Abstract
The family Geminiviridae comprises seven genera differentiated by genome organization, sequence similarity, and insect vector. Capulavirus, an eighth genus, has been proposed to accommodate two newly discovered highly divergent geminiviruses that presently have no known vector. Alfalfa leaf curl virus, identified here as a third capulavirus, is shown to be transmitted by Aphis craccivora. This is the first report of an aphid-transmitted geminivirus.
TEXT
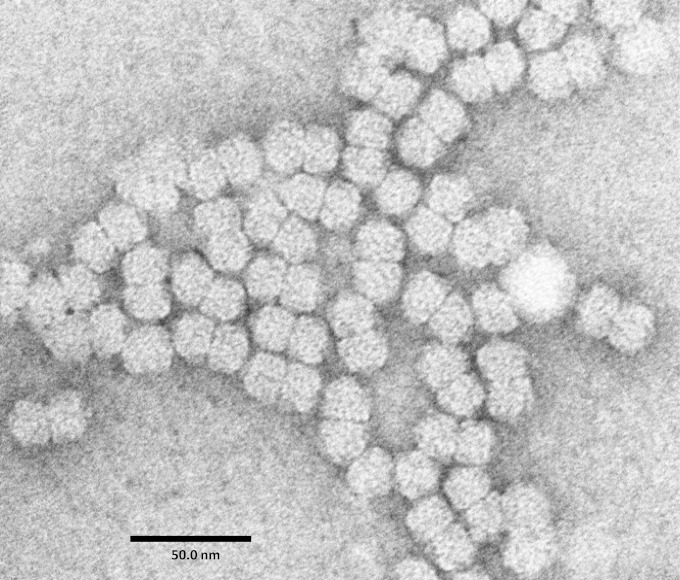
Plant-infecting viruses in the family Geminiviridae have one or two genome components of less than 3.0 kb composed of circular single-stranded DNA (ssDNA) that are encapsidated in typical geminate particles. The seven approved geminivirus genera are Begomovirus, Curtovirus, Topocuvirus, Mastrevirus, Becurtovirus, Turncurtovirus, and Eragrovirus (1). Recently, two highly divergent geminiviruses, euphorbia caput-medusae latent virus (EcmLV) and French bean severe leaf curl virus (FbSLCV), were discovered in South Africa and India, respectively (2). Based on genomic sequence divergence and genome organization, EcmLV and FbSLCV have been proposed to belong to a new genus named Capulavirus. The primary reason that the Capulavirus genus has not been formally accepted within the family Geminiviridae is the absence of any evidence that either EcmLV or FbSLCV forms geminate particles. Here, however, we confirm that EcmLV produces twinned quasi-icosahedral particles (Fig. 1) and that the capulaviruses are therefore bona fide geminiviruses.
FIG 1.
Virions of euphorbia caput-medusae latent virus purified from agroinfected Nicotiana benthamiana plants as described previously (10) with the following modifications. The liquid N2-powdered tissues were resuspended in the grinding buffer and homogenized in a blender for 1 min. The aqueous phase was recovered from the chloroform treatment by centrifugation, filtered through three layers of cheesecloth, and pelleted through a 10% sucrose cushion. The resuspended pellets were clarified with one low-speed centrifugation and loaded on a 10% to 40% sucrose gradient which was centrifuged at 35,000 rpm for 3 h 45 min in a Beckman SW41 rotor. The gradients were fractionated in 1-ml samples, and the fractions containing the opalescent zone of the gradient were pelleted for 2.5 h at 55,000 rpm in a Beckman Ti70 rotor. The micrograph was produced with a Jeol JEM-1400Plus transmission electron microscope.
Besides genome relatedness and genome organization, the insect vector is an important criterion for defining new virus genera within the family Geminiviridae. Viruses of the genus Begomovirus are transmitted by whiteflies, those of the genera Mastrevirus, Curtovirus, Becurtovirus, Turncurtovirus, and probably Eragrovirus are transmitted by leafhoppers, and those of the genus Topocuvirus are transmitted by treehoppers.
Since the discovery of EcmLV and FbSLCV (2), a third capulavirus has been discovered in leaf curl-exhibiting alfalfa (Medicago sativa, Fabaceae) plants collected from 2010 to 2014 during large-scale sampling surveys in the south of France (Rhône delta, Tour du Valat).
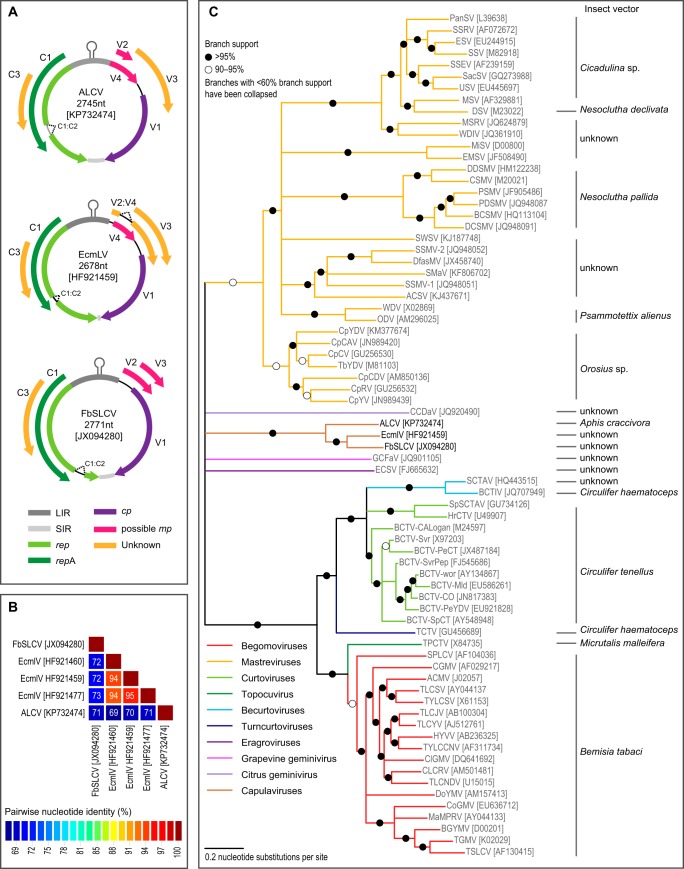
Capulavirus-related sequences were identified in one of these plants (44-1E) using a virion-associated nucleic acids (VANA)-based metagenomics approach (3). Total DNA extracted from 44-1E was subjected to Phi29 DNA polymerase treatment (TempliPhi; GE Healthcare, USA), and a linear DNA fragment of about 2.8 kbp was generated with DraI, NcoI, or NdeI digestion. No fragment falling within the size ranges of reported geminivirus satellite sequences was produced with any of these restriction enzymes. A viral genomic DNA fragment of about 2.8 kbp was amplified from the Phi29 polymerase product with the partially overlapping PCR primers Dar-1981F and Dar-1966R, previously designed to amplify full-length EcmLV genomes (2). The amplification product was cloned (clone 44-1E) and sequenced as previously reported (2). Although the 2,745-nucleotide (nt) viral insert displayed an arrangement of open reading frames (ORFs) (Fig. 2A) that was most similar to those reported for capulaviruses, the arrangement of virion sense open reading frames differed slightly from those of EcmLV and FbSLCV (2).
FIG 2.
(A) Genome organization of capulaviruses: alfalfa leaf curl virus (ALCV) from France, euphorbia caput-medusae latent virus (EcmLV) from South Africa, and French bean severe leaf curl virus (FbSLCV) from India. GenBank accession numbers are given in brackets. (B) Genome-wide percentage pairwise identities of capulaviruses calculated using SDT version 1.2 (11). (C) Neighbor-joining phylogenetic tree of representative geminiviruses based on full genomes. Given that it is not possible to generate a credible alignment of the genomes of all geminiviruses and that interspecies and intragenus recombination has played a significant role in geminivirus diversification, this phylogenetic tree is not intended to be accurate but is rather provided as a guide to display the degrees of sequence similarity shared by the different major groups of geminiviruses. Branches with less than 60% bootstrap support have been collapsed, and the tree is midpoint rooted.
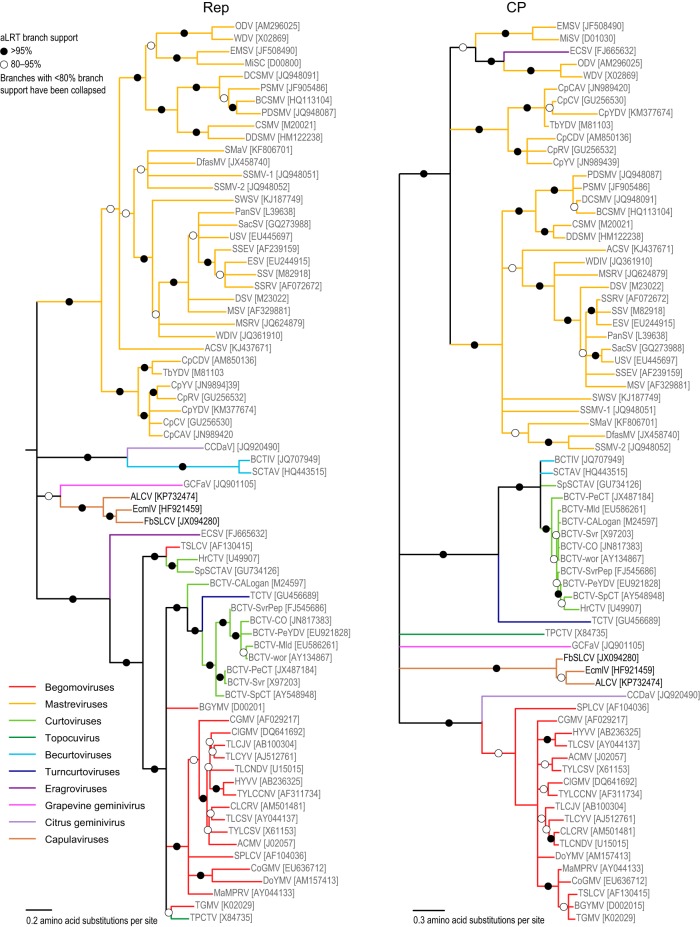
The capulavirus genomes were aligned with representative geminivirus genomes using MUSCLE (4) (see the supplemental material), and a neighbor-joining tree was constructed with the Jukes-Cantor nucleotide substitution model and 1,000 bootstrap replicates (Fig. 2C). Additionally, predicted replication-associated protein (Rep) and capsid protein (CP) amino acid sequences were aligned with Rep and CP sequences of representative geminiviruses using MUSCLE (see the supplemental material). Maximum likelihood phylogenetic trees of the Rep and CP were inferred using PhyML (5), with the rtREV+G+I+F amino acid substitution model chosen as the best fit, using ProtTest (6). Approximate likelihood ratio tests (aLRT) (7) were used to infer relative supports for branches. In all these trees (Fig. 2C and 3), the alfalfa geminivirus clearly clusters with the other capulaviruses with which it shares 69% to 71% genome-wide pairwise identity (Fig. 2B).
FIG 3.
Maximum likelihood phylogenetic trees of the Rep and CP amino acid sequences of representative geminiviruses together with the three known capulavirus species. Branches with less than 80% aLRT branch support have been collapsed. The Rep maximum likelihood phylogenetic tree is rooted with gemycircularviruses (12), whose Rep sequences are distantly related to those of geminiviruses, whereas that of the CP is midpoint rooted.
An agroinfectious clone was prepared from the viral genome 44-1E as previously described for EcmLV (2) and inoculated into 22 Nicotiana benthamiana plants. Although the plants were symptomless, the virus was detected in 10 of them 26 days postinoculation using PCR with primers targeting a 785-nt fragment of the CP gene of the cloned virus (Luz-CP-F, 5′-TGGAATATTGTGCTGCTTGG-3′, and Luz-CP-R, 5′-ATTTTGGGACTTGTGCTCCA-3′). The same purification procedure used to obtain EcmLV particles (Fig. 1) was used for the agroinfected N. benthamiana plants. However, despite spherical, possibly degraded, structures being observed from the sucrose gradient fraction within which geminivirus particles were expected (according to EcmLV purification), twinned icosahedral particles typical of geminiviruses could not be detected by transmission electron microscopy. Nevertheless, these fractions were determined to be positive for viral DNA by Southern blotting with P32-labeled probes prepared by random priming using the full-length genome of the 44-1E clone as a template. As the genome-wide pairwise identity with other geminiviruses was below the lowest of the geminivirus species demarcation thresholds (8, 9), a new capulavirus species was proposed and provisionally named alfalfa leaf curl virus (ALCV).
Since geminiviruses are transmitted by phloem-feeding insects in a persistent circulative manner, the potential vector of ALCV was expected to be an alfalfa phloem-feeding insect. Moreover, as viruses of four geminivirus genera are leafhopper transmitted, it was assumed that leafhoppers might also transmit capulaviruses. However, the CP is the only protein which is involved in transmission specificity, and since capulavirus CPs were not closely related to those of whitefly- or leafhopper-transmitted geminiviruses (Fig. 3), we also tested an alternative hypothesis: we selected aphids because they were frequently detected in alfalfa fields, whereas whiteflies were not detected. Although no aphid-transmitted geminivirus has ever been reported, aphids were considered possible geminivirus vectors because they have been reported as vectors of other circulatively transmitted viruses, including nanoviruses, which, like geminiviruses, also have ssDNA genomes. Thus, the leafhopper Anaceratagallia laevis (Ribaut) and the aphid Aphis craccivora (Fig. 4B) were collected from alfalfa fields in the Rhône delta and Montpellier regions, respectively. Leafhoppers were reared on alfalfa and aphids on alfalfa and faba bean (Vicia faba) with 16 h of light at 24°C and 8 h of darkness at 21°C.
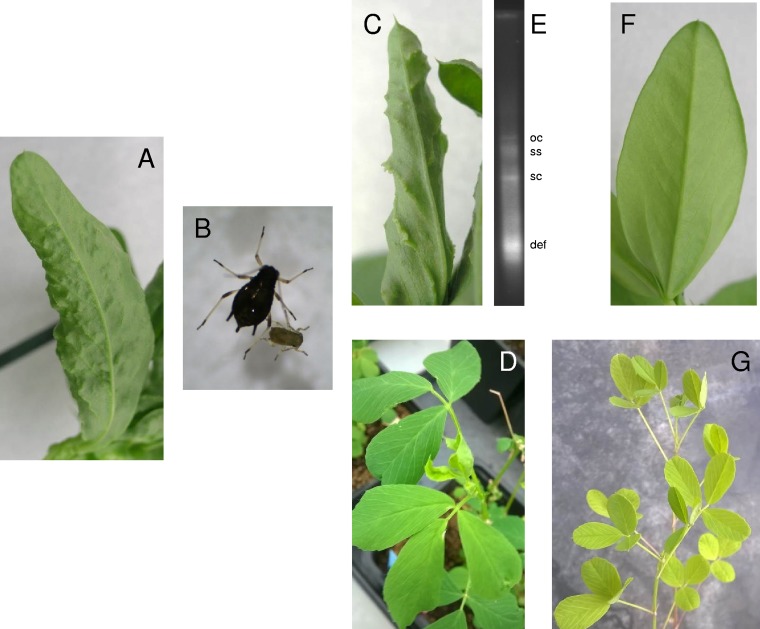
FIG 4.
Faba bean and alfalfa plants infected with Alfalfa leaf curl virus (ALCV). Agroinfected faba bean plants exhibiting vein thickening (A) were used as source plants for transmitting ALCV to faba bean and alfalfa plants using adult Aphis craccivora. (B) Adult and larval A. craccivora. (C and D) The aphid-infected faba plants exhibited vein thickening (C) and the infected alfalfa plants, leaf curl (D). (E) A typical geminivirus DNA profile was revealed by Southern blotting of total DNA extracted from an aphid-infected faba bean plant, using P32-labeled ALCV probes. ss, single-stranded; sc, supercoiled; oc, open circular; def, defective heterogeneous-length DNA. (F and G) Leaves of noninoculated faba bean (F) and alfalfa (G) plants are shown as controls.
Twenty faba bean plants were agroinoculated with the infectious ALCV clone. Three weeks after inoculation, symptoms of vein thickening were detected on the apical leaves of six inoculated plants (Fig. 4A) but not on the mock-inoculated plants. Four weeks after inoculation, 13 inoculated plants exhibited vein thickening and tested positive by PCR with the Luz-CP primers, whereas the other plants remained symptomless and were negative by PCR. A group of 80 aphids was released into a cage containing two of these agroinfected plants for a 2-day acquisition access period (AAP). The two infected plants were replaced with four faba bean seedlings for a 5-day inoculation access period (IAP). A similar transmission test was carried out with 40 leafhoppers given a 2-day AAP on two agroinfected faba bean plants and a 5-day IAP on three faba bean seedlings. Vein thickening symptoms were detected on two aphid-inoculated plants 3 weeks after the IAP, and all four of the plants were positive for the virus by PCR at 4 weeks. One of the 785-nt PCR products was sequenced and was 100% identical to the cognate sequence of the agroinfectious clone. The three leafhopper-inoculated plants did not exhibit vein-thickening symptoms and were negative by PCR. A second transmission test confirmed A. craccivora as a vector (3/7 virus-positive plants with 5 aphids/plant) (Fig. 4C). Southern blotting of total DNA of an aphid-infected faba bean plant using P32-labeled ALCV probes revealed the typical geminivirus DNA profile (Fig. 4E). ALCV also proved transmissible from agroinfected faba bean plants to alfalfa using the same transmission conditions as above except that the IAP was 6 days. Two of four inoculated alfalfa plants were determined by PCR to be positive for the virus. The detection of leaf curl symptoms on the alfalfa plants that were positive by PCR confirmed the etiology of the disease (Fig. 4D). To our knowledge, this is the first report of an aphid-transmitted geminivirus. Further work is required to investigate the vector specificity of this transmission and to test whether other capulaviruses, such as EcmLV, are also transmissible by aphids so as to determine whether, as is expected, aphid transmissibility is a discriminating feature of the proposed Capulavirus genus.
Nucleotide sequence accession number.
The sequence for alfalfa leaf curl virus has been deposited in GenBank under accession number KP732474.
Supplementary Material
ACKNOWLEDGMENTS
We thank Armelle Coeur d'acier (INRA, CBGP, Montpellier) for the identification of A. craccivora and Jean François Germain (ANSES, LSV, Montpellier) for A. laevis identification. We thank Daniel Gargani (CIRAD, BGPI, Montpellier) for electron microscopy observations and Marie-Josée Darroussat, Florence Barthod, Jean-Heinrich Daugrois, Catherine Fenouillet, and Rémy Habas (CIRAD, BGPI, Montpellier) for collecting insects from alfalfa fields. We thank Manade Alain (Assas) for allowing aphid samplings in their alfalfa fields.
This work was supported by INRA, Département Santé des Plantes et Environnement (grant AAP SPE 2015). D.P.M. and A.V. are supported by the National Research Foundation of South Africa.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00453-15.
REFERENCES
- 1.Varsani A, Navas-Castillo J, Moriones E, Hernandez-Zepeda C, Idris A, Brown JK, Zerbini FM, Martin DP. 2014. Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 159:2193–2203. doi: 10.1007/s00705-014-2050-2. [DOI] [PubMed] [Google Scholar]
- 2.Bernardo P, Golden M, Akram M, Naimuddin Nadarajan N, Fernandez E, Granier M, Rebelo AG, Peterschmitt M, Martin DP, Roumagnac P. 2013. Identification and characterisation of a highly divergent geminivirus: evolutionary and taxonomic implications. Virus Res 177:35–45. doi: 10.1016/j.virusres.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Candresse T, Filloux D, Muhire B, Julian C, Galzi S, Fort G, Bernardo P, Daugrois JH, Fernandez E, Martin DP, Varsani A, Roumagnac P. 2014. Appearances can be deceptive: revealing a hidden viral infection with deep sequencing in a plant quarantine context. PLoS One 9:e102945. doi: 10.1371/journal.pone.0102945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 6.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 7.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 8.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X. 2008. Geminivirus strain demarcation and nomenclature. Arch Virol 153:783–821. doi: 10.1007/s00705-008-0037-6. [DOI] [PubMed] [Google Scholar]
- 9.Muhire B, Martin DP, Brown JK, Navas-Castillo J, Moriones E, Zerbini FM, Rivera-Bustamante R, Malathi VG, Briddon RW, Varsani A. 2013. A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol 158:1411–1424. doi: 10.1007/s00705-012-1601-7. [DOI] [PubMed] [Google Scholar]
- 10.Caciagli P, Piles VM, Marian D, Vecchiati M, Masenga V, Mason G, Falcioni T, Noris E. 2009. Virion stability is important for the circulative transmission of Tomato yellow leaf curl Sardinia virus by Bemisia tabaci, but virion access to salivary glands does not guarantee transmissibility. J Virol 83:5784–5795. doi: 10.1128/JVI.02267-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraberger S, Farkas K, Bernardo P, Booker C, Argueello-Astorga GR, Mesleard F, Martin DP, Roumagnac P, Varsani A. 2015. Identification of novel Bromus- and Trifolium-associated circular DNA viruses. Arch Virol 160:1303–1311. doi: 10.1007/s00705-015-2358-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.