ABSTRACT
Poxviruses are large DNA viruses of vertebrates and insects causing disease in many animal species, including reptiles, birds, and mammals. Although poxvirus-like particles were detected in diseased farmed koi carp, ayu, and Atlantic salmon, their genetic relationships to poxviruses were not established. Here, we provide the first genome sequence of a fish poxvirus, which was isolated from farmed Atlantic salmon. In the present study, we used quantitative PCR and immunohistochemistry to determine aspects of salmon gill poxvirus disease, which are described here. The gill was the main target organ where immature and mature poxvirus particles were detected. The particles were detected in detaching, apoptotic respiratory epithelial cells preceding clinical disease in the form of lethargy, respiratory distress, and mortality. In moribund salmon, blocking of gas exchange would likely be caused by the adherence of respiratory lamellae and epithelial proliferation obstructing respiratory surfaces. The virus was not found in healthy salmon or in control fish with gill disease without apoptotic cells, although transmission remains to be demonstrated. PCR of archival tissue confirmed virus infection in 14 cases with gill apoptosis in Norway starting from 1995. Phylogenomic analyses showed that the fish poxvirus is the deepest available branch of chordopoxviruses. The virus genome encompasses most key chordopoxvirus genes that are required for genome replication and expression, although the gene order is substantially different from that in other chordopoxviruses. Nevertheless, many highly conserved chordopoxvirus genes involved in viral membrane biogenesis or virus-host interactions are missing. Instead, the salmon poxvirus carries numerous genes encoding unknown proteins, many of which have low sequence complexity and contain simple repeats suggestive of intrinsic disorder or distinct protein structures.
IMPORTANCE Aquaculture is an increasingly important global source of high-quality food. To sustain the growth in aquaculture, disease control in fish farming is essential. Moreover, the spread of disease from farmed fish to wildlife is a concern. Serious poxviral diseases are emerging in aquaculture, but very little is known about the viruses and the diseases that they cause. There is a possibility that viruses with enhanced virulence may spread to new species, as has occurred with the myxoma poxvirus in rabbits. Provision of the first fish poxvirus genome sequence and specific diagnostics for the salmon gill poxvirus in Atlantic salmon may help curb this disease and provide comparative knowledge. Furthermore, because salmon gill poxvirus represents the deepest branch of chordopoxvirus so far discovered, the genome analysis provided substantial insight into the evolution of different functional modules in this important group of viruses.
INTRODUCTION
Poxviruses are large, complex viruses with linear, double-stranded DNA (dsDNA) genomes that replicate entirely in the cytoplasm and infect insects (Entomopoxvirinae) and vertebrates (Chordopoxvirinae). About half of the approximately 100 conserved genes of chordopoxviruses are also found in entomopoxviruses (1). The known chordopoxviruses have been divided into 10 genera and 1 unassigned genus by the International Committee on Taxonomy of Viruses. Vertebrate hosts include reptiles, birds, and mammals. There are no reports on the occurrence of poxviruses in wild fish populations. However, poxvirus-like particles have been found by transmission electron microscopy (TEM) in gills sampled during serious mortality of farmed koi carp (Cyprinius carpio L.) (2), ayu (Plecoglossus altivelis Temminck & Schlegel) (3), and Atlantic salmon (Salmo salar L., referred to here as salmon) (4). In addition, a poxvirus-like sequence has been reported from koi carp (5). Diagnostic use of a PCR assay based on this sequence suggests that the virus has spread through Europe and that the common grass carp (Ctenopharyngodon idella Valencienes) is also susceptible (6–8). There appear to be two disease manifestations: carp edema in very small fry in winter (2) and koi sleepy disease in larger juveniles in summer (9). In carp edema, the entire fish is swollen and the fish swim close to the surface, akin to fish suffering from hypoxia. In koi sleepy disease, the fish lie on the bottom in a lethargic state but swim away when touched. The gills are always affected, and skin and eye lesions may also occur. Both manifestations are strongly alleviated by immersing the fish in 0.5% saline, but as infectivity is retained, the treatment could be merely symptomatic (9, 10). The ayu suffers a severe proliferative gill disease with large basophilic inclusions that correspond to poxvirus-like particles on TEM. In other cases where poxvirus-like particles have been found in fish, no obvious inclusion bodies have been found using light microscopy (3).
Poxvirus infection in Atlantic salmon was suspected in the 1990s in cases of acute, high-mortality events in freshwater farms with juvenile fish (O. B. Dale and A. Kvellestad, unpublished data). Later, TEM images from diseased Atlantic salmon showed poxvirus-like particles that appeared to be distinct from those in carp, although both have a single lateral body like that found in entomopoxviruses instead of two lateral bodies like those found in other chordopoxviruses (4). Based on material from fish with TEM findings similar to those described above, we present the first complete poxvirus genome from a fish and compare it with that of other chordopoxviruses. Diagnostic tools derived from the sequence have allowed us to analyze the gill disease associated with salmon gill poxvirus (SGPV) in terms of pathology and the location of infection.
MATERIALS AND METHODS
Sample material.
Samples were collected from three different Norwegian salmon farms in which the fish had suspected SGPV-related disease (gill apoptosis) (Table 1) and at the following clinical stages: premortality (n = 20; samples were taken 1 to 3 days before mortality was observed), mortality (n = 60; samples were taken from tanks in which mortality occurred and lethargic fish crowded on the bottom), and postmortality (n = 10; samples were taken from tanks in which mortality was observed a week prior to sampling). The average weight was 27 g (range, 10 to 40 g).
TABLE 1.
Overview of material from Norwegian salmon farms
| Fish | No. of fish with gill apoptosis |
No. of fish/no. of farms |
||||
|---|---|---|---|---|---|---|
| Archival cases, 1995–2006 | Controls (no gill apoptosis) |
|||||
| Farm A | Farm B | Farm C | Diseased fish | Healthy fish | ||
| Archival cases and controls | 39/14 | 48/8 | 3/1 | |||
| Diseased fish | ||||||
| Premortality | 20 | |||||
| Mortalitya | 30 | 25 | 5 | |||
| Postmortality | 10 | |||||
Sampling was performed on 5 dead and 25 moribund fish. Except for the 5 dead fish, all fish used in this study were sampled while still alive.
Archived, formalin-fixed, paraffin-embedded (FFPE) gill tissue was identified from 14 cases with records of gill disease and apoptotic gill epithelial cells. These cases were geographically spread in both fresh- and seawater sites in Norway (Table 1). Included were 12 fish from the first known outbreak of so-called amoebic gill disease in Norway (11). A separate TEM study also demonstrated poxvirus-like particles in those 12 fish (4). In addition, samples from 48 fish with other gill diseases (without gill epithelial apoptosis) and 3 healthy fish were included as controls (Table 1; see also Table 3).
TABLE 3.
Overview of gill lesions in the controls (no gill apoptosis)
| Farm | No. of fish | Gill histopathology and agents visible on light microscopy |
|---|---|---|
| D | 9 | Moderate adherences of lamellae and parasitic flagellates (Ichthyobodo spp.) |
| E | 4 | Moderate detachment of lamellar epithelial cells |
| F | 4 | Moderately thickened lamellae due to epithelial hypertrophy |
| G | 4 | Severe epithelial proliferation |
| H | 4 | Moderate lifting of epithelial cells and fungal infection |
| I | 3 | Moderate epithelial hypertrophy |
| J | 4 | Severe epithelial proliferation |
| K | 10 | Focal detachment and necrosis of lamellar epithelial cells and bacteria colonizing the apical surface of epithelium |
| L | 6 | Moderate hypertrophy and necrosis of epithelial cells; mucous cell proliferation |
Tissue sampling for histology, TEM, and PCR.
All fish were anesthetized and autopsied, and gill tissues were fixed in neutral phosphate-buffered 10% formalin for histology and in RNAlater (Qiagen Inc., Valencia, CA, USA) for quantitative PCR (qPCR). Additional organs sampled for histology were heart, liver, intestine, spleen, kidney, muscle, and skin. Additional organs sampled for PCR were spleen, kidney, and skin from five fish in the premortality stage and five dead fish from farm A (Table 1). Formalin-fixed gill tissue from one fish in the premortality stage and one fish in the mortality stage was prepared for TEM as described previously (12).
In situ staining methods.
Paraffin-embedded and hematoxylin and eosin (H&E)-stained sections were made for histology. For a subset of the samples, a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) in situ cell death detection kit AP (Roche, Basel, Switzerland) was used to confirm apoptosis. Prussian blue staining was used to verify hemosiderosis in the spleen and kidney. Staining for osmoregulatory chloride cells and proliferating cell nuclear antigen (PCNA) was performed as described previously (13, 14).
Immunohistochemistry (IHC) for SGPV.
Sections from gills were dewaxed, rehydrated, treated to demask antigen, and blocked with 5% bovine serum albumin in Tris-buffered saline to prevent nonspecific binding. Sections were incubated at 4°C overnight with a rabbit antibody (Pacific Immunology, Ramona, CA, USA) generated against the synthesized peptide GVNVDVKEFMQKFESNLSN-Cys (which is part of the L1 protein, a transmembrane protein expressed on the surface of the intracellular mature virion). Visualization was performed using an EnVision kit (Dako, Glostrup, Denmark) with horseradish peroxidase and 3-amino-9-ethylcarbazole as the chromogen.
DNA isolation and qPCR detection.
DNA was isolated from various tissues using a QIAcube system and a QIAamp DNA minikit according to the manufacturer's recommendations (Qiagen Nordic, Oslo, Norway). For archive material, a QIAamp DNA FFPE tissue kit was used (Qiagen Inc., Valencia, CA, USA). A qPCR assay based on the SGPV genomic sequence was designed for the molecular detection of virus DNA. The target locus was the homolog of the vaccinia virus (VACV) D13L open reading frame (ORF), which has been suggested to be a unique feature of poxviruses (15). The assay comprised forward primer ATCCAAAATACGGAACATAAGCAAT, reverse primer CAACGACAAGGAGATCAACGC, and the minor groove binding (MGB) probe CTCAGAAACTTCAAAGGA labeled with 6-carboxyfluorescein and a minor groove binding nonfluorescence quencher (MGBNFQ). The assay was run using a Platinum quantitative PCR SuperMix-uracil DNA glycosylase (UDG) kit (Life Technologies AS, Oslo, Norway) and the following PCR parameters: 50°C for 2 min (for UDG incubation), 95°C for 15 min (for UDG inactivation), and 50 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 15 s. Reactions with threshold cycle (CT) values above 40 were repeated for confirmation of the results. All results with CT values above 45 were considered negative. In this study, there were no CT values between 35 and 45.
When sections were cut from archival FFPE tissue, healthy fish gill tissue samples were interspersed between the samples from different disease outbreaks to control for carryover contamination. Isolation of DNA from FFPE tissue sections was done with a QIAamp DNA FFPE tissue kit according to the manufacturer's instructions (Qiagen Inc., Valencia, CA, USA).
RNA and DNA sequencing.
On the basis of the findings of TEM analyses, an Atlantic salmon gill tissue specimen containing poxvirus-like particles was selected for high-throughput sequencing. Total RNA was isolated from gill tissue fixed in RNAlater (Qiagen Norge, Oslo, Norway) using an RNeasy kit (Qiagen) and treated with Turbo DNA-free DNase (Life Technologies AS, Oslo, Norway) according to the manufacturer's recommendations. After DNase inactivation, 50 ng of total RNA was reverse transcribed and amplified using a QuantiTect whole-transcriptome kit (Qiagen). An initial round of total RNA pyrosequencing was done using a Roche 454 GS-FLX system and Titanium chemistry (454 Life Sciences, a Roche Company, Branford, CT, USA). All reads (521,710) from all reading frames and both strands were translated into protein sequences, and searches for sequence similarity to all poxvirus sequences available in GenBank were performed using the tblastx program (16). Two reads with weak similarity to known poxvirus sequences were identified, and one of these reads was used to design a qPCR assay (forward primer, ATCCAAAATACGGAACATAAGCAAT; reverse primer CAACGACAAGGAGATCAACGC; MGB probe, CTCAGAAACTTCAAAGGA; all sequences are written 5′ to 3′). Using the assay, a gill tissue sample with a high viral DNA content was selected for a subsequent round of sequencing. Total DNA was prepared using a DNeasy kit (Qiagen) and sequenced directly using a paired-end strategy and an Illumina HiSeq 2500 system (Illumina, Inc., San Diego, CA, USA). Reads were assembled de novo using a Velvet sequence assembler (17). Primers for PCR-based gap closing were designed using the software Primer Express (version 2.0.0; Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA), and PCR was performed using a HotStarTaq master mix kit (Qiagen). Amplification products were sequenced directly using Sanger sequencing.
The sequencing with the Illumina system gave a total of 169,083,705 pairs of 101-bp reads. Using the Velvet sequence assembler, a total of 68,968 high-confidence contigs could be generated (coverage, >10 times; length, >100 bp). A contig containing the two reads originally identified as being poxvirus-like was found, and pairs of reads where one partner mapped uniquely to one contig and the other mapped to a different contig were extracted using the poxvirus-like contig as a starting point. Using this information, 24 of the contigs produced by the Velvet sequence assembler could be arranged into a tentative scaffold of the genome. PCR was successful across all gaps, but for a small number of loci, the exact number of low-complexity repeats could not be established using Sanger sequencing due to length and base compositional bias. Instead, the approximate lengths of repeat regions were determined using a 2100 Bioanalyzer and a DNA 1000 kit (Agilent Technologies, Santa Clara, CA, USA) to analyze the gap PCR products. Only 5 of the original 521,710 reads from the 454 sequencing data set could be mapped back to the final version of the virus genome. The five reads ranged in length from 52 to 528 bases, and the longest read was identical to the one that was used to design the PCR assay.
Genome annotation.
The SGPV genome was translated by GeneMarkS software (http://exon.biology.gatech.edu/) (18); long (>80-nucleotide) intergenic regions were checked for the presence of ORFs, and ORFs ranging from 50 to 100 codons were annotated to be predicted protein-coding genes if they showed significant sequence similarity to other proteins or to a conserved domain in the National Center for Biotechnology Information Conserved Domains Database (19) or contained predicted transmembrane helices and/or a signal peptide. Transmembrane helices were predicted using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/) (20), and signal peptides were predicted using the SignalP (version 4.1) server (http://www.cbs.dtu.dk/services/SignalP/) (21). Tandem direct repeats were detected using the Tandem Repeats Finder program (22).
Protein sequence analysis and phylogenetic trees.
For detection of protein sequence similarity, the nonredundant protein sequence database at the National Center for Biotechnology Information (NIH, Bethesda, MD) was searched using the PSI-BLAST program (23). Predicted proteins of SGPV were assigned to clusters of nucleocytoplasmic virus orthologous genes (NCVOGs) using the PSI-COGNITOR program as previously described (24, 25). For phylogenetic analysis, protein sequences were aligned using the MUSCLE program (26) (http://www.ncbi.nlm.nih.gov/pubmed/15034147), and columns containing a large fraction of gaps (greater than 30%) and columns with low information content (27) were removed from the alignment. The alignment was used to construct an initial maximum likelihood (ML) phylogenetic tree with the FastTree program (http://www.ncbi.nlm.nih.gov/pubmed/20224823) with default parameters (28). The initial tree and the alignment were fed to the ProtTest program (29) to select the best substitution matrix. For each protein family, the best matrix found by ProtTest was used to construct the final ML tree with the TreeFinder program (30).
For the construction of the phylogenetic tree of poxviruses, multiple-sequence alignments of the sequences of 13 core genes present in all Poxviridae and African swine fever virus (ASFV) were employed (see Fig. S1 in the supplemental material). These genes belong to the following NCVOGs: NCVOG0022, major capsid protein; NCVOG0023, a D5-like helicase-primase; NCVOG0031, unclassified DEAD/SNF2-like helicases; NCVOG0038, DNA polymerase elongation subunit family B; NCVOG0076, DNA or RNA helicases of superfamily II; NCVOG0249, packaging ATPase; NCVOG0261, poxvirus early transcription factor (VETF), large subunit; NCVOG0262, poxvirus late transcription factor VLTF-3-like; NCVOG0267, RNA helicase DExH-NPH-II; NCVOG0271, DNA-directed RNA polymerase subunit beta; NCVOG0274, DNA-directed RNA polymerase subunit alpha; NCVOG1117, mRNA capping enzyme; NCVOG1164, A1L late transcription factor VLTF-2.
Reconstruction of gene content evolution.
The tree reconstructed from the concatenated alignment of 13 conserved proteins and the pattern of the presence-absence of SGPV proteins in the current version of the NCVOGs (24) were used to infer the gene loss and gene gain events and to obtain an ML reconstruction of the ancestral gene sets using COUNT software (31), as previously described (25).
Genome synteny analysis.
Genome synteny was visualized either with the Artemis genome comparison tool (32) or as dot plots of orthologous gene hits ordered by their positions in the genome (33). The synteny distance between viral genomes was calculated as previously described (33), with minor modifications, and a synteny-based neighbor-joining tree of the Poxviridae was constructed using the Neighbor program in the Phylip package (34).
Nucleotide sequence accession numbers.
The complete sequence of the SGPV genome was deposited in GenBank under accession number KT159937.
RESULTS
Evidence for poxvirus infection in farmed salmon.
The RNA isolated from salmon gill tissue containing poxvirus-like particles included sequences encoding putative proteins with significant similarity to those of poxviruses. A PCR probe was made using one such sequence in order to identify tissue with a high viral DNA content for direct paired-end sequencing. De novo assembly was performed, and gaps were filled in by PCR to generate a unique genome of 241,565 bp, excluding the termini, which were presumed to be covalently closed hairpins, as in other poxviruses. The relationship of the SGPV genome to the genomes of other poxviruses is detailed below.
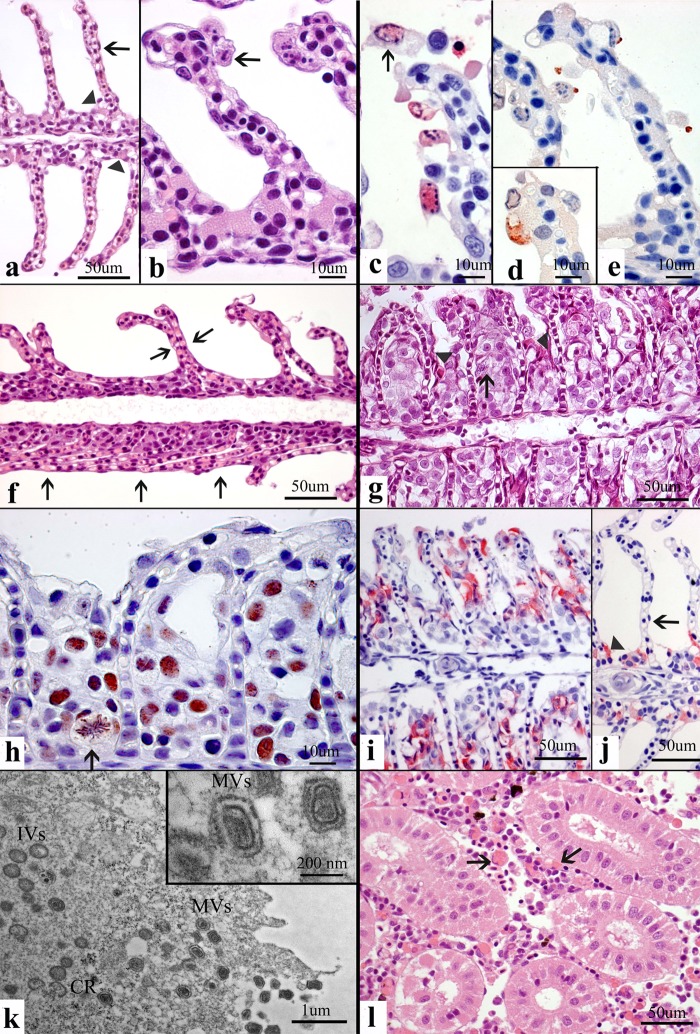
PCR and peptide antibody probes were constructed from SGPV homologs of the highly conserved vaccinia virus D13L gene and L1R virion membrane protein, respectively. Poxvirus DNA was detected by PCR in the gills from all fish sampled from the three outbreak farms, with a trend of increasing CT values over the disease course being detected (Table 2). In fish removed from tanks 1 to 3 days before death occurred (premortality stage), no lesions were found on autopsy, but most fish had no food in the gut, indicating appetite loss. On histopathology, changes were found only in the gills. Already at this stage, before clinical disease, apoptosis of lamellar epithelial cells was consistently found (Fig. 1b and c). Also, a general, moderate hypertrophy of this simple squamous epithelium was present, but no major blocking of the respiratory surfaces was found (Fig. 1b). A sparse fusion of lamellae due to epithelial proliferation and a moderate increase in the number of chloride cells were found in a few fish. All gills but one were IHC positive (Table 2). Only apoptotic epithelial cells either in the cytoplasm or in budding processes stained positive for poxvirus antigen (Fig. 1d and e). PCR of spleen and kidney tissue gave no CT value for four fish, while one fish had a CT value of 33.8 for spleen tissue and a CT value of 34.4 for kidney tissue. All skin tissue PCRs were positive, with the median CT value being 29.7 (range, 23.2 to 32.6).
TABLE 2.
Overview of results
| Clinical stage | No. of fish | Median (range) CT value for poxvirus in gills by qPCR | % of fish with: |
|
|---|---|---|---|---|
| IHC of gills | Hemophagocytosis | |||
| Premortality | 220 | 18.1 (15.8–22.4) | 95 | 0 |
| Mortality | 660 | 20.5 (15.7–28.9) | 91.4a | 66.7 |
| Postmortality | 110 | 24.7 (18.9–30.7) | 20 | 20 |
Two dead fish were not suited for IHC because of autolysis.
FIG 1.
Normal tissues and pathology in SGPV-infected Atlantic salmon. (a) A normal gill with thin lamellae (arrows) ensures efficient gas exchange. Chloride cells are present in normal numbers and at the normal location (arrowheads). (b and c) Detaching apoptotic cells with central clearing of chromatin (arrows) in the nuclear seen by H&E staining (b) and confirmed by red TUNEL staining (c). (d and e) IHC staining of poxvirus (brown) as cytoplasmic granules (d) and apical budding processes from apoptotic gill epithelial cells (e). (f) H&E staining of collapsed, adherent (arrows) thin lamellae losing apoptotic epithelial cells, creating an atelectasis-like condition hindering gas exchange. (g) H&E staining of proliferating (the arrow indicates metaphase), pale, foamy epithelial cells occluding the normally water-filled interlamellar space for gas exchange. Chloride cells are displaced and degenerated (arrowhead). (h) The lesion in panel g stained by IHC for PCNA showing brown nuclei, including proliferating cells in metaphase (arrow). (i and j) The lesion in panel g stained by IHC for chloride cells (red) that are displaced and enlarged (i) compared to the chloride cells in a normal gill (j). (k) TEM showing virus particles consistent with poxvirus in size and shape. Note the presence of crescents (CR), immature virions (IVs), and mature virions (MVs). (l) H&E staining of prominent hemophagocytosis (arrows) in the hematopoietic interrenal tissue. Methods included H&E staining (a, b, f, g, l); IHC staining for TUNEL (c), salmon gill poxvirus (d, e), PCNA (h), and chloride cells (i, j); and TEM (k).
In tanks in which fish were lethargic and there was some mortality (mortality stage), the main autopsy findings were swollen and slightly pale gills. Internal organs were also often pale, some spleens were enlarged, and no feed was found in the gut. Histopathology showed gill apoptosis in all fish at this stage, as described in the premortality stage (Fig. 1b to e). In addition, more severe gill changes obstructing the respiratory area were present in two different ways. First, in the phase with the severe detachment of apoptotic epithelial cells, the widespread adherence of the thin gill lamellae closed the water-filled space for gas exchange in an atelectasis-like manner (Fig. 1f). Second, the water-filled space between lamellae was solidified by proliferating epithelial cells (Fig. 1g), as demonstrated by PCNA staining (Fig. 1h). The proliferation also disrupted the tissue organization of the chloride cells (Fig. 1i), and apoptosis of chloride cells was also found. Histopathological lesions were also present in the spleen, kidney, and liver. A pronounced hemophagocytosis by scavenger endothelial cells and macrophages was found in the hematopoietic tissue of the spleen and kidney (Fig. 1l). Tissues with hemophagocytosis stained positive for Prussian blue Fe(III), demonstrating hemosiderosis (Table 2). Degenerative liver changes were variable but consistently present in dead fish. On IHC, over 90% of the gills were positive (Table 2), and labeling appeared as it did in the early stages (Fig. 1d and e). Furthermore, TEM demonstrated poxvirus-like particles in apoptotic cells (Fig. 1k). Crescents, spherical immature virions, and mature virions were seen in the cytoplasm, and these were also present in the extracellular space. Spleen and kidney were PCR negative in 3/5 fish (CT value range, 32.6 to 35.8). All skin samples were PCR positive, with the median CT value being 27.1 (range, 22.1 to 31.1).
In tanks in which mortality was observed a week prior to sampling (postmortality stage), most fish had no lesions on autopsy, except one fish had pale gills and four fish had enlarged spleens. Only a minor proliferation of gill epithelial cells, a very few apoptotic cells, and no IHC-positive cells were detected in all except two fish (Table 2). These two fish had pathology similar to that at the premortality stage, showing prevalent apoptosis, IHC-positive cells, and markedly lower CT values (18.9 and 19.5) than the other fish. We also observed hemophagocytosis in the spleen and kidney at this stage, although to a much lower degree and in fewer fish than in the mortality stage (Table 2). All samples from the control cases with no signs of gill epithelial apoptosis were PCR negative, but a wide range of other gill pathologies as well as evidence for bacterial, fungal, and parasitic infections were present in the unhealthy fish (Table 3).
From each of the 14 archived, formalin-fixed, paraffin-embedded case series, at least one positive gill tissue sample was found by PCR. The 39 diseased fish had a median CT value of 25.9 (range, 20.1 to 36.2). The interspersed control tissues had either high CT values or no CT value, indicating low or no cross contamination. All 12 samples from the so-called amoebic gill disease case were positive for poxvirus DNA by qPCR.
Genome analysis and evolutionary relationships of SGPV.
The SGPV genome consists of 241,565 bp (excluding the terminal hairpins) with a 37.5% GC content. The genome contains inverted terminal repeats of 5,679 bp each, similar to other poxviruses. Each of the inverted repeats, in turn, encompasses arrays of direct repeats. However, the tandem direct repeat arrays of SGPV, located at the very ends of the available genomic sequence, consist of only two 89-bp repeat units with 90% identity matches (each of these units consists of two 45-bp repeats with 88% identity). Thus, these direct repeat arrays are much smaller than those detected in other chordopoxviruses, although the possibility that they extend beyond the sequenced portion of the genome cannot be ruled out. Indeed, the highly conserved concatemer resolution sequence that is located between the repeat array and the apex of the terminal hairpin in other poxvirus genomes was not detected at the ends of the available SGPV sequence. The SGPV genome encompasses 206 unique predicted protein-coding genes (4 of these are contained within the terminal repeats and, accordingly, are present in the genome in two copies each; for details on gene prediction, see Materials and Methods). Comparison of the protein sequences encoded by these predicted genes to the sequences in the nonredundant protein sequence database at the National Center for Biotechnology Information (NIH, Bethesda, MD) using PSI-BLAST identified homologs with significant sequence similarity (E values, <10−4) for only 60 genes (for several additional predicted proteins, hits with apparently significant E values were identified as originating from regions of low sequence complexity and, accordingly, were dismissed as spurious). In addition to the standard database search, the predicted SGPV protein sequences were compared to the sequences of the NCVOGs (25), clusters of orthologous genes of nucleocytoplasmic large DNA viruses (NCLDVs), using a sequence profile search (see Materials and Methods). This comparison resulted in the assignment of 68 SGPV genes to NCVOGs, including 6 genes that showed no significant similarity to other proteins in BLAST searches. Additionally, a search for conserved domains led to a functional prediction in yet another protein (SGPV102). In total, specific, sequence conservation-based annotations were obtained through these procedures for 71 (34%) SGPV genes (Table 4 and Fig. 2). Among the genes without detectable homologs, 23 contained predicted transmembrane segments and/or a signal peptide, whereas 111 genes (55%) remained completely uncharacterized. Among the predicted products of these uncharacterized genes, many primarily consisted of low-complexity sequences and/or contained simple amino acid repeats (Table 4). These proteins are likely to be structurally disordered or assume unusual tertiary structures.
TABLE 4.
Predicted genes of SGPVa
| SGPV gene | Genome coordinates (protein lengthb) | NCVOG no. | Representation among NCLDVs | VACV gene name | Best hit (GI|Eval|% identity|aln_len|organism) | Predicted TM and SP | Functional annotation, comments, or inferred origin |
|---|---|---|---|---|---|---|---|
| 001 | 1248–298 (317) | Hypothetical protein | |||||
| 002 | 2205–1288 (306) | Hypothetical protein | |||||
| 003 | 3227–2241 (329) | Hypothetical protein; low sequence complexity | |||||
| 004 | 4788–3361 (476) | Hypothetical protein | |||||
| 005 | 5884–4934 (317) | Hypothetical protein | |||||
| 006 | 7314–6361 (318) | Hypothetical protein | |||||
| 007 | 7770–7438 (111) | Hypothetical protein | |||||
| 008 | 8702–7830 (291) | Hypothetical protein | |||||
| 009 | 9383–9054 (110) | 1 TM (C) | Hypothetical type I membrane protein, heptad repeats | ||||
| 010 | 9911–9546 (122) | Hypothetical protein; low sequence complexity | |||||
| 011 | 11295–10021 (425) | Hypothetical protein; low sequence complexity | |||||
| 012 | 12365–11373 (331) | Hypothetical protein | |||||
| 013 | 13629–12421 (403) | Hypothetical protein | |||||
| 014 | 15099–13681 (473) | Hypothetical protein | |||||
| 015 | 15912–15166 (249) | Hypothetical protein | |||||
| 016 | 16318–16007 (104) | Hypothetical protein | |||||
| 017 | 17728–16409 (440) | Hypothetical protein | |||||
| 018 | 18401–17922 (160) | Hypothetical protein | |||||
| 019 | 18870–18373 (166) | Hypothetical protein | |||||
| 020 | 19147–18863 (95) | Hypothetical protein | |||||
| 021 | 19468–19196 (91) | Hypothetical protein | |||||
| 022 | 19721–19533 (63) | 1 TM (M) | Hypothetical membrane protein | ||||
| 023 | 20272–19751 (174) | Hypothetical protein | |||||
| 024 | 21336–20332 (335) | Hypothetical protein | |||||
| 025 | 21953–21345 (203) | Hypothetical protein | |||||
| 026 | 23067–21967 (367) | Hypothetical protein | |||||
| 027 | 24336–23167 (390) | Hypothetical protein | |||||
| 028 | 25255–24395 (287) | Hypothetical protein | |||||
| 029 | 25986–25294 (231) | Hypothetical protein | |||||
| 030 | 26353–26033 (107) | Hypothetical protein | |||||
| 031 | 26850–26389 (154) | 1 TM (C) | Hypothetical type I membrane protein | ||||
| 032 | 27837–26851 (329) | 0017 | Phy, Mimi, Ent | 401825817|3.E−11|28|180|Encephalitozoon hellem ATCC 50504 | N-Myristoyl transferase; probable independent acquisition in different viruses; CACQ | ||
| 033 | 28446–27847 (200) | Hypothetical protein | |||||
| 034 | 28512–30752 (747) | 0330 | Most NCLDVs, all families except Asf | 660515722|3.E-07|26|242|Armadillidium vulgare iridescent virus | Divergent RING finger protein, potential E3 subunit of ubiquitin ligase; uncharacterized N-terminal domain upstream of RING domain; RING proteins in different NCLDVs likely have different origins; this SGPV protein is most similar to homologs from Iri and Phy; CACQ | ||
| 035 | 31420–30749 (224) | Hypothetical protein | |||||
| 036 | 32306–31434 (291) | Hypothetical protein | |||||
| 037 | 34172–32325 (616) | Hypothetical protein | |||||
| 038 | 34466–34263 (68) | SP | Hypothetical protein | ||||
| 039 | 34829–34503 (109) | Hypothetical protein | |||||
| 040 | 35364–34792 (191) | 0202 | Pox, Iri | 617520525|5.E-06|30|123|Poecilia formosa | 1 TM (C), SP | Ig domain type I membrane protein; not closely related to Ig domain-containing proteins of other NCLDVs; CACQ | |
| 041 | 35957–35583 (125) | Hypothetical protein | |||||
| 042 | 36110–36592 (161) | 1 TM | Hypothetical protein | ||||
| 043 | 36828–37679 (284) | 0284 | Some representatives of most NCLDVs except Asf | 511086842|1.E-59|39|276|Entamoeba histolytica | Ser/Thr protein kinase; probable eukaryotic origin; putative ribosomal protein S6K, mTOR pathway component; not closely related to any other NCLDV kinase, likely independent origin; CACQ | ||
| 044 | 38096–37665 (144) | 1 TM (M) | Hypothetical membrane protein | ||||
| 045 | 39115–38135 (327) | 1068 | Scattered distribution in all NCLDV families | F2L | 254568556|8.E-14|33|141|Komagataella pastoris GS115 | 1 TM (N) | Trimeric dUTPase highly similar to homologs from Phy but not poxviruses; contains uncharacterized N-terminal domain with a predicted TM; ANC |
| 046 | 39674–39102 (191 | Hypothetical protein | |||||
| 047 | 40162–39689 (158) | Hypothetical protein | |||||
| 048 | 40352–40882 (177) | Hypothetical protein | |||||
| 049 | 41988–40891 (366) | Hypothetical protein | |||||
| 050 | 42947–42033 (305) | Hypothetical protein | |||||
| 051 | 45667–42950 (906) | 502875360|8.E-4|24|403|Planctomyces limnophilus | Metalloendopeptidase of the M60-like family; UAQ | ||||
| 052 | 46422–45664 (253) | 1 TM (C) | Hypothetical type I membrane protein | ||||
| 053 | 46580–46422 (53) | 1 TM (C) | Hypothetical type I membrane protein | ||||
| 054 | 47190–46636 (185) | 1 TM (N) | Hypothetical type II membrane protein | ||||
| 055 | 48431–47325 (369) | Hypothetical protein | |||||
| 056 | 49116–48292 (275) | Hypothetical protein | |||||
| 057 | 50255–49101 (385) | Hypothetical protein | |||||
| 058 | 52022–50265 (586) | Hypothetical protein | |||||
| 059 | 52040–52516 (159) | 1122 | All Pox, some Iri, Mimi | J5 | 1 TM | Myristylated membrane protein, entry-fusion complex subunit; ANC; TAAATG | |
| 060 | 53257–52439 (273) | Hypothetical protein | |||||
| 061 | 53859–53299 (187) | 0258 | All Chor | J4R | 13876678|1E−08|31|186|lumpy skin disease virus | DNA-dependent RNA polymerase subunit Rpo22; CPOX | |
| 062 | 54821–53886 (312) | 1152 | All Pox, Pith, some Mimi | J3R | 41057489|3E−42|36|276|bovine papular stomatitis virus | Poly(A) polymerase small subunit, cap O-methyltransferase; ANC | |
| 063 | 55426–54788 (213) | Hypothetical protein | |||||
| 064 | 55824–55390 (145) | 1 TM (M) | Hypothetical membrane protein | ||||
| 065 | 56600–55842 (253) | 1063 | All Pox | L4R | DNA-binding virion core protein VP8; POX; TAAATG | ||
| 066 | 56631–57599 (323) | 1168 | All Pox | L3L | 659488262|5.E-08|24|308|penguinpox virus | Virion protein required for early transcription; POX | |
| 067 | 59023–57596 (476) | 0295 | Most NCLDV families except Asf and Pan | F10L | 544837|6.E-26|28|396|variola virus VAR, India-1967, peptide, 439 amino acids | Protein kinase involved in early stages of virion morphogenesis; ANC | |
| 068 | 59043–59897 (285) | 0249 | All NCLDVs | A32L | 12085104|1.E-17|28|264|Yaba-like disease virus | DNA packaging ATPase; ANC; TAAATG | |
| 069 | 59901–60317 (139) | Hypothetical protein | |||||
| 070 | 62029–60323 (569) | 1165 | All Pox | E1L | 9631476|2.E-14|23|381|Melanoplus sanguinipes entomopoxvirus | Poxvirus poly(A) polymerase catalytic subunit POX | |
| 071 | 62691–62071 (207) | 0272 | All Pox, most other NCLDVs | E4L | 38229198|5.E-18|29|174|Yaba monkey tumor virus | Transcription factor S-II (TFIIS); ANC; TAAATG | |
| 072 | 63448–62681 (256) | Hypothetical protein | |||||
| 073 | 64802–63516 (429) | Hypothetical protein | |||||
| 074 | 64862–66538 (559) | 1173 | All Pox | E6R | 40556061|3E-07|19|547|canarypox virus | Virion protein required for the formation of mature virions; POX; TAAATG | |
| 075 | 66539–67645 (369) | Hypothetical protein; low sequence complexity | |||||
| 076 | 70831–67634 (1,066) | 0038 | All NCLDVs | E9L | 659488229|5E−142|32|1027|penguinpox virus | DNA polymerase ANC | |
| 077 | 71147–70848 (100) | 0052 | All NLCDVs | E10R | 40556058|3E−24|48|92|canarypox virus | 1 TM (false positive) | Disulfide (thiol) oxidoreductase (Erv1/Alr family) involved in disulfide bond formation during virion morphogenesis; ANC; TAAATG |
| 078 | 71159–71788 (210) | Hypothetical protein | |||||
| 079 | 72152–71760 (131) | Hypothetical protein | |||||
| 080 | 73310–72168 (381) | 1160 | All Chor | I1L | 5830616|4E−13|27|281|variola minor virus | DNA-binding virion core protein; CPOX | |
| 081 | 74930–73677 (418) | Hypothetical protein | |||||
| 082 | 76048–74933 (372) | 1171 | All Chor | I6L | Telomere-binding protein involved in viral DNA encapsidation; CPOX; TAAATG | ||
| 083 | 77179–76049 (377) | Hypothetical protein | |||||
| 084 | 77295–78926 (544) | Hypothetical protein | |||||
| 085 | 79246–78857 (130) | Hypothetical protein; low sequence complexity | |||||
| 086 | 81234–79657 (526) | Hypothetical protein; low sequence complexity | |||||
| 087 | 81913–81221 (231) | Hypothetical protein; low sequence complexity | |||||
| 088 | 81912–82319 (136) | 4 TM | Protein consists of hydrophobic decamer repeats; TM prediction could be spurious | ||||
| 089 | 86171–82275 (1,299) | 0190 | Hypothetical protein | ||||
| 090 | 88408–86198 (737) | 0031 | Nearly all NCLDVs | D6R | 345107280|1E−156|41|657|Yoka poxvirus | SNF2-like helicase involved in early transcription; ANC | |
| 091 | 90927–88453 (825) | 0023 | All NCLDVs | D5R | 571798002|5E−93|28|768|squirrelpox virus | Primase-helicase; ANC | |
| 092 | 91684–90920 (255) | 0211 | All Chor | F9L | 9634782|8E−06|33|123|fowlpox virus | 1 TM (C) | Myristylated IMV envelope protein; CPOX; TAAATG |
| 093 | 92207–91647 (187) | Hypothetical protein | |||||
| 094 | 92262–92876 (205) | 1067 | Mimi | 494264790|3E−09|28|178|Marinobacter algicola | Deoxynucleotide monophosphate kinase shared with Mimi, probable bacterial origin; CACQ | ||
| 095 | 96496–93050 (1,149) | 0037 | Phy, Mimi, Mar, CrPV (multiple paralogs) but not other Pox | 5121|2E−53|25|933| Schizosaccharomyces pombe | DNA topoisomerase II; ANC | ||
| 096 | 96825–96496 (110) | 1 TM (M) | Hypothetical membrane protein | ||||
| 097 | 97628–96828 (267) | 0211 | Most NCLDVs, all Pox | L1R | 12085043|2E−29|31|225|Yaba-like disease virus | 1 TM (C) | Myristylated IMV envelope protein; ANC; TAAATG |
| 098 | 99427–97661 (589) | 0022 | All NCLDVs except Pan | D13L | 345107288|8E−50|28|570|Yoka poxvirus | Major capsid protein (involved in morphogenesis but not incorporated into virions in poxviruses); ANC | |
| 099 | 99845–99456 (130) | 1164 | All NCLDV | A1L | 289183841|1E−12|29|123|pseudocowpox virus | Late transcription factor VLTF-2; ANC | |
| 100 | 100665–99853 (271) | 0262 | All NCLDVs except Pith | A2L | 571798015|7E−8|38|195|squirrelpox virus | Late transcription factor VLTF; ANC | |
| 101 | 103192–100967 (742) | 1162 | All Pox, Mimi | A3L | 115531788|1E−49|24|697|Nile crocodilepox virus | Poxvirus P4B major core protein; POX | |
| 102 | 103950–103237 (238) | MCV, some Mimi, Phy | J domain-containing protein, putative cochaperonin; distantly related to J domains of other NCLDVs; CACQ | ||||
| 103 | 103956–104498 (181) | 1377 | All Chor | A5R | 40556180|3E−14|33|172|canarypox virus | DNA-dependent RNA polymerase subunit Rpo19; CPOX | |
| 104 | 105738–104503 (412) | 1179 | All Chor | A6L | Virion core protein required for membrane biogenesis and formation of mature virions; CPOX; TAAATG | ||
| 105 | 107964–105751 (738) | 0261 | All Pox, scattered in other NCLDVs | A7L | 659488305|2E−104|32|734|penguinpox virus | VETF, large subunit; ANC | |
| 106 | 107945–109252 (436) | 1176 | All Chor | A8R | 40556183|2E−08|24|248|canarypox virus | Poxvirus intermediate transcription factor VITF-3 subunit; CPOX; TAAATG | |
| 107 | 109518–109261 (86) | 2 TM | Hypothetical membrane protein | ||||
| 108 | 113053–109535 (1,173) | 0257 | All Pox | A10L | 157939724|4E−13|20|561|tanapox virus | Virion core protein P4; POX; TAAATG | |
| 109 | 113084–113923 (280) | Hypothetical protein | |||||
| 110 | 113964–114296 (111) | 1 TM (N) | Hypothetical type II membrane protein | ||||
| 111 | 114326–114619 (98) | Hypothetical protein; low sequence complexity | |||||
| 112 | 114935–114600 (112) | Hypothetical protein containing serine-rich repeats | |||||
| 113 | 115188–115556 (123) | Hypothetical protein | |||||
| 114 | 115768–115556 (71) | 1 TM (N) | Hypothetical type II membrane protein | ||||
| 115 | 117013–115769 (415) | 1045 | Some Iri and Mimi | 339906034|2E−07|30|145|Wiseana iridescent virus | 5′-3′ exoribonuclease of the XRN family; NCLDV proteins appear to be monophyletic; ANC | ||
| 116 | 117383–117045 (113) | 2 TM | Hypothetical protein | ||||
| 117 | 117721–117401 (107) | Hypothetical protein | |||||
| 118 | 118830–117736 (365) | 1122 | All Pox, Mimi, some Iri | A16L | 41057529|2E−15|29|204|bovine papular stomatitis virus | 1 TM | Myristylated protein, entry-fusion complex subunit; ANC; TAAATG |
| 119 | 119936–118848 (363) | 2 TM | Hypothetical membrane protein | ||||
| 120 | 119988–121418 (477) | 0076 | All Pox, in many other NCLDVs | A18R | 115531805|1E−54|29|424|Nile crocodilepox virus | DNA helicase of superfamily 2, transcript release factor; ANC | |
| 121 | 121419–122474 (352) | 2643 | Some Mimi | 504603808|3E−15|30|151|Ornithobacterium rhinotracheale | Apurinic-apyrimidinic endonuclease of the exonuclease III family; probable bacterial origin; CACQ | ||
| 122 | 122812–122465 (116) | 1370 | All Pox | A21L | 506498863|2E−06|24|111|Choristoneura rosaceana entomopoxvirus L. | 1 TM (C) | Type I membrane protein, entry-fusion complex subunit; POX; TAAATG |
| 123 | 122842–125133 (764) | 0035 | CrPV, Ent, some Iri, Mimi | NAD+-dependent DNA ligase; poorly conserved sequence but contains intact catalytic residues and shows the closest sequence similarity to NAD+-dependent ligases of Ent; ANC | |||
| 124 | 125105–125602 (166) | 0278 | All Pox, majority of other NCLDVs | A22R | 659488557|6E−16|32|149|pigeonpox virus | RuvC family Holliday junction resolvase; ANC | |
| 125 | 125599–126816 (406) | 0263 | All Pox | A23R | 9634858|3E−25|27|395|fowlpox virus | Intermediate transcription factor; POX | |
| 126 | 126817–130305 (1,163) | 0271 | All NCLDVs except some Phy | A24R | 225194776|0|47|1169|skunkpox virus | DNA-directed RNA polymerase subunit beta; TAAATG | |
| 127 | 130720–130310 (137) | 1418 | All Pox | A28L | 51317191|3E−17|33|128|Diachasmimorpha longicaudata entomopoxvirus | 1 TM (N) | Type I membrane protein, entry-fusion complex subunit beta; ANC; TAAATG |
| 128 | 131699–130725 (325) | 0260 | All Pox | A29L | 148912996|9E−08|27 |181|goatpox virus Pellor | DNA-directed RNA polymerase, 35-kDa subunit; POX | |
| 129 | 131870–132817 (316) | Hypothetical protein | |||||
| 130 | 132821–133525 (235) | Hypothetical protein | |||||
| 131 | 133536–135035 (500) | Hypothetical protein | |||||
| 132 | 135013–135495 (161) | Hypothetical protein | |||||
| 133 | 135919–135470 (150) | Hypothetical protein | |||||
| 134 | 136606–135941 (222) | 1115 | All Pox, scattered in other NCLDVs | D4R | 9634732|1E−15|28|216|fowlpox virus | UDG; ANC | |
| 135 | 136671–138380 (570) | Hypothetical protein; low sequence complexity | |||||
| 136 | 138373–139212 (280) | 0259 | All Pox | D7R | 9629029|4E−17|30|145|molluscum contagiosum virus subtype 1 | DNA-directed RNA polymerase, 18-kDa subunit; POX | |
| 137 | 139235–139879 (215) | 0236 | All Pox, most other NCLDVs | D10R | 9629031|1E−15|29 |161|molluscum contagiosum virus subtype 1 | Nudix hydrolase, decapping enzyme; ANC | |
| 138 | 141785–139887 (633) | 0027 | All Pox, some Mimi | D11L | 115531782|2E−174|43|635|Nile crocodilepox virus | Superfamily 2 helicase D11; POX; TAAATG | |
| 139 | 141949–142902 (318) | 0330 | All NCLDVs except Asco and Pith | 658035022|2E−06|31|75|Malus domestica | RING finger-containing E3 ubiquitin ligase; probably independent acquisition in different NCLDV families; CACQ | ||
| 140 | 143889–142951 (313) | 1169 | All Pox | D12L | 9629033|3E−31|30|289|molluscum contagiosum virus subtype 1 | Poxvirus mRNA capping enzyme, small subunit; POX; TAAATG | |
| 141 | 144893–143889 (335) | 1122 | All Pox, some Mimi, Iri | G9R | 9634797|7E−06|36|78|fowlpox virus | 1 TM | Myristylated protein, entry-fusion complex subunit; ANC; TAAATG |
| 142 | 145769–144894 (292) | 1369 | All Chor | G8R | 41057481|1E−06|26|171|bovine papular stomatitis virus | Protein containing a derived PCNA domain; VLTF-1; CPOX; TAAATG | |
| 143 | 145819–147291 (491) | Hypothetical protein | |||||
| 144 | 147884–147288 (199) | 1182 | All Pox | G6R | Predicted hydrolase or acyltransferase of the NlpC/P60 superfamily; weak sequence similarity to orthologs in other poxviruses; POX; TAAATG | ||
| 145 | 148111–147914 (66) | 1368 | All Chor, one Ent, Asf | G5.5R | 289183806|2E−04|24|65|pseudocowpox virus | RNA polymerase, subunit 10 (a very small protein, possibly missed during genome annotation of other viruses); POX | |
| 146 | 149892–148072 (607) | 1060 | All Pox, scattered in other NCLDVs | G5R | 539191060|6E−13|36|176|myxoma virus | Flap endonuclease required for poxvirus genome replication; ANC | |
| 147 | 149931–150485 (185) | 505137967|1E−05|41|59|Methanomethylovorans hollandica | Thioredoxin; no close homologs in other viruses; UAQ | ||||
| 148 | 150507–150884 (126) | 1 TM (M) | Hypothetical membrane protein | ||||
| 149 | 150881–152773 (631) | 1170 | All Pox | G1L | 115531736|6E−35|31|233|Nile crocodilepox virus | Metalloprotease essential for virion morphogenesis; POX; TAAATG | |
| 150 | 154796–152760 (679) | 0267 | All Pox, Asf, Mimi | I8R | 41057099|5E−121|37|597|orf virus | RNA helicase of superfamily 2 implicated in early transcription termination; ANC; TAAATG | |
| 151 | 154823–156076 (418) | 1161 | All Pox, most other NCLDVs | I7L | 115531734|7E−15|21|429| | Virion core cysteine protease involved in virion protein maturation; ANC; TAAATG | |
| 152 | 156073–156567 (165) | Hypothetical protein | |||||
| 153 | 156623–157354 (244) | 1 TM (C), SP | Hypothetical protein | ||||
| 154 | 157464–164144 (2,227) | 0269 | All Chor; disrupted in some, including VACVs | (B22R VARV) | 422933904|3E−120|29|1049|cyprinid herpesvirus 2 | 1 TM (C), SP | Giant type I membrane protein with homologs also in cyprinid herpesviruses, suggestive of gene transfer from SGPV to the herpesviruses (see the phylogenetic tree in Fig. 8); implicated in T cell inactivation; paralog of SGPV159 and SGPV162; CPOX |
| 155 | 164257–168030 (1,258) | 1 TM (C), SP | Hypothetical type I membrane protein | ||||
| 156 | 168031–169008 (326) | Hypothetical protein | |||||
| 157 | 168995–169900 (302) | Hypothetical protein | |||||
| 158 | 170583–169939 (215) | Hypothetical protein | |||||
| 159 | 170638–173652 (1,005) | 0269 | All Chor; disrupted in some, including VACV | 0 | 9634792|5E−11|24|462|fowlpox virus | 1 TM (C), SP | Giant type I membrane protein with homologs also in cyprinid herpesviruses, suggestive of gene transfer from SGPV to the herpesviruses (see the phylogenetic tree in Fig. 8); implicated in T cell inactivation; paralog of SGPV154 and SGPV162; CPOX |
| 160 | 173910–173665 (82) | 1 TM (N) | Hypothetical type II membrane protein containing pentapeptide repeats | ||||
| 161 | 173870–181351 (2,494) | SP | Hypothetical secreted protein | ||||
| 162 | 181528–185433 (1,302) | 0269 | All Chor; disrupted in some, including VACVs | 0 | 9628967|5E−25|25|413|molluscum contagiosum virus subtype 1 | 1 TM (C), SP | Giant type I membrane protein with homologs also in cyprinid herpesviruses, suggestive of gene transfer from SGPV to the herpesviruses (see the phylogenetic tree in Fig. 8); implicated in T cell inactivation; paralog of SGPV154 and SGPV159; CPOX |
| 163 | 185473–186558 (362) | Hypothetical protein | |||||
| 164 | 186693–188648 (652) | SP | Hypothetical secreted protein | ||||
| 165 | 188749–192687 (1,313) | 0274 | All NCLDVs except for some Phy | J6R | 115531763|0|41|1311|Nile crocodilepox virus | DNA-directed RNA polymerase subunit alpha; ANC | |
| 166 | 193271–192684 (196) | 1 TM (C) | Hypothetical type I membrane protein | ||||
| 167 | 193287–194597 (437) | SP | Hypothetical secreted protein, pentapeptide repeats | ||||
| 168 | 195155–194586 (190) | 0253 | All Pox | H2R | 594019595|2E−35|40|151|avipoxvirus OKr-2014 | 1 TM (N) | Type II membrane protein, fusion-entry complex subunit; POX; TAAATG |
| 169 | 197626–195161 (822) | 1163 | All Pox | H4L | 6969751|3E−67|30|583|vaccinia virus Tian Tan | Pox_Rap94, RNA polymerase-associated transcription specificity factor, Rap94; POX; TAAATG | |
| 170 | 197724–198404 (227) | Hypothetical protein | |||||
| 171 | 198405–199343 (313) | 0036 | All Pox, Mimi | H6R | 345107272|5E−60|40|310|Yoka poxvirus | DNA topoisomerase IB; ANC; TAAATG | |
| 172 | 199715–199329 (129) | SP | Hypothetical secreted protein | ||||
| 173 | 199747–202368 (874) | 1451 | All NCDLVs except Asco and Pan | D1R | 225194732|4E−110|33|867|volepox virus | mRNA capping enzyme large subunit; ANC; TAAATG | |
| 174 | 204943–202382 (854) | Hypothetical protein | |||||
| 175 | 205237–204956 (94) | Hypothetical protein | |||||
| 176 | 205654–205238 (139) | Hypothetical protein | |||||
| 177 | 205659–207647 (663) | Hypothetical protein | |||||
| 178 | 207690–209033 (448) | Hypothetical protein | |||||
| 179 | 209178–209951 (258) | Hypothetical protein | |||||
| 180 | 210027–211280 (418) | Hypothetical protein | |||||
| 181 | 211532–213193 (554) | Hypothetical protein | |||||
| 182 | 213211–213954 (248) | Hypothetical protein | |||||
| 183 | 213947–214258 (104) | Hypothetical protein | |||||
| 184 | 214236–214847 (204) | Hypothetical protein | |||||
| 185 | 215300–214851 (150) | Hypothetical protein | |||||
| 186 | 215396–216664 (423) | 167525479|6E−18|28|228|Monosiga brevicollis MX1 | DNA or RNA methyltransferase; UAQ | ||||
| 187 | 216775–217242 (156) | 209734208|9E−29|46|127|Salmo salar | Macrodomain, most similar to O-acetyl-ADP-ribose deacetylase; UAQ | ||||
| 188 | 217294–218286 (331) | Hypothetical protein | |||||
| 189 | 218360–219514 (385) | Hypothetical protein | |||||
| 190 | 219572–220492 (307) | Hypothetical protein; low sequence complexity; partly consists of tetrapeptide repeats | |||||
| 191 | 220576–221535 (320) | Hypothetical protein | |||||
| 192 | 221579–222580 (334) | Hypothetical protein | |||||
| 193 | 222676–223716 (347) | Hypothetical protein | |||||
| 194 | 224007–224258 (84) | Hypothetical protein; hydrophobic; 12-mer repeats | |||||
| 195 | 224390–225718 (443) | Hypothetical protein; low sequence complexity | |||||
| 196 | 226126–226542 (139) | Hypothetical protein | |||||
| 197 | 226596–227135 (180) | Hypothetical protein; cysteine rich; low sequence complexity | |||||
| 198 | 227202–228812 (537) | Hypothetical protein; low sequence complexity | |||||
| 199 | 228872–229897 (342) | Hypothetical protein | |||||
| 200 | 229951–230268 (106) | Hypothetical protein | |||||
| 201 | 230293–230985 (231) | Hypothetical protein | |||||
| 202 | 231098–231802 (235) | Hypothetical protein | |||||
| 203 | 232049–233533 (495) | Hypothetical protein | |||||
| 204 | 233892–233491 (134) | 4 TM | Hypothetical protein; hydrophobic, consists mostly of hexapeptide repeats; TM prediction might be false positive | ||||
| 205 | 234526–235545 (340) | Hypothetical protein | |||||
| 206 | 235633–236631 (333) | Hypothetical protein | |||||
| 207 | 236777–238204 (476) | Inverted terminal repeat; identical to SGPV001 gene | |||||
| 208 | 238338–239324 (329) | Inverted terminal repeat; identical to SGPV002 gene | |||||
| 209 | 239360–240277 (306) | Inverted terminal repeat; identical to SGPV003 gene | |||||
| 210 | 240317–241267 (317) | Inverted terminal repeat; identical to SGPV004 gene |
In the first column, “SGPV” is omitted from the gene identifiers for brevity; in the last column “SGPV” is included; GI, GenInfo Identifier sequence identification number); aln_len, the length of pairwise protein alignment produced by BLASTP searches; SP, (predicted) signal peptide; TM, (predicted) transmembrane helix (C, M, and N denote the C-terminal, middle, and N-terminal location of the predicted transmembrane helix in the protein, respectively); the percent identity and alignment length are taken directly from BLASTP searches. IMV stands for intracellular mature virions; VARV stands for variola virus. The inferred origin of genes is indicated as follows: ANC, ancestral to NCLDV; POX, ancestral to poxviruses; CPOX, ancestral to chordopoxviruses; CACQ, convergent acquisition (with other NCLDVs); UAQ, unique acquisition. The transcription start element TAAAT is shown for those SGPV genes that have orthologs from other chordopoxviruses (the sequence TAAATG includes the translation start codon of the respective gene). Abbreviations for groups of viruses: Asco, Ascoviridae; Asf, Asfarviridae; Chor; Chordopoxvirinae; CrPV, crocodile poxvirus; Ent, Entomopoxvirinae; Iri, Iridoviridae; Mar, Marseilleviridae; MCV, molluscum contagiosum virus; Mimi, Mimiviridae; Pan, Pandoravirus; Phy, Phycodnaviridae; Pith, Pithovirus; Pox, poxviruses.
Protein lengths are in numbers of amino acids.
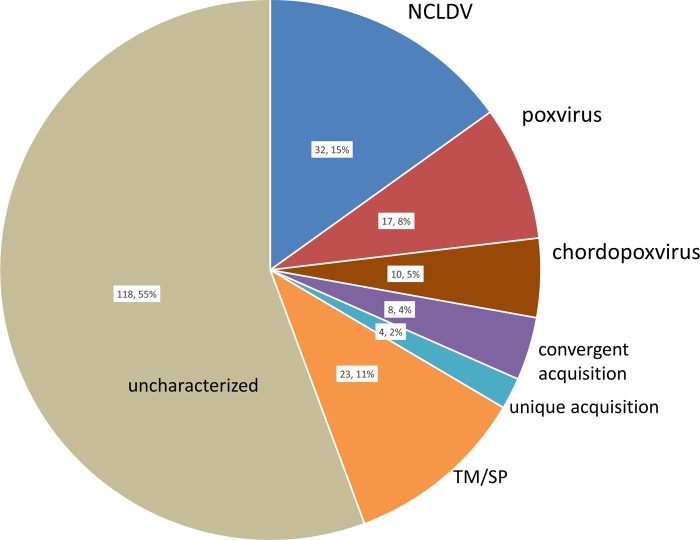
FIG 2.
Distribution of SGPV genes by tiers of inferred origin. The number of genes in each tier and the percentage of the total are indicated. NCLDV, genes inferred to have been present in the common ancestor of all NCLDVs; poxvirus, genes that originated in the common ancestor of the poxviruses; chordopoxvirus, genes that originated in the common ancestor of chordopoxviruses; TM/SP, transmembrane helix/signal peptide.
Among the predicted gene products of SGPV, homologs in other chordopoxviruses were detected for 59 proteins (Table 4 and Fig. 2). Among these conserved chordopoxvirus proteins, 32 belong to the previously inferred ancestral NCLDV gene set (24, 35), 17 are represented in all poxviruses (including entomopoxviruses), and 10 are specific for chordopoxviruses (Table 4 and Fig. 2). Eight genes have homologs in other NCLDVs but most likely were acquired independently (convergently), as suggested by sequence similarity and phylogenetic analysis, and only 4 genes appear to represent unique genes (with respect to the NCLDVs) captured from cellular organisms (Table 4 and Fig. 2; see Discussion).
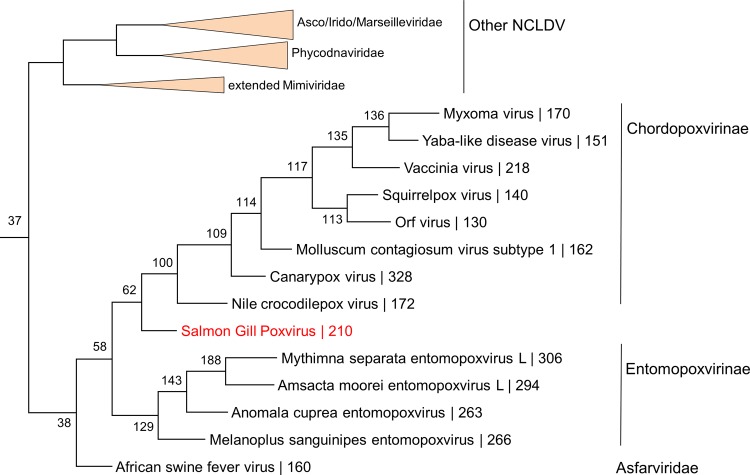
The conserved gene set includes most of the essential genes involved in virus DNA replication and expression as well as the morphogenesis and structure of the virion core and the capsid (see the discussion of some notable exceptions in “Shared and distinct gene functions between SGPV and other chordopoxviruses and unexpected evolutionary patterns among SGPV genes” below). All conserved genes of SGPV showed the highest sequence similarity to the orthologs from chordopoxviruses, with only 3 exceptions, where the highest similarity (albeit by a small margin) was observed with entomopoxvirus orthologs (Table 4). These observations imply that the conserved SGPV genes share an evolutionary history, at least within the poxviruses. Accordingly, we used concatenated multiple-sequence alignments of the sequences of 13 highly conserved genes from this ancestral gene set to construct a maximum likelihood (ML) phylogenetic tree in which the root was placed between ASFV and the poxviruses, given that ASFV and poxviruses are sister groups in the overall NCLDV phylogeny (25). In the resulting tree, SGPV was placed at the root of the chordopoxvirus branch with unequivocal bootstrap support (Fig. 3). Thus, the phylogeny of chordopoxviruses generally follows the phylogeny of their hosts.
FIG 3.
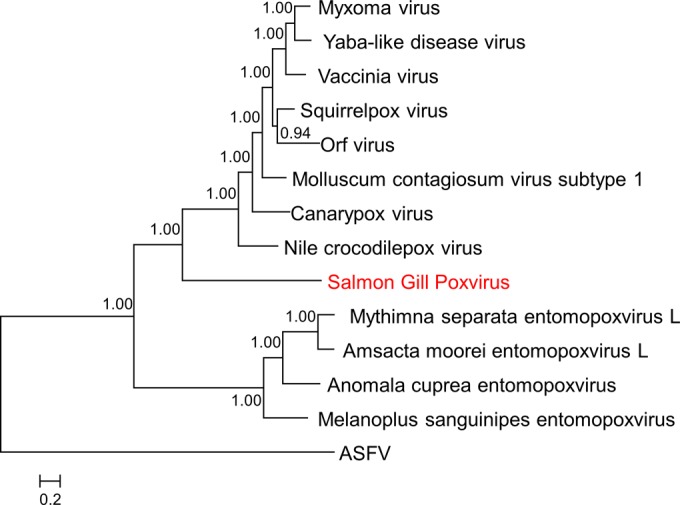
Phylogenetic tree of poxviruses. The tree was constructed from a multiple-sequence alignment of 13 proteins that are conserved in all poxviruses and ASFV (NCVOG0022, major capsid protein; NCVOG0023, D5-like helicase-primase; NCVOG0031, unclassified DEAD/SNF2-like helicases; NCVOG0038, DNA polymerase elongation subunit family B; NCVOG0076, DNA or RNA helicases of superfamily II; NCVOG0249, packaging ATPase; NCVOG0261, poxvirus early transcription factor [VETF], large subunit; NCVOG0262, poxvirus late transcription factor VLTF-3-like; NCVOG0267, RNA helicase DExH-NPH-II; NCVOG0271, DNA-directed RNA polymerase subunit beta; NCVOG0274, DNA-directed RNA polymerase subunit alpha; NCVOG1117, mRNA capping enzyme; NCVOG1164, A1L transcription factor VLTF-2). The root position was forced between the two families. Numbers at internal nodes indicate bootstrap support (on a scale of from 0 to 1). Figure S1 in the supplemental material contains the alignments used to generate the tree.
In order to gain further insight into the evolution of the gene complement of SGPV, we performed an ML reconstruction of the ancestral gene sets using the poxvirus phylogenetic tree (Fig. 3) as a guide. The inferred ancestral gene sets showed an unexpected pattern (Fig. 4): 58 genes were mapped to the common ancestor of all poxviruses, and 62 genes were mapped to the common ancestor of chordopoxviruses. Thus, taken in their entirety, chordopoxviruses possess almost the same conserved gene set as the entire family Poxviridae, with very few additional conserved genes appearing after the divergence from the common ancestor with entomopoxviruses. In contrast, 38 additional genes were mapped to the common ancestor of the chordopoxviruses infecting tetrapods; i.e., these genes were gained along the tree branch between SGPV and crocodile poxvirus (CrPV). Thus, the reconstruction reveals a dramatic difference in the conserved gene repertoires between the common ancestor of all chordopoxviruses and the tetrapod poxvirus ancestor (Fig. 4). This difference likely reflects a major biological transition, the possible nature of which is discussed in “Shared and distinct gene functions between SGPV and other chordopoxviruses and unexpected evolutionary patterns among SGPV genes” below.
FIG 4.
Reconstruction of the evolution of the gene repertoire of the NCLDVs. The numbers at internal branches (shown only for the ASFV-Poxviridae branch and for the root) indicate the maximum likelihood estimates of the number of genes mapped to the respective ancestral form. The numbers after the virus names indicate the number of annotated genes. The NCLDV families used as outgroups are shown by triangles. The NCLDV tree topology is from reference 25.
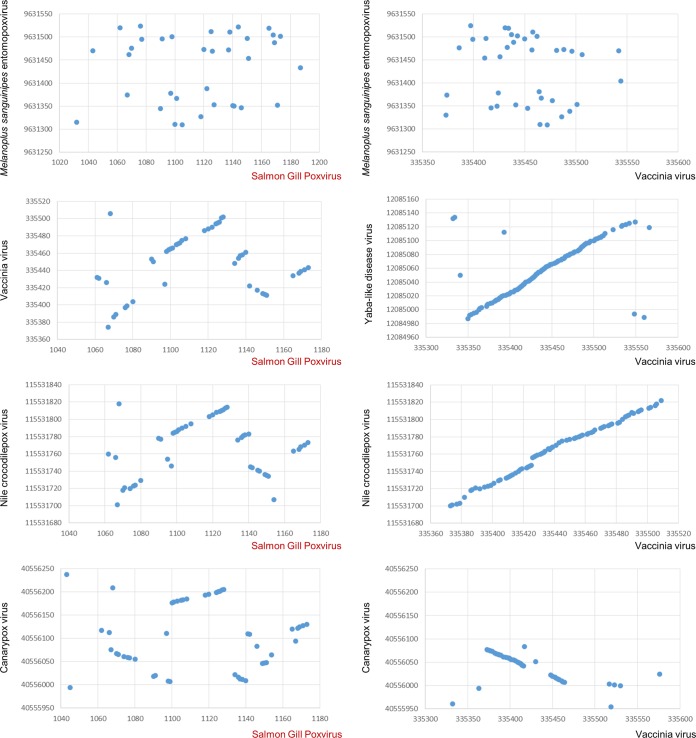
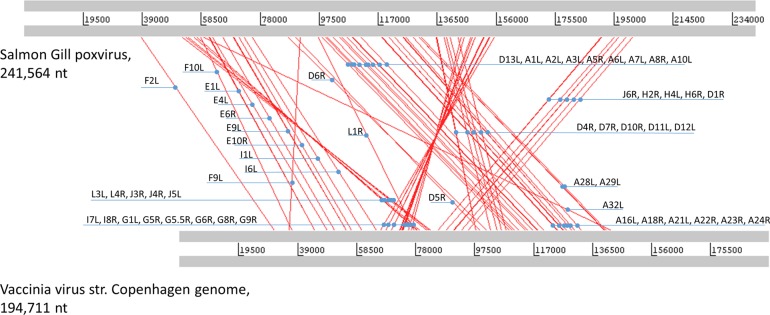
The tetrapod chordopoxviruses, except for avipoxviruses, are characterized by a distinct genome architecture whereby the central portion of the genome shows a nearly perfect conservation of gene synteny and the terminal regions are highly divergent and often contain unique genes (36, 37), as depicted in the dot plots of Fig. 5. In contrast, genome-wide comparison of the gene orders between SGPV and other chordopoxviruses shows the extensive decay of synteny in SGPV and the complete disappearance of synteny between chordopoxviruses and entomopoxviruses (Fig. 5). Examination of the genomic dot plots (Fig. 5) and a genome architecture alignment (Fig. 6) between SGPV and other chordopoxviruses reveals several conserved gene blocks in the central part of the genome that are separated by strings of nonhomologous genes of variable length, along with at least two inversions of conserved genomic segments. To assess the evolution of the poxvirus genome architecture in more quantitative terms, we calculated the matrix of genome rearrangement distances and used it to construct an evolutionary tree of genome architectures (Fig. 7). This tree shows that the decay of synteny roughly follows the evolution of gene sequences (compare the trees in Fig. 7 and 3), but the rate of disruption of the ancestral gene order is nonuniform, with the major change mapping to the branch between SGPV and the rest of the chordopoxviruses.
FIG 5.
Dot plot comparison of poxvirus gene orders. Each dot corresponds to a pair of orthologous genes. The horizontal axis shows the SGPV genes, and the vertical axis shows the GenInfo Identifier sequence identification numbers for genes of the respective viruses.
FIG 6.
Alignment of the genome architectures of SGPV and VACV. The alignment was generated using the Artemis tool and the table of gene orthology derived from the NCVOG assignments obtained in this work. The orthologous genes are connected by red lines, and the names of the respective vaccinia virus genes are indicated. nt, nucleotides.
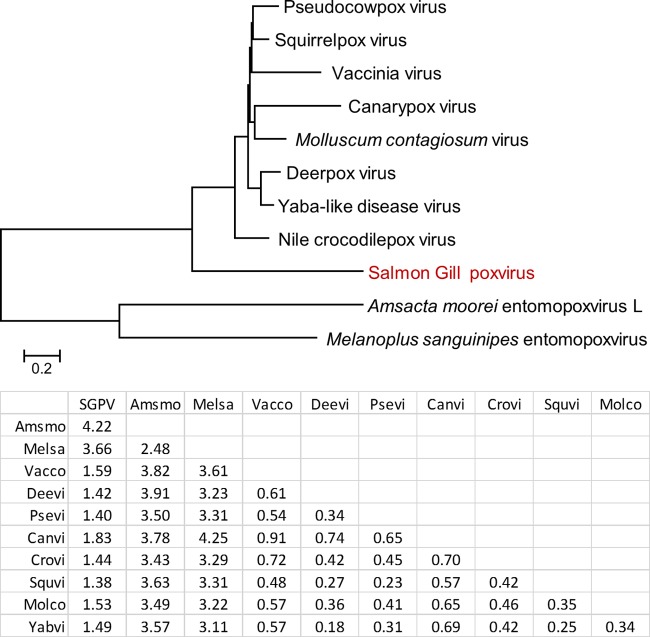
FIG 7.
Synteny-based evolutionary tree of poxviruses. The root between chordopoxviruses and entomopoxviruses was forced. The tree was constructed using the neighbor-joining method, and the distances between the genome architectures of the respective viruses that were estimated as described previously (33) are shown in the table underneath the tree; a unit distance means that the fraction of orthologous gene pairs that belong to synteny blocks is equal to e−1. Amsmo, Amsacta moorei entomopoxvirus; Melsa, Melanoplus sanguinipes entomopoxvirus; Vacco, vaccinia virus; Deevi, deerpox virus; Psevi, pseudocowpox virus; Canvi, canarypox virus; Crovi, crocodilepox virus; Squvi, squirrelpox virus; Molco, molluscum contagiosum virus; Yabvi, Yaba-like disease virus.
Shared and distinct gene functions between SGPV and other chordopoxviruses and unexpected evolutionary patterns among SGPV genes.
Here we discuss the predicted functions and some unusual aspects of evolution of the SGPV genes in the order of the tiers of ancestry, i.e., the point of gene origin for (acquisition by) this virus (Fig. 2). The ancestral NCLDV genes retained by SGPV encode the principal functions required for genome replication and expression, with no genes having been lost since the common ancestor of all poxviruses. However, two genes merit special mention in the context of poxvirus evolution, namely, the genes for DNA topoisomerase II (Topo II; SGPV095) and NAD-dependent DNA ligase (SGPV123). These (putative) ancestral NCLDV genes are uncharacteristic of chordopoxviruses, being present only in CrPV and SGPV, whose genomes encode both Topo II (which has multiple paralogs in CrPV) and Topo IB, which is conserved in the rest of the chordopoxviruses. The evolution of topoisomerases in NCLDVs appears to have been quite complex, involving both differential gene loss and the apparent independent acquisition of homologous genes (38). Thus, SGPV and CrPV could represent either the ancestral state with two distinct topoisomerase genes or an intermediate state after the Topo IB gene had been acquired but the Topo II gene had not been lost as it was in chordopoxviruses and entomopoxviruses independently.
The NAD-dependent ligase also appears to be an ancestral NCLDV gene but was replaced by the distinct, ATP-dependent ligase in several groups of viruses, including most of the chordopoxviruses, after the divergence from the common ancestor with CrPV (39). The finding that SGPV encodes an NAD-dependent ligase but not an ATP-dependent ligase is compatible with this scenario. The predicted NAD-dependent ligase of SGPV shows an unexpectedly low sequence similarity to homologs from other NCLDVs (Table 4), suggestive of some peculiarity in the DNA replication process of this virus.
The next evolutionary tier of the SGPV genes, those that are conserved in all poxviruses (Table 4) (1, 24), includes components of the transcription apparatus, such as several RNA polymerase subunits and the poly(A) polymerase catalytic subunit; several components of the virion core and proteins involved in virion morphogenesis, such as the metalloprotease G1; and six subunits of the fusion-entry complex (40, 41) (homologs of three paralogous subunits of this complex, A16, G9, and J5, are also detectable in mimiviruses and iridoviruses, suggesting that some form of this complex might be ancestral in NCLDVs). Of note is the presence in SGPV of a highly diverged ortholog of the G6 protein, a predicted amidase or acyltransferase that is thought to be important for the virus-host interaction but whose specific function remains obscure (42).
The genes that are conserved in chordopoxviruses, to the exclusion of entomopoxviruses, follow the same general functional themes, including RNA polymerase subunits, such as A5 and J4; the late-stage transcription factor G8 containing a highly diverged PCNA domain; and proteins involved in core morphogenesis, e.g., the telomere-binding protein I6 and the protein A6 required for virus membrane biogenesis. Particularly notable in this group are three paralogous genes (SGPV154, SGPV159, SGPV162), located near the right end of the genome, that encode homologs of variola virus B22R, a giant type I membrane protein implicated in the virus-induced shutdown of the host adaptive immunity, specifically, inhibition of T lymphocytes (43). Of these three paralogous genes, SGPV154 is similar in length to homologs from other poxviruses, whereas SGPV159 and SGPV162 are considerably shorter and, thus, have apparently been truncated during the evolution of the SGPV lineage. However, the conservation of the predicted signal peptide and the C-terminal transmembrane helix in all three proteins (Table 4) suggests that they remain functional. The proliferation of this gene in SGPV, which parallels its independent triplication in CrPV (44), implies an important role of this route of counterdefense. Interestingly, homologs of this gene were also detected in cyprinid herpesviruses, suggesting transfer of this gene, involved in virus-host interaction from SGPV (or its relative), to unrelated viruses within the same host (Fig. 8). Moreover, the cluster of SGPV genes that encompasses the three B22R paralogs also contains two other proteins with a similar, very large size (SGPV155 and SGPV161; Table 4) that are predicted to be, respectively, membrane associated and secreted. The sequences of these proteins show no similarity to the B22R sequence, but the proteins might perform roles similar to the role performed by B22R via a distinct mechanism.
FIG 8.
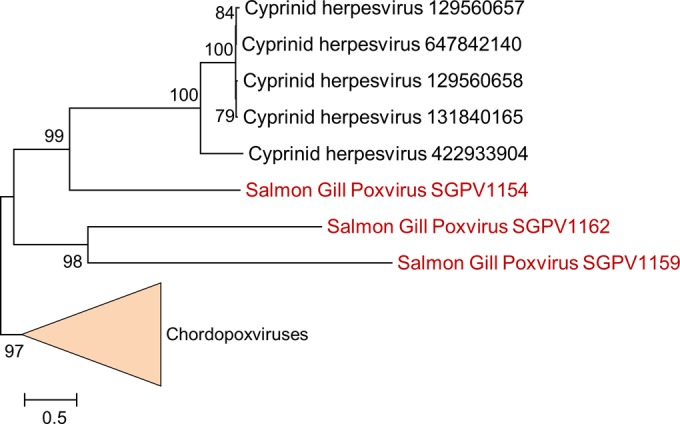
Phylogenetic tree of the viral B22R-like genes. The numbers on the left show bootstrap values as percentages. The bar shows the scale as the estimated number of amino acid substitutions per site. For the cyprinid herpesviruses, the GI numbers are indicated on the right. The three paralogs from SGPV are shown in red. The chordopoxvirus sequences are collapsed and shown as a triangle. Figure S2 in the supplemental material contains the alignments used for construction of the tree.
The late-stage genes of chordopoxviruses, as well as most intermediate and some early genes, contain a distinct sequence element within which transcription starts. This element has the sequence TAAAT, where the second T usually corresponds to the second nucleotide of the translation initiation ATG codon of the respective gene (45–47). In the process of transcription initiation, the complement of this element serves as the template for the formation of the 5′-terminal poly(A) sequence that is present in many chordopoxvirus transcripts and is produced by RNA polymerase slippage (48, 49). This TAAAT element is conserved in nearly all SGPV homologs of the respective chordopoxvirus genes (Table 4), suggesting that the main features of transcription initiation are shared by all chordopoxviruses.
Eight genes of SGPV have homologs in other NCLDVs and, thus, are formally assigned to NCVOGs, but as indicated by sequence similarity searches and phylogenetic analysis results (Table 4), they have probably been independently acquired by different viruses, which implies that they play important roles in virus-host interactions. Two of these genes (SGPV034 and SGPV139) encode RING finger proteins that could function as either specialized E3 subunits of ubiquitin ligases or inhibitors of ubiquitin pathways. RING finger-containing E3 proteins are encoded by many NCLDVs, including some of the orthopoxviruses, in which they are essential for pathogenicity (50–52). However, viral RING finger domains, including those encoded by the SGPV genome, show limited sequence similarity to each other and have probably been acquired independently. This independent acquisition was likely driven by the selection for virus interaction with the host ubiquitin networks. A similar trend of likely independent acquisition by diverse viruses is apparent for the DnaJ (J) domain, which was detected in the SGPV102 protein sequence. The J domain is present in mimiviruses, some phycodnaviruses, a single chordopoxvirus (molluscum contagiosum virus [36]), as well as polyomaviruses. The polyomavirus J domain has been shown to function as a cochaperonin, enhancing the activity of the Hsc70 chaperone in the infected cells (53). A similar role in viral protein folding could be played by SGPV102.
Of special interest is the SGPV043 protein, a predicted serine/threonine protein kinase that is highly similar to the eukaryotic ribosomal protein S6 kinase (S6K), with which it shows up to 60% amino acid sequence identity. S6K is a component of the mTOR pathway and, more specifically, of the TORC1 complex, an environmental sensor that promotes anabolic pathways and inhibits catabolic pathways (54). Thus, this gene, which seems to have been convergently captured by SGPV and several other NCLDVs, could act as a regulator of the global metabolic state of virus-infected cells.
Only four SGPV genes appear to be unique acquisitions from cellular organisms, as they have no homologs in other NCLDVs. These are the metalloendopeptidase SGPV051, the thioredoxin SGPV147, the predicted DNA or RNA methyltransferase SGPV186, and the macrodomain-containing protein SGPV187, a putative O-acetyl-ADP-ribose deacetylase. Each of these proteins showed a high level of divergence from cellular homologs, presumably due to the high rate of evolution upon transfer to the viral genome, precluding a convincing inference of origin by phylogenetic analysis (not shown). The presence of the macrodomain is of special interest. Previously, this domain has been detected in several groups of animal positive-strand RNA viruses (55) and has been shown to inhibit double-strand RNA-dependent phosphorylation of the interferon regulatory factor 3 (IRF-3), a key transcription factor for interferon induction (56). The macrodomain of SGPV, to our knowledge, is the first domain of this family to be discovered in a DNA virus, and it might play a similar role as an inhibitor of the interferon pathway.
Conserved poxvirus genes that are missing in SGPV: distinct pathways of membrane biogenesis?
As pointed out above, SGPV lacks numerous genes that are (nearly) fully conserved among the tetrapod-infecting chordopoxviruses, with the implication being that they are lost in SGPV (Table 5). Only three ancestral poxvirus genes appear to have been lost in SGPV; one of these, the gene for the protein phosphatase H1, was, apparently, also independently lost in some entomopoxviruses. The absence in SGPV of the A11, L2, A14, and A17 genes highlights a central functional theme that extends into the longer list of conserved chordopoxvirus genes that are missing in SGPV, namely, membrane biogenesis (57) (Table 5). At least half of the missing genes (14 of 28) encode proteins implicated in this process. Of the seven subunits of a distinct protein complex involved in the association of the viroplasm with membranes, which is required for immature virion formation (58), only one, the protein kinase F10, an ancestral NCLDV protein that is likely to perform multiple functions (59), is represented by an ortholog in SGPV (Table 5; two complex subunits, D2 and A15, are not listed because they appear to have been lost in some other chordopoxviruses as well). Taken together, these findings imply that SGPV employs a pathway of membrane biosynthesis that is distinct from that of other chordopoxviruses. Several uncharacterized SGPV proteins contain predicted transmembrane segments (Table 4) but show no detectable sequence similarity to the sequences of proteins of other poxviruses shown to participate in membrane biogenesis; it appears likely that at least some of these SGPV proteins belong to the putative alternative pathway.
TABLE 5.
Conserved chordopoxvirus genes missing in SGPV
| Conserved genea | VACV gene | Known or predicted function | Essentialb | Comment |
|---|---|---|---|---|
| Genes conserved in chordopoxviruses and entomopoviruses | ||||
| 1178 | L5R | Membrane protein, fusion-entry complex subunit | Yes | |
| 1181 | A11R | Membrane-associated protein implicated in endoplasmic reticulum recruitment for virion morphogenesis | Yes | |
| 0040 | H1L | Dual-specificity (Ser/Thr and Tyr) protein phosphatase | Yes | Conserved in only two entomopoxviruses |
| Genes conserved only in chordopoxviruses | ||||
| 1185 | A20R | DNA polymerase processivity factor | Yes | |
| 1385 | I3L | Single-stranded DNA-binding protein essential for replication | Yes | |
| 1184 | G2R | Late transcription elongation factor | Yes | |
| 1172 | A12L | Virion core protein | Yes | |
| 1177 | F17R | DNA-binding virion core protein | Yes | |
| 1043 | G7L | Virion core protein required for immature virion formation | Yes | |
| 1398 | A19L | Virion core protein | Yes | |
| 0060 | G4L | Glutaredoxin involved in the pathway for cytoplasmic disulfide bond formation | Yes | |
| 1396 | A2.5L | Thioredoxin-like protein involved in the pathway for cytoplasmic disulfide bond formation | Yes | |
| 0012 | A33R | C-type lectin involved in extracellular virion morphogenesis | No | |
| 0268 | A25/A26L | A-type inclusion body-like protein | No | |
| 0255 | O1L | Poorly characterized protein, activator of the extracellular signal-regulated kinase pathway | No | |
| 1167 | F12L | Protein involved in intracellular enveloped virion maturation and cytoskeleton-dependent virion export | No | Inactivated derivative of DNA polymerase, possibly of bacteriophage origin |
| 1367 | G3L | Fusion-entry complex subunit | Yes | |
| 1376 | H7R | Protein involved in MVc membrane biogenesis | Yes | |
| 1380 | A14L | Protein involved in MV membrane biogenesis | Yes | |
| 1411 | A17L | Protein involved in MV membrane biogenesis | Yes | |
| 1395 | L2R | Protein involved in MV membrane biogenesis | Yes | |
| 1366 | I5L | MV membrane protein | No | |
| 1383 | I2L | Membrane protein essential for virus entry | Yes | |
| 1391 | J1R | Protein involved in MV formation, assembly complex subunit | Yes | |
| 1416 | D3R | Protein involved in MV formation, assembly complex subunit | Yes | |
| 1412 | A30L | Protein involved in MV formation, assembly complex subunit | Yes | |
| 1392 | A9L | Protein involved in MV morphogenesis | Yes | |
| 0256 | H3L | MV membrane protein involved in cell attachment | No | |
| 1415 | A14.5L | MV membrane protein that enhances virulence | No |
NCVOG number.
Essentiality was determined for vaccinia virus.
MV, mature virion.
Among the other conspicuous gaps in the gene repertoire of SGPV are the single-stranded DNA-binding protein I3 (60, 61) and the DNA polymerase processivity factor A20 (62), two proteins that are essential for VACV DNA replication. Among the predicted SGPV gene products, there are no obvious candidates that could replace these proteins, so the involvement of functionally analogous host proteins seems to be a distinct possibility.
Also missing in SGPV are two components of the thiol-disulfide oxidoreductase pathway, which is essential for the formation of the disulfide bonds in the subunits of the VACV fusion-entry complex, as well as envelope proteins L1 and F9 (63–65). Orthologs of the L1 protein along with the upstream component of the thiol-disulfide oxidoreductase pathway, the E10 protein, are conserved in nearly all NCLDVs (24), with the implication being that the pathway as such is essential. The two missing subunits, the glutaredoxin G4 (66) and the thioredoxin-like protein A2.5 (67), have no orthologs in other NCLDV families either, indicating that the complete oxidoreductase pathway characterized in VACV evolved only after the divergence of SGPV and the rest of the chordopoxviruses. The predicted SGPV thioredoxin (SGPV147) might be responsible, at least in part, for the missing portion of the pathway.
DISCUSSION
Poxvirus infection in salmon was suspected in the 1990s, as TEM showed apoptotic gill epithelial cells with poxvirus-like particles in samples submitted to the Norwegian Veterinary Institute from acute, high-mortality events in freshwater farms with juvenile fish (Dale and Kvellestad, unpublished). Typical poxvirus structures were further characterized in a TEM study of gill disease in Atlantic salmon (4), but no taxonomic assignment was possible in the absence of sequence data. In this study, we confirmed the presence of poxvirus particles and determined the sequence and phylogeny of salmon gill poxvirus, developed qPCR and IHC methods, and analyzed the disease from current as well as archival samples, including the outbreak previously studied by TEM (4).
As there was no experimental model for SGPV disease, we obtained samples from fish with spontaneous cases of the suspect apoptotic gill disease in two hatcheries without other significant disease problems. As controls we included samples from fish involved in several gill disease outbreaks without apoptosis of gill epithelium as well as healthy fish. We found the SGPV infection by qPCR only in the disease cases and could link the SGPV infection in situ to the apoptotic respiratory epithelium by IHC. We found that the infection was widespread in the gills at least 3 days before the onset of severe clinical disease. Mortality coincided with blocking of the respiratory gill surfaces by two different mechanisms. The SGPV infection seems to induce massive apoptosis and detachment of the epithelium, resulting in the acute adherence of the thin gill lamellae. In other fish, an excessive proliferation of the epithelium blocked the respiratory surfaces. These findings indicate that viral replication precedes the gill pathologies that can be likened to atelectasis and solidification of the lungs, respectively. Hypoxia and osmoregulatory disturbances are the expected pathophysiological consequences from such lesions in fish. This is in keeping with the clinical experience that stopping feeding, raising oxygen levels, and avoiding all stress minimize mortality, which otherwise may approach 100% within hours in a tank of fish. Although the classical Koch's postulates remain to be fulfilled, our findings indicate that SGPV causes a distinct disease primarily affecting the gills in salmon. In fish, localized gill infection seems to be the rule for the suspected poxviruses (2–4, 9).
With regard to systemic pathology and infection, we found that hemophagocytosis was associated with severe disease. In infectious salmon anemia, a severe orthomyxoviral disease of salmon, hemophagocytosis is due to virus attachment to the erythrocyte surfaces (68). However, only high or no CT values were found in organs of the poxvirus-infected salmon with hemophagocytosis, while low CT values were obtained from the gills in these fish. Hemophagocytosis is possibly a sign of circulatory disturbances aggravating the SGPV disease. It is noteworthy that lethargy, gill pathology, and hemophagocytosis are also reported in koi sleepy disease, associated with a pox virus of gills and seemingly not internal organs (9). Poxviruses are generally epitheliotropic, and skin tissue samples were positive by PCR both in this study and in the study of koi carp (9). However, we found no skin lesions in the salmon, and at present, we cannot exclude the possibility that skin tissue samples carry just virus shed from the gills. IHC seemed to indicate a very narrow cell tropism, since the simple, squamous lamellar epithelium of the gill was infected, while the adjacent stratified epithelium did not show signs of infection. The high mortality due to respiratory SGPV infection appears to be different from that in poxvirus infections in air-breathing vertebrates, where lung pathology is usually seen as part of a generalized infection. This could be related to the fundamental anatomical differences in the respiratory systems in the two vertebrate groups, where fish have their respiratory surface much more exposed to the exterior.
All salmon sampled a week after mortality had subsided in a tank were still infected, as shown by PCR. However, virus levels were generally lower and hemophagocytosis was much less prominent. These findings suggest that although recurrent, acute outbreaks in a tank are not reported by the farmers, SGPV infection may persist. For how long we do not know, and as the reservoir of infection is also unknown, we cannot rule out the possibility of reinfections. However, our archival samples do show that the infection is found not only in freshwater hatcheries but also in the seawater farms that receive the salmon for additional growth. In the cases of combined amoebic gill disease and SGPV disease that we confirmed here, 82% of the fish died (11). This is in agreement with the findings of other studies demonstrating that most gill diseases in the seawater rearing phase are complex, with multiple agents being present (69). Investigation of the role of SGPV in mixed infections will be an important future task, as the gill problems caused by these mixed infections cause considerable losses. To this end, we now have two new diagnostic methods, qPCR and IHC, for the detection of the SGPV in fish tissues, and these are useful for screening and resolving the complex pathology, respectively.
Judging from the archival samples, the SGPV disease emerged in the mid-1990s in Norwegian salmon farms over a wide geographical range. However, SGPV is distinct from other chordopoxviruses that have been analyzed, and its reservoir is unknown. High mortality, like that caused by SGPV in salmon, can be a sign of a new host-agent system with low compatibility; on the other hand, intensive farming may have changed an old host-agent balance. Further studies are needed to clarify the reservoir and host range of SGPV and other aquatic poxviruses beyond salmon farming. The level of production of farmed food fish grew from 13 million to 66 million tons during the period between 1990 and 2012 (70). Epidemics of orthomyxoviral disease virtually stopped salmon production in Chile (71) and demonstrate the importance of disease control in fish farming. Also, for feral fish populations, introduction of new viruses may have serious consequences, as shown by the mass mortalities associated with rhabdoviral disease in the Great Lakes (72). We urgently need more knowledge of fish poxviruses, as the global trade and movement of aquaculture animals are growing. Research on the poxviruses of fishes may also bring cures for the aquaculture industry in the form of vaccines and the development of vectors.
Genome analysis of SGPV established the position of this fish-infecting virus at the base of the chordopoxvirus tree, as could be expected under the assumption of virus-host coevolution. However, the differences between the gene complements of SGPV and those of the rest of the chordopoxviruses are extensive, with SGPV lacking 38 genes otherwise conserved in chordopoxviruses. This difference in gene content is spread unevenly across the functional classes of viral genes. Most of the genes involved in genome replication and expression, as well as core and capsid structure and morphogenesis, are shared by SGPV and other chordopoxviruses. In sharp contrast, the majority of the chordopoxviruses genes implicated in viral membrane biogenesis are missing in SGPV. Chordopoxviruses employ a unique pathway of viral membrane derivation from the membranes of the endoplasmic reticulum of the infected cells that requires the participation of multiple viral proteins (57). The most parsimonious explanation for the absence of most of these proteins in SGPV is that this pathway, at least in its complete form, evolved in viruses infecting tetrapods; however, the alternative scenario, in which the pathway evolved in the ancestral chordopoxvirus but was subsequently lost in SGPV, cannot be ruled out. In addition, the multiprotein complex that in vaccinia virus is involved in viroplasm association with membranes, that is essential for virion maturation, and that so far appears to be conserved in all chordopoxviruses (58) is missing in SGPV. Nevertheless, recognizable poxvirus crescent membranes, immature virions, and mature virions are formed. Thus, SGPV appears to employ a pathway of viral membrane biogenesis similar to that of other chordopoxviruses, despite the absence of key conserved proteins; multiple predicted membrane proteins of SGPV without homologs in other viruses could contribute to this alternative version of membrane biogenesis.
SGPV also lacks the proteins that are involved in the interaction of other chordopoxviruses with host defense systems, such as multiple paralogous genes that encode proteins containing kelch and ankyrin repeats as well as proteins involved in the suppression of host immune mechanisms. The conspicuous exception is the conserved B22R-like giant membrane proteins. However, the SGPV genome encodes numerous uncharacterized genes, many of which encode predicted membrane and secreted proteins as well as predicted nonglobular proteins with low-complexity sequences and simple repeats. Many, if not most, of the protein products of these genes are likely involved in currently unknown interactions with the immunity systems of the fish host. Experimental study of these uncharacterized proteins of SGPV could help the study of the pathogenesis of the gill disease in salmon and, more generally, the mechanisms of the poxvirus-host interaction.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Research Council of Norway grant 234037 and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank the many field veterinarians and fish health biologists that contributed samples to the Norwegian Veterinary Institute. Agnar Kvellestad is acknowledged for unpublished electron microscopy of diagnostic SGPV cases in the 1990s.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01174-15.
REFERENCES
- 1.Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol 77:7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono S, Nagai A, Sugai N. 1986. A histopathological study on juvenile colorcarp, Cyprinus carpio, showing edema. Fish Pathol 9:9. [Google Scholar]
- 3.Wada S, Kurata O, Hatai K, Ishii H, Kasuya K, Watanabe Y. 2008. Proliferative branchitis associated with pathognomonic, atypical gill epithelial cells in cultured ayu Plecoglossus altivelis. Fish Pathol 43:89–91. doi: 10.3147/jsfp.43.89. [DOI] [Google Scholar]
- 4.Nylund A, Watanabe K, Nylund S, Karlsen M, Saether PA, Arnesen CE, Karlsbakk E. 2008. Morphogenesis of salmonid gill poxvirus associated with proliferative gill disease in farmed Atlantic salmon (Salmo salar) in Norway. Arch Virol 153:1299–1309. doi: 10.1007/s00705-008-0117-7. [DOI] [PubMed] [Google Scholar]
- 5.Oyamatsu T, Matoyama H, Yamamoto K-Y, Fukuda H. 1997. A trial for the detection of carp edema virus by using polymerase chain reaction. Aquaculture Sci 45:247–251. [Google Scholar]
- 6.Haenen O, Way K, Stone D, Engelsma M. 2014. ‘Koi sleepy disease’ found for the first time in koi carps in the Netherlands. Tijdschr Diergeneeskd 139:26–29. (In Dutch.) [PubMed] [Google Scholar]
- 7.Lewisch E, Gorgoglione B, Way K, El-Matbouli M. 2015. Carp edema virus/koi sleepy disease: an emerging disease in Central-East Europe. Transbound Emerg Dis 62:6–12. doi: 10.1111/tbed.12293. [DOI] [PubMed] [Google Scholar]
- 8.Way K, Stone D. 2013. Emergence of carp edema virus-like (CEV-like) disease in the UK. Finfish News 15:32–34. [Google Scholar]
- 9.Miyazaki T, Isshiki T, Katsuyuki H. 2005. Histopathological and electron microscopy studies on sleepy disease of koi Cyprinus carpio koi in Japan. Dis Aquat Organ 65:197–207. doi: 10.3354/dao065197. [DOI] [PubMed] [Google Scholar]
- 10.Seno R, Hata N, Fukuda H. 2003. Curative effect of 0.5% salt water treatment on carp, Cyprinius carpio, infected with carp edema virus (CEV) results mainly from reviving the physiological condition of the host. Suisanzoshoku 51:123–124. [Google Scholar]
- 11.Steinum T, Kvellestad A, Ronneberg LB, Nilsen H, Asheim A, Fjell K, Nygard SM, Olsen AB, Dale OB. 2008. First cases of amoebic gill disease (AGD) in Norwegian seawater farmed Atlantic salmon, Salmo salar L, and phylogeny of the causative amoeba using 18S cDNA sequences. J Fish Dis 31:205–214. doi: 10.1111/j.1365-2761.2007.00893.x. [DOI] [PubMed] [Google Scholar]
- 12.Gjessing MC, Falk K, Weli SC, Koppang EO, Kvellestad A. 2012. A sequential study of incomplete Freund's adjuvant-induced peritonitis in Atlantic cod. Fish Shellfish Immunol 32:141–150. doi: 10.1016/j.fsi.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Seidelin M, Madsen SS, Blenstrup H, Tipsmark CK. 2000. Time-course changes in the expression of Na+, K+-ATPase in gills and pyloric caeca of brown trout (Salmo trutta) during acclimation to seawater. Physiol Biochem Zool 73:446–453. doi: 10.1086/317737. [DOI] [PubMed] [Google Scholar]
- 14.Haugarvoll E, Bjerkas I, Nowak BF, Hordvik I, Koppang EO. 2008. Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J Anat 213:202–209. doi: 10.1111/j.1469-7580.2008.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborne RJ, Symonds TM, Sriskantha A, Lai-Fook J, Fernon CA, Dall DJ. 1996. An entomopoxvirus homologue of the vaccinia virus D13L-encoded ‘rifampicin resistance’ protein. J Gen Virol 77:839–846. doi: 10.1099/0022-1317-77-5-839. [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Systems 8:581–599. doi: 10.1142/S0129065797000537. [DOI] [PubMed] [Google Scholar]
- 22.Benson G. 1999. Tandem Repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yutin N, Wolf YI, Koonin EV. 2014. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol 25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jobb G, von Haeseler A, Strimmer K. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Csuros M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 32.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 33.Novichkov PS, Wolf YI, Dubchak I, Koonin EV. 2009. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol 191:65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felsenstein J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266:418–427. doi: 10.1016/S0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 35.Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senkevich TG, Koonin EV, Bugert JJ, Darai G, Moss B. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 37.Moss B. 2001. Poxviridae: the viruses and their replication, p 2849–2884. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 38.Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yutin N, Koonin EV. 2009. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: a case of hidden complexity. Biol Direct 4:51. doi: 10.1186/1745-6150-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senkevich TG, Ojeda S, Townsley A, Nelson GE, Moss B. 2005. Poxvirus multiprotein entry-fusion complex. Proc Natl Acad Sci U S A 102:18572–18577. doi: 10.1073/pnas.0509239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moss B. 2012. Poxvirus cell entry: how many proteins does it take? Viruses 4:688–707. doi: 10.3390/v4050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senkevich TG, Wyatt LS, Weisberg AS, Koonin EV, Moss B. 2008. A conserved poxvirus NlpC/P60 superfamily protein contributes to vaccinia virus virulence in mice but not to replication in cell culture. Virology 374:506–514. doi: 10.1016/j.virol.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alzhanova D, Hammarlund E, Reed J, Meermeier E, Rawlings S, Ray CA, Edwards DM, Bimber B, Legasse A, Planer S, Sprague J, Axthelm MK, Pickup DJ, Lewinsohn DM, Gold MC, Wong SW, Sacha JB, Slifka MK, Fruh K. 2014. T cell inactivation by poxviral B22 family proteins increases viral virulence. PLoS Pathog 10:e1004123. doi: 10.1371/journal.ppat.1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonso CL, Tulman ER, Delhon G, Lu Z, Viljoen GJ, Wallace DB, Kutish GF, Rock DL. 2006. Genome of crocodilepox virus. J Virol 80:4978–4991. doi: 10.1128/JVI.80.10.4978-4991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosel JL, Earl PL, Weir JP, Moss B. 1986. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol 60:436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanggi M, Bannwarth W, Stunnenberg HG. 1986. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcription initiation. EMBO J 5:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Martens CA, Bruno DP, Porcella SF, Moss B. 2012. Pervasive initiation and 3′-end formation of poxvirus postreplicative RNAs. J Biol Chem 287:31050–31060. doi: 10.1074/jbc.M112.390054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwer B, Visca P, Vos JC, Stunnenberg HG. 1987. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell 50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ink BS, Pickup DJ. 1990. Vaccinia virus directs the synthesis of early mRNAs containing 5′ poly(A) sequences. Proc Natl Acad Sci U S A 87:1536–1540. doi: 10.1073/pnas.87.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senkevich TG, Koonin EV, Buller RM. 1994. A poxvirus protein with a RING zinc finger motif is of crucial importance for virulence. Virology 198:118–128. doi: 10.1006/viro.1994.1014. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Villa NY, McFadden G. 2009. Interplay between poxviruses and the cellular ubiquitin/ubiquitin-like pathways. FEBS Lett 583:607–614. doi: 10.1016/j.febslet.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 52.Shchelkunov SN. 2010. Interaction of orthopoxviruses with the cellular ubiquitin-ligase system. Virus Genes 41:309–318. doi: 10.1007/s11262-010-0519-y. [DOI] [PubMed] [Google Scholar]
- 53.Salma A, Tsiapos A, Lazaridis I. 2007. The viral SV40 T antigen cooperates with dj2 to enhance hsc70 chaperone function. FEBS J 274:5021–5027. doi: 10.1111/j.1742-4658.2007.06019.x. [DOI] [PubMed] [Google Scholar]
- 54.Magnuson B, Ekim B, Fingar DC. 2012. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 55.Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. 2014. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol 88:11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moss B. 2015. Poxvirus membrane biogenesis. Virology 479-480:619–626. doi: 10.1016/j.virol.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szajner P, Jaffe H, Weisberg AS, Moss B. 2004. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology 330:447–459. doi: 10.1016/j.virol.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 59.Punjabi A, Traktman P. 2005. Cell biological and functional characterization of the vaccinia virus F10 kinase: implications for the mechanism of virion morphogenesis. J Virol 79:2171–2190. doi: 10.1128/JVI.79.4.2171-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rochester SC, Traktman P. 1998. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J Virol 72:2917–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greseth MD, Boyle KA, Bluma MS, Unger B, Wiebe MS, Soares-Martins JA, Wickramasekera NT, Wahlberg J, Traktman P. 2012. Molecular genetic and biochemical characterization of the vaccinia virus I3 protein, the replicative single-stranded DNA binding protein. J Virol 86:6197–6209. doi: 10.1128/JVI.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boyle KA, Stanitsa ES, Greseth MD, Lindgren JK, Traktman P. 2011. Evaluation of the role of the vaccinia virus uracil DNA glycosylase and A20 proteins as intrinsic components of the DNA polymerase holoenzyme. J Biol Chem 286:24702–24713. doi: 10.1074/jbc.M111.222216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Senkevich TG, White CL, Koonin EV, Moss B. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc Natl Acad Sci U S A 99:6667–6672. doi: 10.1073/pnas.062163799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senkevich TG, Ward BM, Moss B. 2004. Vaccinia virus A28L gene encodes an essential protein component of the virion membrane with intramolecular disulfide bonds formed by the viral cytoplasmic redox pathway. J Virol 78:2348–2356. doi: 10.1128/JVI.78.5.2348-2356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Townsley AC, Senkevich TG, Moss B. 2005. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J Virol 79:10988–10998. doi: 10.1128/JVI.79.17.10988-10998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White CL, Senkevich TG, Moss B. 2002. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J Virol 76:467–472. doi: 10.1128/JVI.76.2.467-472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Senkevich TG, White CL, Weisberg A, Granek JA, Wolffe EJ, Koonin EV, Moss B. 2002. Expression of the vaccinia virus A2.5L redox protein is required for virion morphogenesis. Virology 300:296–303. doi: 10.1006/viro.2002.1608. [DOI] [PubMed] [Google Scholar]
- 68.Aamelfot M, Dale OB, Weli SC, Koppang EO, Falk K. 2012. Expression of the infectious salmon anemia virus receptor on Atlantic salmon endothelial cells correlates with the cell tropism of the virus. J Virol 86:10571–10578. doi: 10.1128/JVI.00047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kvellestad A. 2013. Gill inflammation in Norwegian seawater-farmed Atlantic salmon: a study of aetiology and manifestation. vol 154 Unipub, Oslo, Norway. [Google Scholar]
- 70.Food and Agriculture Organization of the United Nations Fisheries Department. 2014. The state of world fisheries and aquaculture. Food and Agriculture Organization of the United Nations, Rome, Italy. [Google Scholar]
- 71.Mardones FO, Martinez-Lopez B, Valdes-Donoso P, Carpenter TE, Perez AM. 2014. The role of fish movements and the spread of infectious salmon anemia virus (ISAV) in Chile, 2007-2009. Prev Vet Med 114:37–46. doi: 10.1016/j.prevetmed.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 72.Faisal M, Shavalier M, Kim RK, Millard EV, Gunn MR, Winters AD, Schulz CA, Eissa A, Thomas MV, Wolgamood M, Whelan GE, Winton J. 2012. Spread of the emerging viral hemorrhagic septicemia virus strain, genotype IVb, in Michigan, USA. Viruses 4:734–760. doi: 10.3390/v4050734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.