signal-integration logic from DNA sequence an outstanding challenge77.
In order to describe the circuit functions of all possible three-gene motifs under all possible regulatory programs and all possible initial gene expression states, we relied on a computational model of gene regulatory circuits. Specifically, we chose to study a Boolean model35 and to focus our attention on fixed-point gene expression patterns. These two decisions come with two caveats. First, the discrete nature of the model prohibits the analysis of protein production and degradation rates, which is a common theme in motif analyses6. We therefore cannot speak of a circuit as, for example, a “response accelerator”12 or as a “sign-sensitive delay”14. Second, our focus on fixed-point expression patterns necessarily excludes oscillating gene expression patterns, which are important for several biological functions, such as the cell cycle48. We were willing to accept these two caveats because they do not interfere with our ability to show that a circuit structure may have many functions, that a circuit function may be realized by many circuit structures, or that the number of circuit structures capable of driving multiple functions decreases with the number of functions. We therefore draw our three main conclusions: In gene regulatory circuits, function does not follow form, form rarely follows function, and form is severely constrained by multifunctionality.
Additional Information
How to cite this article: Payne, J. L. and Wagner, A. Function does not follow form in gene regulatory circuits. Sci. Rep. 5, 13015; doi: 10.1038/srep13015 (2015).
Supplementary Material
Acknowledgments
J.L.P. acknowledges support through the Forschungskredit program at the University of Zurich and through the Ambizione program of the Swiss National Science Foundation. A.W. acknowledges support through Swiss National Science Foundation grant 31003A_146137, as well as through the University Priority Research Program in Evolutionary Biology at the University of Zurich.
Footnotes
Author Contributions J.L.P. and A.W. designed the research. J.L.P. performed the research and analyzed the data. J.L.P. and A.W. wrote the paper.
References
- Carroll S. B., Grenier J. K. & Weatherbee S. D. From DNA to Diversity. Molecular Genetics and the Evolution of Animal Design (Blackwell, 2001). [Google Scholar]
- Davidson E. H. The Regulatory Genome (Academic Press, 2006). [Google Scholar]
- Erwin D. H. & Davidson E. H. The evolution of hierarchical gene regulatory networks. Nat. Rev. Genet. 10, 141–148 (2009). [DOI] [PubMed] [Google Scholar]
- Alon U., Surette M. G., Barkai N. & Leibler S. Robustness in bacterial chemotaxis. Nature 397, 168–171 (1999). [DOI] [PubMed] [Google Scholar]
- Raspopovic J., Marcon L., Russo L. & Sharpe J. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science 345, 566–570 (2014). [DOI] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Gen. 8, 450–461 (2007). [DOI] [PubMed] [Google Scholar]
- Milo R. et al. Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002). [DOI] [PubMed] [Google Scholar]
- Shen-Orr S. S., Milo R., Mangan S. & Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31, 64–68 (2002). [DOI] [PubMed] [Google Scholar]
- Neph S. et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell 150, 1274–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S. & Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100, 11980–11985 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram P. J., Stumpf M. P. H. & Stark J. Network motifs: structure does not determine function. BMC Genomics 7, 108 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S., Itzkovitz S., Zaslaver A. & Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J. Mol. Biol. 356, 1073–1081 (2006). [DOI] [PubMed] [Google Scholar]
- Cotterell J. & Sharpe J. An atlas of gene regulatory networks reveals multiple three-gene mechanisms for interpreting morphogen gradients. Mol. Syst. Biol. 6, 425 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S., Zaslaver A. & Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol. Biol. 334, 197–204 (2003). [DOI] [PubMed] [Google Scholar]
- Ghosh B., Karmakar R. & Bose I. Noise characteristics of feed forward loops. Phys. Biol. 2, 36–45 (2005). [DOI] [PubMed] [Google Scholar]
- Hayot F. & Jayaprakash C. A feedforward loop motif in transcriptional regulation: induction and repression. J. Theor. Biol. 234, 133–143 (2005). [DOI] [PubMed] [Google Scholar]
- Prill R. J., Iglesias P. A. & Levchenko A. Dynamic properties of network motifs contribute to biological network organization. PLoS Biol. 3, e343 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A. & Skotheim J. M. Feedforward regulation ensures stability and rapid reversibility of a cellular state. Mol. Cell 50, 856–868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli Y. et al. A unified design space of synthetic stripe-forming networks. Nat. Comm. 5, 4905 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzy-Randrup Y., Fleishman S. J., Ben-Tal N. & Stone L. Comment on “Network motifs: simple building blocks of complex networks” and “Superfamilies of evolved and designed networks”. Science 305, 1107c (2004). [DOI] [PubMed] [Google Scholar]
- Cordero O. X. & Hogeweg P. Feed-forward loop circuits as a side effect of genome evolution. Mol. Biol. Evol. 23, 1931–1936 (2006). [DOI] [PubMed] [Google Scholar]
- Solé R. V. & Valverde S. Are network motifs the spandrels of cellular complexity? Trends Ecol. Evol. 21, 419–422 (2006). [DOI] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat. Rev. Gen. 8, 803–813 (2007). [DOI] [PubMed] [Google Scholar]
- Wall M. E., Dunlop M. J. & Hlavacek W. S. Multiple functions of a feed-forward-loop gene circuit. J. Mol. Biol. 349, 501–514 (2005). [DOI] [PubMed] [Google Scholar]
- Conrad E., Mayo A. E., Ninfa A. J. & Forger D. B. Rate constants rather than biochemical mechanism determine behaviour of genetic clocks. J. R. Soc. Interface 5, S9–S15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macía J., Widder S. & Solé R. Specialized or flexible feed-forward loop motifs: a question of topology. BMC Syst. Biol. 3, 84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J. J., Hong C. I., Thron C. D. & Novak B. A simple model of circadian rhythms based on dimerization and proteolysis of PER and TIM. Biophys. J. 77, 2411–2417 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo A. E., Setty Y., Shavit S., Zaslaver A. & Alon U. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 4, e45 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan S., Bren A., Zaslaver A., Dekel E. & Alon U. Diverse two-dimensional input functions control bacterial sugar genes. Mol. Cell 29, 786–792 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker A., Tuboly C., Horváth P., Krishna S. & Semsey S. Genetic flexibility of regulatory networks. Proc. Natl. Acad. Sci. USA 107, 12998–13003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne J. L. & Wagner A. Constraint and contingency in multifunctional gene regulatory circuits. PLoS Comput. Biol. 9, e1003071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaskas N. et al. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell 148, 273–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel G. M., Garcia-Ojalvo J., Liberman L. M. & Elowitz M. B. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440, 545–550 (2006). [DOI] [PubMed] [Google Scholar]
- Payne J. L. & Wagner A. Latent phenotypes pervade gene regulatory circuits. BMC Syst. Biol. 8, 64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S. A. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 22, 437–467 (1969). [DOI] [PubMed] [Google Scholar]
- Peter I. S., Faure E. & Davidson E. H. Predictive computation of genomic logic processing functions in embryonic development. Proc. Natl. Acad. Sci. USA 109, 16434–16442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman O. E. et al. Digital clocks: simple Boolean models can quantitatively describe circadian systems. J. R. Soc. Interface 9, 2365–2382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C., Yang S., Albert R. & Shea K. A network model for plant-pollinator community assembly. Proc. Natl. Acad. Sci. USA 108, 197–202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongard J. C. Spontaneous evolution of structural modularity in robot neural network controllers. In Proceedings of the Genetic and Evolutionary Computation Conference, 251–258 (2011). [Google Scholar]
- Ishiura M. et al. Expression of a gene cluster kaiABC as a circadian feedback process in Cyanobacteria. Science 281, 1519–1523 (1998). [DOI] [PubMed] [Google Scholar]
- Tan M. H. et al. An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol. Sys. Biol. 9, 632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. P. et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat. Genet. 45, 1021–1028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E. et al. Probing the effects of promoters on noise in gene expression using thousands of designed sequences. Genome Res. 24, 1698–1706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E. et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 30, 521–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Myers C. A., Corbo J. C. & Cohen B. A. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. USA 110, 11952–11957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Masuno K., Cooper S. B. & Yamamoto K. R. Incoherent feed-forward regulatory logic underpinning glucocorticoid receptor action. Proc. Natl. Acad. Sci. USA 110, 1964–1969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W. & Kay S. A. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702–715 (2001). [DOI] [PubMed] [Google Scholar]
- Pomerening J. R., Kim S. Y. & Ferrell J. E. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell 122, 565–578 (2005). [DOI] [PubMed] [Google Scholar]
- Oppenheim A. B., Kobiler O., Stavans J., Court D. L. & Adhya S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 39, 409–429 (2005). [DOI] [PubMed] [Google Scholar]
- Cataudella I., Sneppen K., Gerdes K. & Mitarai N. Conditional cooperativity of toxin-antitoxin regulation can mediate bistability between growth and dormancy. PLoS Computat. Biol. 9, e1003174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday A. D. et al. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol. Microbiol. 90, 22–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys D. N. et al. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283, 532–534 (1999). [DOI] [PubMed] [Google Scholar]
- Ma W., Trusina A., El-Samad H., Lim W. A. & Tang C. Defining network topologies that can achieve biochemical adaptation. Cell 138, 760–773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong A. E., Tuch B. B., Li H. & Johnson A. D. Evolution of alternative transcriptional circuits with identical logic. Nature 443, 415–420 (2006). [DOI] [PubMed] [Google Scholar]
- Martchenko M., Levitin A., Hogues H., Nantel A. & Whiteway M. Transcriptional rewiring of fungal Galactose-metabolism circuitry. Curr. Biol. 17, 1007–1013 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin O. C. & Wagner A. Multifunctionality and Robustness Trade-Offs in Model Genetic Circuits. Biophys. J. 94, 2927–2937 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberti S., Martin O. C. & Wagner A. Innovation and robustness in complex regulatory gene networks. Proc. Natl. Acad. Sci. USA 104, 13591–13596 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A., Monéger F., Samal A. & Martin O. C. Network function shapes network structure: the case of the Arabidopsis flower organ specification genetic network. Mol. BioSyst. 9, 1726–1735 (2013). [DOI] [PubMed] [Google Scholar]
- Ludwig M. Z., Bergman C., Patel N. H. & Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564–567 (2000). [DOI] [PubMed] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L. & McCallion A. S. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312, 276–279 (2006). [DOI] [PubMed] [Google Scholar]
- Stergachis A. B. et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515, 365–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J. et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano L. A. & Wray G. A. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development 130, 4187–4199 (2003). [DOI] [PubMed] [Google Scholar]
- Baker C. R., Booth L. N., Sorrells T. R. & Johnson A. D. Protein modularity, cooperative binding, and hybrid regulatory states underlie transcription network diversification. Cell 151, 80–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N., Wapinski I., Margalit H., Regev A. & Friedman N. A functional selection model explains evolutionary robustness despite plasticity in regulatory networks. Mol. Syst. Biol. 8, 619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson C. I., Schwimmer D. B. & Barolo S. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr. Biol. 21, 1186–1196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N. & Alon U. Spontaneous evolution of modularity and network motifs. Proc. Natl. Acad. Sci. USA 102, 13773–13778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K.-Y., Ganguli S. & Tang C. Function constrains network architecture and dynamics: A case study on the yeast cell cycle Boolean network. Phys. Rev. E 75, 051907 (2007). [DOI] [PubMed] [Google Scholar]
- Burda Z., Krzywicki A., Martin O. C. & Zagorski M. Motifs emerge from function in model gene regulatory networks. Proc. Natl. Acad. Sci. USA 108, 17263–17268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez A., Dobrin R., Sergi D., Eckmann J.-P., Oltvai Z. N. & Barabási A.-L. The topological relationship between the large-scale attributes and local interaction patterns of complex networks. Proc. Natl. Acad. Sci. USA 101, 17940–17945 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yuan J., Shi Y. & Zagal J. C. Growing scale-free networks with tunable distributions of triad motifs. Physica A 428, 103–110 (2015). [Google Scholar]
- Setty Y., Mayo A. E., Surette M. G. & Alon U. Detailed map of a cis-regulatory input function. Proc. Natl. Acad. Sci. USA 100, 7702–7707 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. D., Shay T., O’Shea E. K. & Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science 325, 429–432 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T. & Gordân R. Protein-DNA binding: complexities and multi-protein codes. Nucleic Acids Res. 42, 2099–2111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S. & Segal E. The grammar of transcriptional regulation. Hum. Genet. 133, 701–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. S. III, Surette M. G. & Elowitz M. B. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3, 145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E. & Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach Nat. Rev. Genet. 10, 443–456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.