ABSTRACT
Rabies virus (RABV) polymerase L together with phosphoprotein P forms the PL polymerase complex that is essential for replication and transcription. However, its exact mechanism of action, interactions with cellular factors, and intracellular distribution are yet to be understood. Here by imaging a fluorescently tagged polymerase (mCherry-RABV-L), we show that L accumulates at acetylated and reorganized microtubules (MT). In silico analysis revealed a dynein light chain 1 (DLC1) binding motif in L that could mediate MT binding through dynein motors. As DLC1 binding by polymerase cofactor P is known, we compared the impact of the DLC1-binding motifs in P and L. Viruses with mutations in the respective motifs revealed that both motifs are required for efficient primary transcription, indicating that DLC1 acts as a transcription enhancer by binding to both P and L. Notably, also the levels of cellular DLC1 protein were regulated by both motifs, suggesting regulation of the DLC1 gene expression by both P and L. Finally, disruption of the motif in L resulted in a cell-type-specific loss of MT localization, demonstrating that DLC1 is involved in L-mediated cytoskeleton reorganization. Overall, we conclude that DLC1 acts as a transcription factor that stimulates primary RABV transcription by binding to both P and L. We further conclude that L influences MT organization and posttranslational modification, suggesting a model in which MT manipulation by L contributes to efficient intracellular transport of virus components and thus may serve as an important step in virus replication.
IMPORTANCE Regulation of rabies virus polymerase complex by viral and cellular factors thus far has not been fully understood. Although cellular dynein light chain 1 (DLC1) has been reported to increase primary transcription by binding to polymerase cofactor phosphoprotein P, the detailed mechanism is unknown, and it is also not known whether the large enzymatic polymerase subunit L is involved. By fluorescence microscopy analysis of fluorescence-tagged rabies virus L, in silico identification of a potential DLC1 binding site in L, and characterization of recombinant rabies virus mutants, we show that a DLC1 binding motif in L is involved in cytoskeleton localization and reorganization, primary transcription regulation by DLC1, and regulation of cellular DLC1 gene expression. By providing evidence for a direct contribution of a DLC1 binding motif in L, our data significantly increase the understanding of rabies virus polymerase regulation and host manipulation by the virus as well.
INTRODUCTION
Rabies virus (RABV) and related lyssaviruses (family Rhabdoviridae) are neurotropic viruses that enter the central nervous system (CNS) by axonal transport along microtubules (1). The negative-sense genomic viral RNA encodes five proteins, of which the 240-kDa large enzymatic subunit L and the 35-kDa cofactor phosphoprotein P form the PL polymerase complex (2). The viral polymerase is a ribonucleoprotein (RNP)-dependent polymerase that only uses the genomic RNA as a template for RNA synthesis when it is encapsidated by nucleoprotein N. The RNP is used as the template for viral RNA replication and transcription, and both RNA synthesis modes are performed by the PL polymerase complex.
To successfully infect a cell, RABV and all other negative-strand RNA viruses must incorporate the viral polymerase complex in the virion to allow essential primary transcription after release of RNPs into the cytoplasm and de novo synthesis of virus proteins. Only after de novo synthesis of virus proteins, subsequent replication of full-length genomic RNAs (3) and secondary transcription can occur. Virion-associated PL polymerase of vesicular stomatitis virus (VSV) is asymmetrically packaged and is located at the blunt site of the bullet-shaped rhabdovirus particles (4), where the 5′ end of the incorporated genome is located (5). How the polymerase gets access to the 3′ end to perform transcription of subgenomic RNAs from the 3′-terminal genome promoter after RNA release into the cytoplasm is unknown.
Primary transcription by RABV polymerase is regulated by dynein light chain 1 (DLC1) (6). By binding of DLC1 to the polymerase cofactor P (7, 8), the efficiency of primary transcription by virion-associated polymerase is increased, indicating that DLC1 serves as a transcription factor that increases processivity of the viral polymerase during primary transcription (6). At later phases of infection, the polymerase is regulated by the viral matrix protein M to allow increased replication (9, 10). RNA polymerase activities of other negative-strand RNA viruses are influenced by tubulin and microtubule-associated proteins (MAPs) (11–13), indicating that cytoskeleton components are involved in polymerase regulation. Besides DLC1-mediated regulation of primary transcription by binding to polymerase cofactor P, no other cellular factors are known that regulate RABV polymerase activities.
In order to investigate the intracellular distribution of RABV-L protein and to analyze to which cellular structures RABV-L binds, we expressed mCherry-tagged RABV-L in different cell lines. Strong accumulation of RABV-L at microtubules in three different cell lines and identification of a conserved DLC1 binding motif in RABV-L involved in microtubule binding led to the generation of recombinant RABVs (rRABVs) in which the DLC1 binding motifs in P, L, or both proteins were mutated. Primary transcription and effects on intracellular DLC1 levels previously described for the DLC1 binding motif in P (6) were compared between the different viruses to assess whether the predicted DLC1 binding motif in RABV-L has similar functions to the DLC1 binding motif in RABV-P. We conclude that DLC1 stimulates primary virus transcription by serving as a transcription factor that binds to both RABV-P and -L and that cellular DLC1 gene expression is regulated by both virus proteins via their DLC1 binding motifs. Accumulation of L at acetylated and reorganized microtubules further demonstrates an intrinsic affinity of L to the cytoskeleton that may play a role in in host cell manipulation and support of efficient intracellular virus transport.
MATERIALS AND METHODS
Cells and viruses.
HEK293 and HEK293T (14) and NA 42/13 neuroblastoma cells were cultivated in minimal essential medium (1:1 Earl's and Hanks' salts) supplemented with nonessential amino acids and 10% fetal calf serum. BSR T7/5 cells were cultivated as described previously (15). HEK293-YFPtub cells were generated by cotransfection of pEYFP-Tub (Clontech) in HEK293 cells and selection of G418-resistant cell clones (1 mg G418/ml cell culture medium). All cells were provided by the Collection of Cell Lines in Veterinary Medicine (CCLV), Friedrich-Loeffler Institut, Riems, Germany.
Recombinant rabies viruses (rRABVs) comprising the nucleotide sequence of attenuated vaccine strain SAD B19 (16) were generated from cDNAs as described previously (17). Mutations in the P and L genes were introduced into full-length cDNA clones by standard techniques. RABV stocks were prepared on BSR T7/5 cells.
Antibodies and sera.
Monospecific rabbit sera N161-5 and P160-5 against RABV-N and -P proteins have been described previously (18). Monoclonal mouse antibodies against β-tubulin (T8328; immunofluorescence, 2 μg/ml in PBS) and acetylated α-tubulin (T7451; immunofluorescence, 0.05 μg/ml in PBS) were purchased from Sigma-Aldrich. Rabbit polyclonal serum DYNLL1/2 (sc-13969; Santa Cruz Biotechnology) was used for Western blot (WB) detection of DLC1. For immunofluorescence, Alexa Fluor 488-conjugated anti mouse IgG (Molecular Probes) was used in a dilution of 1:1,000 in phosphate-buffered saline (PBS).
DNA transfection.
DNA transfections into HEK293T, BSR T7/5, and NA cells were performed with polyethylenimine (PEI) (Sigma-Aldrich). Briefly, for the transfection of 2.5 to 5.0 × 106 cells in 3.5-cm dishes, 6 μg plasmid DNA was mixed with 9 μg PEI in 800 μl Dulbecco's modified Eagle's medium (DMEM). After 20 min of incubation at room temperature, the DNA-PEI mixture was added to the cell cultures. After 3.5 h of incubation, the medium was replaced by fresh medium, and the cells were further cultivated at 37°C and 5% CO2.
Translation inhibition.
To block de novo protein synthesis, 1 h prior to virus infection, cycloheximide (CHX; 150 μg/ml cell culture supernatant) was added to the cell cultures. After 1 h of incubation in the presence of CHX, the cultures were infected with RABVs at a multiplicity of infection (MOI) of 3. One or 24 h after infection, cells were lysed for RNA preparation or Western blot analysis. In the case of 24 h of incubation, 1 h after virus infection, the virus-containing medium was removed and replaced by fresh cell culture medium containing CHX.
Western blots.
Western blots and SDS-PAGE were performed with standard protocols. Proteins were detected with specific sera against viral (N and P) and cellular proteins (β-tubulin and DLC1) and horseradish peroxidase-conjugated secondary antibodies (Dianova). Chemiluminescence signals were detected in a VersaDoc (Bio-Rad) imaging system.
RNA extraction, Northern blots, and qRT-PCR.
RNA was prepared from infected NA cell monolayers with TRIzol reagent (Invitrogen), according to the supplier's instructions.
Virus RNAs were detected by Northern hybridization with an N-gene-specific cDNA probe consisting the PCR-amplified N open reading frame (ORF). [α-32P]dCTP labeling of the probes by nick translation (Invitrogen nick translation system) was performed according to the supplier's instructions.
Prior to reverse transcription 5 μg RNA were treated with RNasin (Promega) and DNase I (Fermentas). Then, 1 μg of the RNA was used for reverse transcription with oligo(dT)12–18 primers (Invitrogen), 40 U/μl RNasin, and Superscript III reverse transcriptase (Invitrogen). All steps were performed according to the supplier's instructions.
For quantitative real-time PCR (qRT-PCR), 2 μl cDNA, 3 μl qPCR primer mixture (2.5 mM each β-actin F/R or R14F/R), 5 μl diethyl pyrocarbonate (DEPC)-H2O and 10 μl SYBR green PCR Mastermix (Applied Biosystems) were mixed, and PCR was performed for 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C with a C-1000 Thermal Cycler (Bio-Rad). All standards and control samples were mixed according to supplier's instructions for SYBR green PCR mastermix. Quantifications were performed with RNAs from three independent infections. Measurement of each RNA was done with three technical replicates.
Oligonucleotides.
β-Actin primers were synthesized by Eurofins MWG, GmbH (Ebersberg, Germany) (β-Actin F, 5′-ACAGCTTCTTTGCAGCTCCTTCG-3′; β-Actin R, 5′-ATCGTCATCCATGGCGAACTGGTG-3′). The RABV-N primers (R14F and R14R) were a gift from Thomas Müller (FLI, Riems, Germany) and published before in reference 19.
Indirect immunofluorescence, confocal laser scanning microscopy, and image processing.
For indirect immunofluorescence detection of microtubules, on glass coverslips, growing cells were fixed with 80% acetone in H2O. After 20 min of incubation at 4°C, acetone was removed and the specimens were blocked by incubation with 0.025% milk powder in PBS for 20 min at room temperature. Immunostainings were performed by 2-h incubation with primary antibody, two wash steps with PBS, and subsequent 45 min of incubation with secondary antibodies.). After 3 further wash steps, specimens were mounted on coverslips. Nuclear chromatin was stained by addition of Hoechst 33342 (1 μg/ml) to the secondary antibody working solution.
Images were acquired with a Leica SP5 confocal laser scan microscope (63× objective; numerical aperture, 1.4) with sequential acquisition of the fluorophores in double fluorescent specimen. Images were processed with the ImageJ software version 1.48b (20). The cells shown in Fig. 1 to 6 exhibit representative phenotypes that have been repeatedly observed in multiple independent experiments (at least 3 independent transfection experiments per figure).
FIG 1.
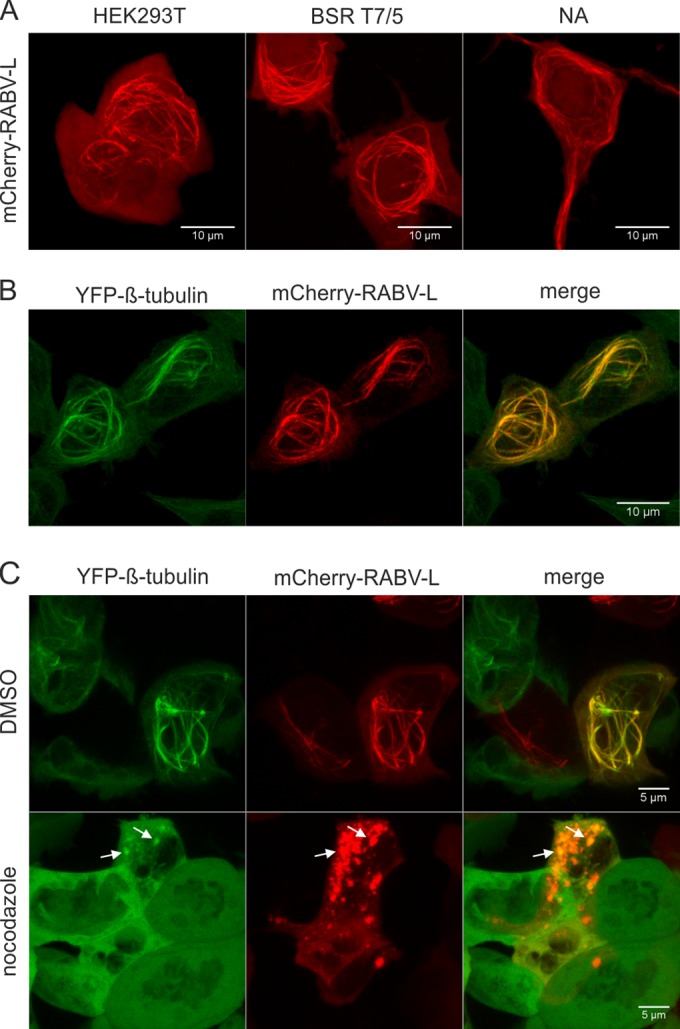
Fluorescence-tagged RABV-L protein accumulates at microtubules. (A) HEK293T, BSR T7/5, and NA cells were transfected with mCherry-RABV-L-coding plasmid. After 16 h, cells were analyzed by live confocal laser scanning microscopy. (B) Localization of mCherry-RABV-L in HEK293-YFPtub cells at 16 h posttransfection. (C) mCherry-RABV-L in HEK293-YFPtub cells at 16 h posttransfection. Three hours prior to live imaging, cells were treated with nocodazole or with dimethyl sulfoxide (DMSO) as a negative control. White arrows show residual colocalization of YFP-tubulin and mCherry-RABV-L. All images are maximum projections of z-stacks acquired by live confocal imaging (optical slice, 0.772 μm; step size, 0.36 μm).
FIG 6.
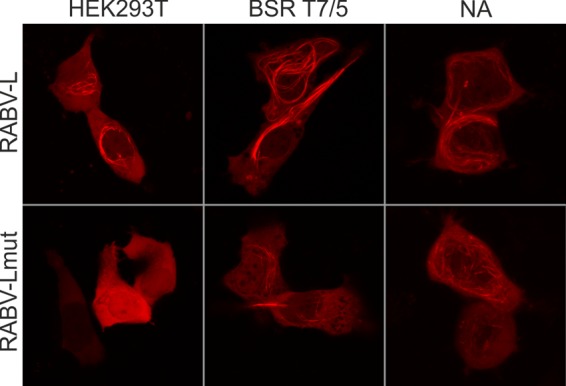
Cell-type-specific effect on microtubule localization. mCherry-tagged RABV-L polymerase and the mutated RABV-Lmut were expressed in HEK293T, BSR T7/5, and NA cells. At 16 h posttransfection, the intracellular distribution of the tagged proteins was analyzed by live confocal microscopy. Images are maximum projections of z-stacks acquired by live confocal imaging (optical slice, 0.772 μm; step size, 0.36 μm).
RESULTS
Fluorescently tagged RABV-L localizes to filaments.
The intracellular distribution of mCherry fluorescence protein-tagged RABV-L (mCherry-RABV-L) was investigated after transfection of expression plasmid into HEK293T, BsrT7/5, and NA neuroblastoma cells. Live confocal laser scanning microscopy of cells at 16 h after transfection revealed mCherry-RABV-L accumulation in cytoplasmic filaments (Fig. 1A), indicating that mCherry-RABV-L associated with the cytoskeleton.
To check whether mCherry-RABV-L accumulated at microtubules, HEK293-YFPtub cells that constitutively express YFP-tagged tubulin were transfected with mCherry-RABV-L expression plasmid and live imaging of tubulin and mCherry-RABV-L filaments was performed after 16 h of incubation (Fig. 1B). Colocalization of YFP-tubulin and mCherry-RABV-L showed that the protein indeed accumulated at yellow fluorescent protein (YFP)-labeled microtubules. A 3-h nocodazole treatment led to the disruption of YFP-labeled microtubules and mCherry-RABV-L filaments, providing further evidence for microtubule localization of mCherry-RABV-L. In the presence of nocodazole, mCherry-RABV-L accumulated in large cytoplasmic aggregates. Only weak residual colocalization of YFP-tubulin and mCherry-RABV-L was observed in the aggregates (Fig. 1C, arrows).
mCherry-RABV-L accumulates at acetylated microtubules.
Accumulation of mCherry-RABV-L at thick microtubules led to the assumption that RABV-L may induce microtubule modifications that can cause microtubule bundling or stabilization. One posttranslational modification that correlates with microtubule stabilization is α-tubulin acetylation. Immunofluorescence detection of acetylated microtubules by acetylated α-tubulin (ac-α-tubulin) in comparison to β-tubulin-labeled microtubules was performed to analyze whether mCherry-RABV-L selectively accumulates at acetylated microtubules.
HEK293T cells were transfected with expression plasmid for mCherry-RABV-L, and 20 h later, cells were fixed with 80% acetone. Immunostaining with antibodies either against β-tubulin or ac-α-tubulin in cells not expressing mCherry-RABV-L showed that the organization of β-tubulin-labeled microtubule network differed from ac-α-tubulin-labeled microtubules. The latter appeared as bundled, were thicker, and were more concentrated at perinuclear regions (Fig. 2, bottom line, right side). In contrast, β-tubulin-labeled microtubules were detected as a network of thinner tubules spreading in the complete cytoplasm (Fig. 2, bottom line, left side).
FIG 2.
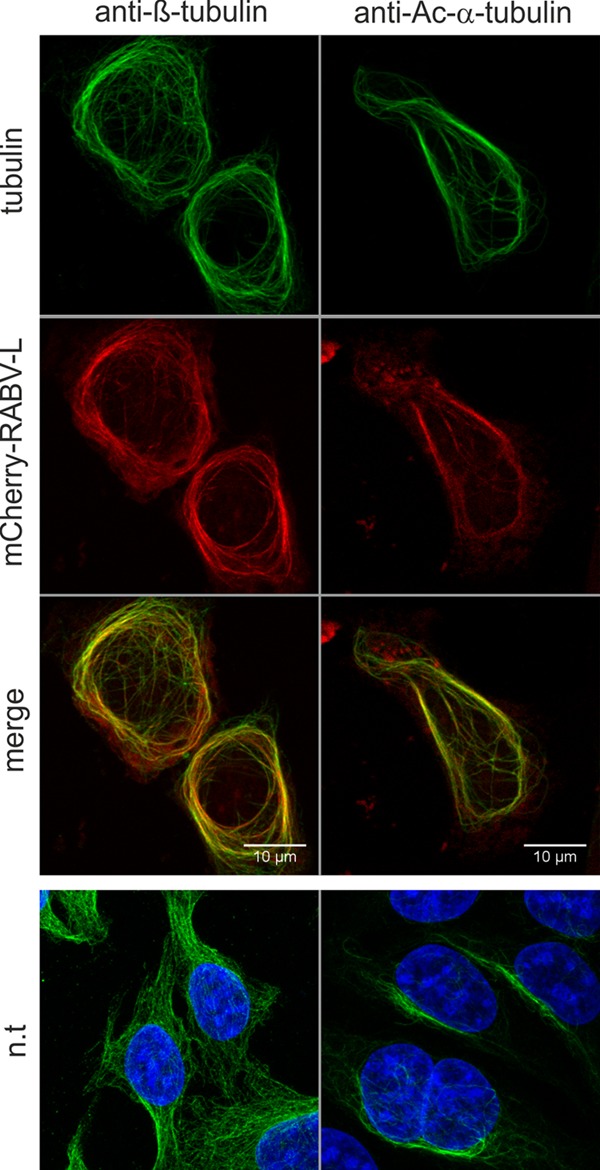
Accumulation of mCherry-RABV-L at acetylated microtubules. mCherry-RABV-L was expressed in HEK293T cells, and immunostainings with β-tubulin- and Ac-α-tubulin-specific antibodies was performed after fixation with 80% acetone. The top three rows show colocalization of mCherry-RABV-L with β-tubulin and acetylated α-tubulin. The bottom row shows β- and acetylated α-tubulin in cells not transfected (n.t.) with mCherry-RABV-L expression plasmid. All images are maximum projections of z-stacks acquired by confocal laser scanning microscopy (optical slice, 0.772 μm; step size, 0.36 μm).
Notably, after staining with anti-β-tubulin and with anti-Ac-α-tubulin antibodies, complete colocalization was observed for mCherry-RABV-L (Fig. 2). These data indicated that in the presence of mCherry-RABV-L, acetylated microtubules no longer formed a subset of the microtubule network. We conclude that mCherry-RABV-L not only bound to microtubules but also increased acetylation of the microtubule network.
Increased microtubule acetylation was further supported by complete colocalization of YFP-labeled β-tubulin, Ac-α-tubulin and mCherry-RABV-L in microtubules (Fig. 3A, top row), whereas in the absence of mCherry-RABV-L, a typical filigree microtubule network was observed and anti-Ac-tubulin staining remained at low levels (Fig. 3A, bottom row). Figure 3B confirmed that acetylated α-tubulin was only found in a subset of microtubules in cells not expressing mCherry-RABV-L.
FIG 3.
Reorganization and increased acetylation of microtubules by mCherry-RABV-L. mCherry-RABV-L was expressed in HEK293-YFPtub cells, and immunostainings with Ac-α-tubulin-specific antibodies were performed after fixation with 80% acetone. (A) Colocalization of YFP-β-tubulin, mCherry-RABV-L, and Ac-α-tubulin and increased acetylation of mCherry-RABV-L-expressing cells. The bottom row shows untransfected HEK293-YFPtub cells imaged with comparable settings. n.t., not transfected. (B) Partial colocalization of YFP-β-tubulin and Ac-α-tubulin. Partial colocalization of YFP-β-tubulin and Ac-α-tubulin in the absence of mCherry-RABV-L was confirmed in nontransfected cells at more sensitive settings for Ac-α-tubulin detection. Note that approximately 50% of cells in the HEK293-YFPtub cultures expressed YFP-tubulin. All images are maximum projections of z-stacks acquired by confocal laser scanning microscopy (optical slice, 0.772 μm; step size, 0.36 μm).
RABV-L and EBLV1-L, but not EBLV2-L accumulate at microtubules.
Microtubule association of and modification by mCherry-RABV-L may be caused by specific binding to tubulins or to microtubule-associated proteins. However, we could not exclude that strong and unpredictable conformational changes caused by the N-terminal fusion of the 27-kDa mCherry tag led to microtubule association. To assess whether L proteins from European bat lyssavirus types 1 and 2 (EBLV1 and EBLV2) that exhibit 86 and 87% amino acid identity to RABV-L also accumulate at microtubules, mCherry-EBLV1-L and mCherry-EBLV2-L were expressed in HEK293-YFPtub cells, and formation of filaments was monitored at 16 h posttransfection. Whereas mCherry-RABV-L and mCherry-EBLV1-L accumulated at microtubules (Fig. 4, 1st and 2nd rows), mCherry-EBLV2-L remained diffuse in the cytoplasm without any detectable accumulation at cytoskeletal structures (Fig. 4, 3rd row). These data indicated that microtubule association was due to specific sequences in the RABV-L and EBLV1-L proteins and that those sequences differ in EBLV2-L.
FIG 4.
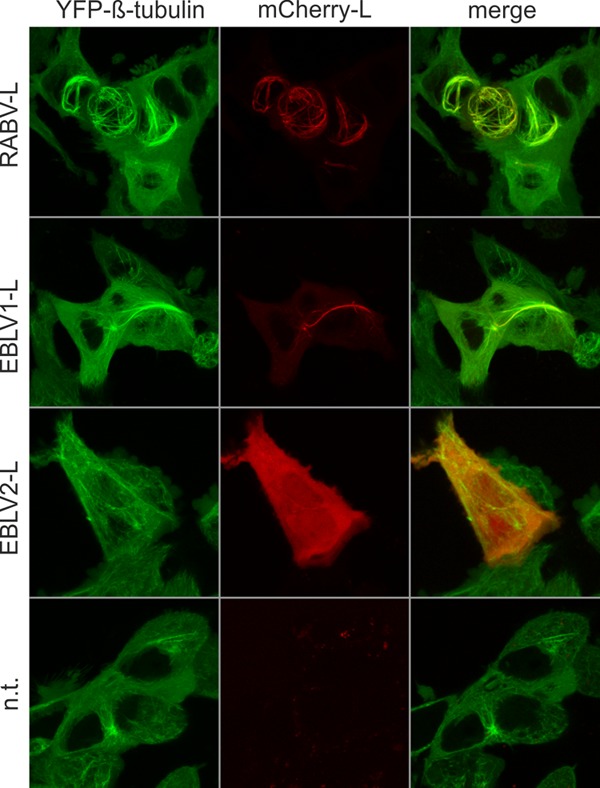
Both RABV-L and EBLV1-L, but not EBLV2-L, accumulate at microtubules. Localization of mCherry-tagged L proteins of RABV (RABV-L), European bat lyssavirus type 1 L (EBLV1-L), and European bat lyssavirus type 2 L (EBLV2-L). Live confocal imaging was performed at 16 h posttransfection of expression plasmids into HEK-YFPtub cells. Not transfected (n.t.) cells were imaged as a negative control. All images are maximum projections of z-stacks acquired by live confocal imaging (optical slice, 0.772 μm; step size, 0.36 μm).
Mutations in a putative DLC1 binding motif interfere with microtubule localization of RABV-L.
Microtubule localization of lyssavirus L proteins could be due to direct binding to microtubules or to microtubule-associated proteins (MAPs). DLC1 is a component of dynein motor complexes that mediate retrograde vesicle transport, and is known to bind RABV polymerase cofactor phosphoprotein P (7, 8). Binding of DLC1 is mediated by a KSTQT motif in P that matches the consensus motif [K/R]xTQT. To study whether similar DLC1 binding motifs also exist in the polymerase L, RABV-L amino acid sequences were screened for linear eukaryotic protein binding motifs (21).
An RMTQT motif in RABV-L at amino acid positions 1079 to 1083 matched the DLC1 binding consensus motif. With a low probability score of 2.26 × 10−5, identification of this putative DLC1 binding motif strongly indicated that RABV-L may interact with DLC1. Comparison of 238 RABV-L sequences from the GenBank database revealed that only 18 sequences differed by one amino acid exchange in the 4th position (RMIQT), indicating that the putative DLC1 binding motif is conserved among members of the RABV species.
Comparison with seven EBLV1-L and three EBLV2-L sequences further showed that in all available European bat lyssavirus sequences, the 5th amino acid position was replaced by V (RMTQV). Differences between EBLV1 and EBLV2 were not detectable in the motif but in surrounding amino acids. All EBLV1 motifs were preceded by serine (SRMTQV). In contrast, the EBLV2-L used here contained aspartic acid at position 1078 (DRMTQV), as present in EBLV2 clone 9018HOL (accession no. EU293114). In two other EBLV2 sequences from GenBank (AGQ16852.1 and YP_001285397) (22, 23), position 1078 was covered by asparagine. Also 92.1% of the analyzed RABV sequences contained asparagine at position 1078, followed by 6.7% serine (as the RABV-L protein used in this study) and 1.3% threonine. These data indicated that the putative DLC1 binding motif is conserved among RABVs and that EBLVs differ in at least one amino acid within the motif and in a flanking amino acid at position −1 relative to the motif.
To test whether the RMTQT motif has an effect on microtubule association of and reorganization by RABV-L, an mCherry-tagged L mutant (mCherry-RABV-Lmut) with three amino acid exchanges at positions 1079, 1081, and 1083 (RMTQT→AMTAA) was expressed in HEK293-YFPTub cells, and the intracellular distribution of microtubules and fluorescence-tagged L was analyzed by live confocal laser scanning microscopy. As shown above, the RMTQT motif-containing mCherry-RABV-L accumulated at reorganized microtubules (Fig. 5, upper row). In contrast, mCherry-RABV-Lmut lost its microtubule localization and was diffusely distributed in the cell (Fig. 5, bottom row).
FIG 5.

Mutations in a putative DLC1 binding motif interfere with microtubule localization of RABV L. Intracellular distribution of the mCherry-tagged RABV-L after mutation of the predicted DLC1 binding motif RMTQT to AMTAA (RABV-Lmut). Live confocal imaging was performed at 16 h posttransfection. White arrows indicate microtubules in cells not expressing mCherry-RABV-L protein. Images are maximum projections of z-stacks acquired by live confocal imaging (optical slice, 0.772 μm; step size, 0.36 μm). The white arrow indicates microtubules in a cell that does not express mCherry-RABV-L.
These data showed that the putative DLC1 binding motif in RABV-L was responsible for microtubule localization of mCherry-RABV-L. Since a clear reorganization of microtubules was not detectable for the L mutant, we further concluded that the RMTQT motif in RABV-L is responsible for microtubule localization and reorganization.
Cell-type-dependent effect of the RMTQT motif on microtubule localization.
To test whether the mutations in the RMTQT motif affect microtubule interaction in different cell lines, the intracellular localizations of mCherry-RABV-L and mCherry-RABV-Lmut were compared in HEK293T, BSR T7/5, and NA cells. Whereas expression of mCherry-RABV-L resulted in the observation of labeled filaments in all three cell lines (Fig. 6A and Fig. 1, 1st row), mCherry-RABV-Lmut lost its filament association in HEK293T cells (Fig. 5). In BSR T7/5 and NA cells, the number of mCherry-positive filaments was decreased, but some remained.
This showed that modifications within the RMTQT motif indeed affected microtubule association of RABV-L. Continued detection of labeled filaments in BSR T7/5 and NA cells (Fig. 6) showed that the influence of the RMTQT motif on microtubule localization was cell type specific and indicated that RABV-L mediates multiple interactions with microtubules, MAPs, or microtubule-associated motor complexes. Cell-specific variations among those cellular structures may determine influence of the RMTQT motif on microtubule localization and modification.
Mutations in the RMTQT motif of RABV-L regulate intracellular DLC1 levels.
Because RABV-L expression after transfection was rather inefficient and the available antisera did not specifically immunoprecipitate expressed L protein, we were not able to directly measure binding of DLC1 to RABV-L and the AMTAA mutant. Instead, as it is known that intracellular DLC1 levels increase after RABV infection (6), the impact of mutant viruses on intracellular DLC1 levels was investigated.
To dissect the effects of both the RMTQT motif in RABV-L and the known KSTQT motif in RABV-P on intracellular DLC1 levels, recombinant viruses with mutations in either P (KSTQT to AAAAA), L (RMTQT to AMTAA), or both proteins were constructed (Fig. 7A). The resultant viruses, rRABV-Pmut, rRABV-Lmut, and rRABV-PLmut, were rescued from cDNAs, and growth curves after infection of NA cells at MOI of 3 and 0.01 were generated. Whereas infection at an MOI of 3 led to similar growth curves for all tested viruses (not shown), after infection at an MOI of 0.01, the titers for rRABV-Pmut and rRABV-Lmut were decreased about 1 log compared to rRABV. Notably, final titers of the double mutant rRABV-PLmut were further decreased (Fig. 7B), indicating an additive effect of the mutated motifs on virus replication.
FIG 7.
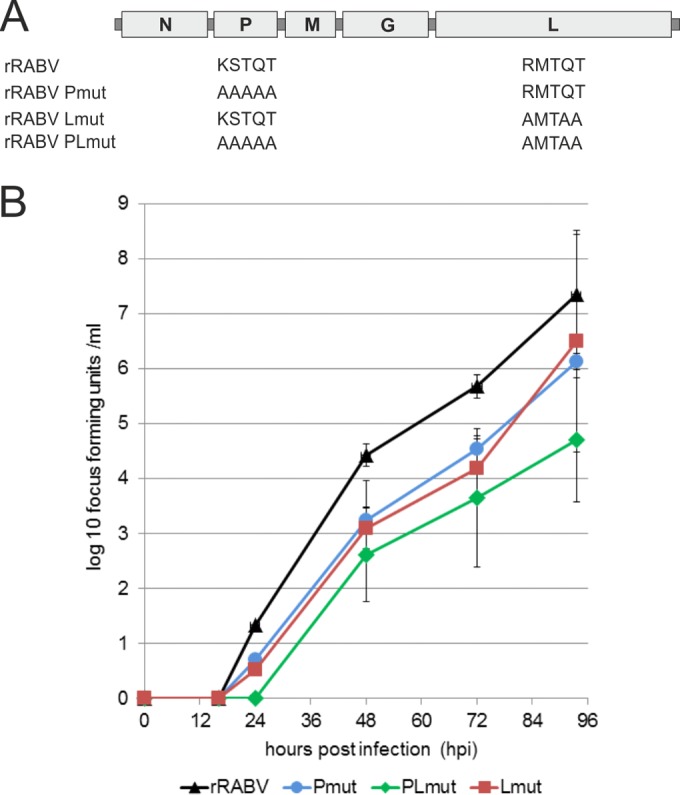
Growth kinetics of recombinant viruses mutated in P and L. (A) Genome organization and mutated motifs. In rRABV, the original motifs KSTQT and RMTQT were present in P and L, respectively. The virus single mutants rRABV-Pmut and rRABV-Lmut and the double mutant rRABV-PLmut contained the indicated amino acid exchanges. (B) Growth curves after infection of NA cells with the indicated viruses at an MOI of 0.01.
To test whether infection by mutant viruses affected the intracellular DLC1 levels, NA cells were infected at an MOI of 5, and cell lysates for Western blot analysis were prepared at 24, 48, and 72 h postinfection (hpi). Western blot detection of cellular DLC1 and β-tubulin (Fig. 8) revealed that DLC1 levels varied among the different virus mutants, whereas β-tubulin remained constant for all four viruses. As previously described (6), DLC1 protein levels were increased compared to those in noninfected cells (Fig. 8, 24 hpi). In contrast, DLC1 levels in rRABV-Pmut-infected cells were not increased. Also, the double mutant rRABV-PLmut did not lead to increased DLC1 levels. Although in rRABV-Lmut-infected cells the DCL1 levels were increased, they did not reach the levels observed for rRABV (Fig. 8, all time points). At 72 hpi, the DLC1 levels for rRABV-Lmut were comparable to those of noninfected cells, whereas a clear increase was still detectable in rRABV-infected cells. Similar to β-tubulin signals, levels of RABV-N and -P proteins were comparable for all tested viruses, confirming comparable infection of the NA cells with the different virus mutants. Similar results were observed on BSR T7/5 cells (not shown), confirming that the effect of the mutations on DLC1 levels was not cell line specific.
FIG 8.
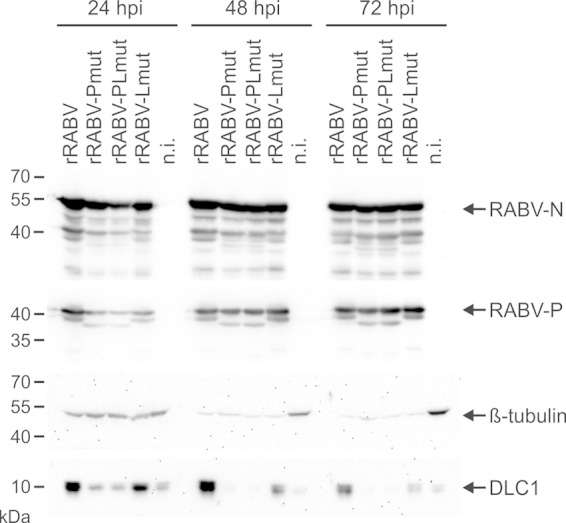
Mutations in the RMTQT motif of RABV-L regulate intracellular DLC1 levels. NA cells were infected at an MOI of 5, and cell lysates for Western blotting were prepared at 24, 48, and 72 hpi. Cell-derived DLC1 and β-tubulin as well as virus proteins N and P were detected with specific antibodies. n.i., not infected.
From these data, we concluded that both the DLC1 binding KSTQT motif in P and the predicted DLC1 binding RMTQT motif in L are involved in DLC1 interactions. By so far unknown mechanisms, DLC1 binding through P and L may lead to activation of DLC1 gene transcription and/or DLC1 protein stabilization.
The RMTQT motif in RABV-L and KSTQT motif in RABV-P are important for efficient primary transcription.
Recombinant RABV with a defect in the KSTQT motif in P exhibited less efficient primary transcription in infected neuroblastoma cells (6). To see whether the RMTQT motif in L is also involved in efficient primary transcription, virus RNA synthesis was investigated by qRT-PCR and Northern blot analysis in the presence of cycloheximide (CHX). Inhibition of protein translation by CHX prevents synthesis of new virus proteins, and RNA synthesis is due to primary transcription by virion-associated polymerase.
One hour prior to infection, NA cells were incubated with CHX, followed by infection with the virus mutants at an MOI of 3. At 1 and 24 hpi, cell lysates were prepared for Western blot analysis and RNA extraction. Western blots with RABV N and β-tubulin-specific antibodies confirmed that CHX blocked RABV protein synthesis (Fig. 9A). In contrast, abundant signals for RABV-N were detectable in the absence of CHX. Constant levels of β-tubulin in all samples indicated that endogenous β-tubulin levels were only marginally affected by CHX treatment.
FIG 9.
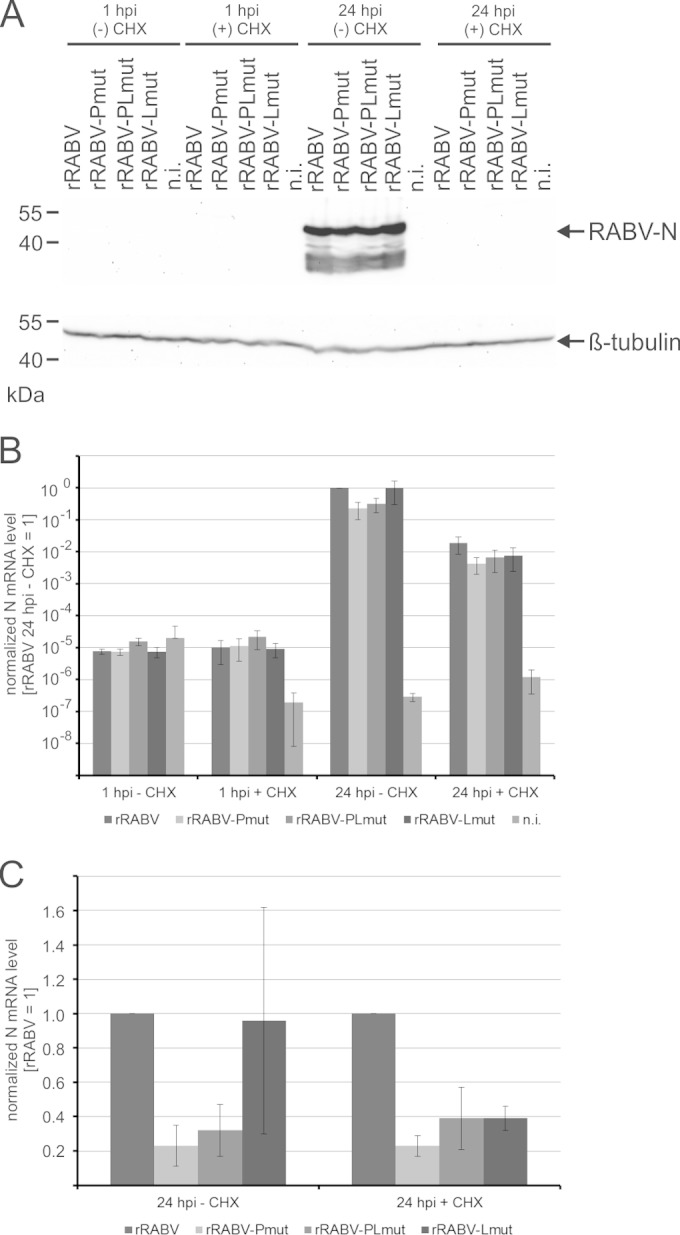
Both the KSTQT motif in P and the RMTQT motif in L are required for efficient primary transcription. (A) Cells were treated with CHX and 1 h later infected with indicated rRABVs. Cell lysates were prepared after 1 or 24 h, and RABV nucleoprotein N and cellular β-tubulin were detected in Western blots. (B) RNA was prepared after 1 and 24 hpi, and N mRNAs were quantified by qRT-PCR. Shown are data after normalization to actin mRNAs and setting the values of rRABV at 24 hpi in the absence of CHX to 1. Standard deviations (n = 3 independent infections) are shown by error bars. (C) For better comparison at 24 hpi, values for rRABV in the absence (− CHX) and presence (+ CHX) of cycloheximide were set to 1. Standard deviations (n = 3 independent infections) are shown by error bars.
Real-time PCR detection at 1 and 24 hpi revealed a substantial increase of N mRNA levels from 1 hpi to 24 hpi (Fig. 9B). Compared to nontreated cells, CHX-treated cells contained 47- to 123-fold less N mRNA at 24 hpi. These primary transcripts made by virion-associated viral polymerase were further compared by normalization to rRABV-N mRNA levels. In the presence of CHX, the N mRNA levels of all three virus mutants were decreased (Fig. 9C), whereas a significant decrease in the absence of CHX only was detectable for rRABV-Pmut and rRABV-PLmut. Results from qRT-PCR analysis were confirmed by Northern blotting. In the presence of CHX, decreased N mRNA levels were verified after infection by all three virus mutants (Fig. 10).
FIG 10.
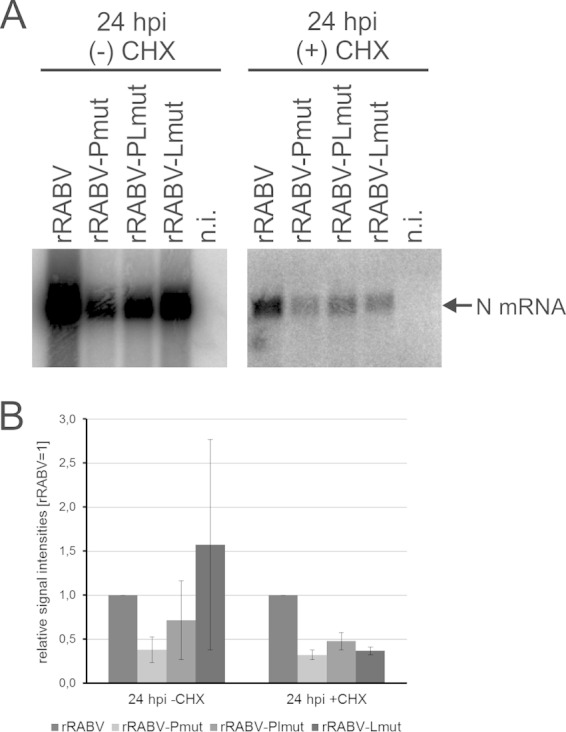
Northern blot detection of mRNAs in the presence (+) and absence (−) of CHX. (A) Representative Northern blots after hybridization with an N gene-specific probe. (B) Signal intensities were quantified after phosphorimaging, and relative signal intensities (rRABV = 1) were calculated from three independent infection experiments. Standard deviations are indicated by error bars.
From qRT-PCR and Northern blot analyses, we conclude that the KSTQT motif in RABV-P and the RMTQT motif in RABV-L play a role in primary transcription. Since primary transcription regulation has been linked to DLC1 binding by RABV-P, we conclude that also the predicted RMTQT motif in RABV-L is a DLC1-interacting motif.
DISCUSSION
We show here that a DLC1 binding motif at positions 1079 to 1083 in RABV-L is involved in (i) microtubule association, (ii) posttranslational modification and reorganization of microtubules, (iii) regulation of intracellular DLC1 levels, and (iv) regulation of primary RABV transcription. These data strongly support a model in which both polymerase cofactor phosphoprotein P and the enzymatic subunit L of the RABV polymerase complex are ligands for DLC1 and that binding of DLC1 to both proteins plays a central role in the regulation of transcription in early phases of infection. Our data further support the idea that RABV not only is hijacking the microtubule network during virus entry but actively manipulates the cytoskeleton to achieve fast and efficient virus transport.
Recent work on p75NTR receptor and RABV cointernalization at axon endings showed that RABV not only hijacks but also accelerates axonal transport mechanisms (24). Whereas p75NTR-dependent transport manipulation is related to early phases of neuronal infections, we show that also posttranslational modification of microtubules can be induced by the expression of a virus protein. Although tagged RABV-L was analyzed in these studies, we exclude tag-dependent effects because microtubule association and reorganization depended on a specific RMTQT motif in RABV-L (Fig. 5). Conservation of the RMTQT motif within the RABV species and the observed microtubule association of mCherry-EBLV1-L encoding an RMTQV sequence at positions 1079 to 1083 indicated that predicted DLC1 binding through L and downstream effects occur in a broader range of lyssaviruses. Accordingly, a perfect RxTQT or EBLV-like RxTQV motif can be found even in phylogenetically distant lyssavirus species, such as Lagos bat virus, Mokola virus, Duvenhage virus, or Khujand virus (not shown).
Interestingly, the loss of microtubule association observed for mCherry-EBLV2-L, also comprising the EBLV-1-like RMTQV motif (Fig. 4), indicated that other sequences in L may also contribute to the observed phenotype. Nevertheless, abrogation of the RMTQT motif in RABV L and the resulting loss of microtubule localization in HEK293T cells (Fig. 5) clearly showed that the motif is substantially involved in the observed interaction with the microtubule cytoskeleton. According to reports about the importance of flanking amino acids for DLC1 binding (25), both a D residue upstream of the RMTQV motif in EBLV2-L as well as an S residue upstream of the respective motifs in the EBLV1- and RABV-L used in this study could allow DLC1 binding and appear unlike to explain the observed differences between EBLV2-L and the other L proteins in microtubule localization. Thus, it remains to be analyzed whether sequence variations in the −1 position to the predicted motif were responsible for the observed differences in microtubule localization or whether other sequence differences contributed to the observed phenotype. As EBLV1 and EBLV2 are considered to represent independent lyssavirus species with many different biological characteristics (26, 27), this may further contribute to deeper understanding of lyssavirus adaptation and evolution.
Acetylation is a modification of microtubules that often is associated with microtubule stabilization and bundling (28). Viruses may affect microtubule stability and polymerization to support virus infection. HIV-1 induces formation of stable microtubules to enhance early virus infection (29), and Ebola virus VP40 protein has been shown to localize at microtubule bundles and to directly increase tubulin polymerization (30).
Indeed, accumulation of mCherry-RABV-L protein at acetylated microtubules (Fig. 2) and conversion of microtubules to an acetylated form (compare Fig. 2 and 3) showed that RABV-L not only colocalized with microtubules but also supported microtubule acetylation. Although the mechanism is not yet clear, DLC1 appears to have a central role, as mutations in the predicted DLC1-binding motif led to the loss of microtubule localization of mCherry-RABV-L (Fig. 5). Unfortunately we could not test for direct interaction of L and DLC1 because of an intrinsic instability of transiently expressed rhabdovirus L proteins (31). However, comparison of DLC1 levels between rRABV- and rRABV-Lmut-infected cells (Fig. 8) strongly indicated that the predicted motif RMTQT in RABV-L is involved in an interaction with DLC1 and may affect subsequent regulation of DLC1 gene transcription (described below).
Moreover, mutations in the RMTQT motif in rRABV-Lmut affected primary transcription to an extent that was comparable to effects observed for the corresponding P mutant. This not only indicated binding of DLC1 to both proteins but also the contribution of both motifs to identical mechanisms of transcription regulation in early phases of virus infection.
Microtubule stabilization by RABV-L-induced acetylation supports an attractive model in which RNP transport after replication in cytoplasmic inclusion bodies to peripheral sites of virus budding is stimulated by more stable and reorganized microtubules. Moreover, efficiency of longer-distance transport in neuronal dendrites and even recently characterized anterograde transport in axons (32) may be crucial for virus spread in neurons. However, no data are available so far that show accelerated virus transport at acetylated microtubules, and the degree of microtubule modification in virus-infected cells also has not yet been clarified. As the majority of L and P proteins are trapped in viral inclusion bodies (33), a less dominant effect on microtubules is expected in the context of a virus infection.
Interestingly, DLC1 has been recently shown to act also as a microtubule-associated protein (MAP) by direct binding of DLC1 dimers to tubulin. Moreover, DLC1 overexpression led to microtubule stabilization and increased acetylation (34). In view of the dose-dependent effect on microtubule stabilization, the presence of increased DLC1 levels in RABV-infected cells (Fig. 8) (6) may support microtubule stabilization. In RABV-infected cells, however, accumulation of DLC1 in viral inclusion bodies (IBs) may serve as a decoy and could decrease levels of free DLC1, although overall DLC1 levels are increased. Moreover, it is also conceivable that binding of DLC1 by P and L proteins may interfere with the stabilizing MAP function of DLC1. In mCherry-RABV-L-expressing cells, the absence of other virus proteins and the lack of trapping of IBs may maintain the polymerase at microtubules without destruction of microtubule-DLC1 dimer interactions. This may lead to the observed colocalization of mCherry-RABV-L and microtubules (Fig. 1 and 2). Further experiments on the degree of microtubule acetylation in virus-infected cells should clarify whether interaction of L and maybe also of P with DLC1 decreases or increases stability and acetylation of cellular microtubules in infected cells.
Although the exact outcome of the observed affinity of RABV-L to microtubules remains to be clarified, the dependence of microtubule association on a short DLC1 binding consensus motif and the observed effects on microtubule acetylation strongly indicate that this molecular interaction plays an important role in virus replication and its communication with cellular pathways.
Evidence for an interaction of the identified RMTQT motif in RABV-L with DLC1 was provided by the observation that DLC1 levels were decreased in infected cells after destruction of either the RMTQT motif in P, the RMTQT motif in L, or both (Fig. 8). A lesser effect of rRABV-Lmut on DLC1 levels indicated that effects on the DLC1 levels correlated with the relative virus protein levels in the infected cell. Virus mRNA synthesis is regulated by a transcription gradient (35) that leads to lower expression of L compared to the upstream-encoded P gene.
Regulation of intracellular DLC1 levels may either occur through DLC1 stabilization in complexes with virus proteins or by activation of DLC1 gene transcription after free DLC1 has been trapped in viral protein complexes. Indeed, the DLC1 promoter is regulated by the transcription activator ATM substrate Chk2-interacting Zn2+ finger (ASCIZ), and DLC1 itself negative regulates ASCIZ by binding to the transcription activation domain (36). Trapping of DLC1 by P and L may decrease the level of free DLC1, resulting in the release of DLC1 from ASCIZ and transcription activation of the DLC1 gene.
Comparable effects of the mutated KSTQT motif in P and of the RMTQT motif in L on virus RNA synthesis by virion-associated polymerase (Fig. 8 and 9) showed that both motifs are required for efficient primary transcription. Comparable N mRNA levels in rRABV-Pmut- and rRABV-Lmut-infected cells, without an additional decrease in the double mutant rRABV-PLmut, further indicated that the previously predicted function of DLC1 as a polymerase processivity stimulation factor (6) requires cooperative action of P and L. From these data, we conclude that DLC1 is a transcription factor of the RABV polymerase whose regulatory effect depends on binding sites in P and L. However, transcription regulation through the two motifs in L and P is not essential, and virus primary transcription can occur even in the absence of both DLC1 binding motifs.
The use of cellular proteins as transcription factors to support RNA synthesis of nonsegmented negative-strand RNA viruses (NNSV) has been discussed previously. Tubulin serves as a positive transcription factor of VSV, Sendai virus, and measles virus (11, 12). Tubulin-associated proteins are also able to stimulate virion-associated VSV polymerase (13), indicating that polymerase stimulation by the cytoskeleton or cytoskeleton-associated proteins is a common theme in NNSV. In particular, the role of DLC1 in supporting primary transcription of RABV (6) and a recent report about the regulation of Ebola virus RNA synthesis by an interaction of DLC1 with VP35 (37) indicate a common mechanism, which not only targets the phosphoprotein cofactors in RABV and Ebola virus but also requires interaction with the polymerase subunit L to act as a transcription factor, at least for RABV.
ACKNOWLEDGMENTS
We thank Angela Hillner and Dietlind Kretzschmar for technical assistance and are grateful to Thomas C. Mettenleiter for helpful discussions and critical comments on the manuscript.
This study was supported by the German Israeli Foundation for Scientific Research and Development (grant no. 1107-73.1/2010) and by an intramural collaborative research grant at the Friedrich-Loeffler-Institut.
REFERENCES
- 1.Tsiang H, Ceccaldi PE, Lycke E. 1991. Rabies virus infection and transport in human sensory dorsal root ganglia neurons. J Gen Virol 72:1191–1194. doi: 10.1099/0022-1317-72-5-1191. [DOI] [PubMed] [Google Scholar]
- 2.Finke S, Conzelmann KK. 2005. Replication strategies of rabies virus. Virus Res 111:120–131. doi: 10.1016/j.virusres.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Patton JT, Davis NL, Wertz GW. 1984. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol 49:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodges J, Tang X, Landesman MB, Ruedas JB, Ghimire A, Gudheti MV, Perrault J, Jorgensen EM, Gerton JM, Saffarian S. 2013. Asymmetric packaging of polymerases within vesicular stomatitis virus. Biochem Biophys Res Commun 440:271–276. doi: 10.1016/j.bbrc.2013.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. 2010. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan GS, Preuss MA, Williams JC, Schnell MJ. 2007. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc Natl Acad Sci U S A 104:7229–7234. doi: 10.1073/pnas.0701397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raux H, Flamand A, Blondel D. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74:10212–10216. doi: 10.1128/JVI.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacob Y, Badrane H, Ceccaldi PE, Tordo N. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J Virol 74:10217–10222. doi: 10.1128/JVI.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finke S, Mueller-Waldeck R, Conzelmann KK. 2003. Rabies virus matrix protein regulates the balance of virus transcription and replication. J Gen Virol 84:1613–1621. doi: 10.1099/vir.0.19128-0. [DOI] [PubMed] [Google Scholar]
- 10.Finke S, Conzelmann KK. 2003. Dissociation of rabies virus matrix protein functions in regulation of viral RNA synthesis and virus assembly. J Virol 77:12074–12082. doi: 10.1128/JVI.77.22.12074-12082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyer SA, Baker SC, Horikami SM. 1990. Host cell proteins required for measles virus reproduction. J Gen Virol 71:775–783. doi: 10.1099/0022-1317-71-4-775. [DOI] [PubMed] [Google Scholar]
- 12.Moyer SA, Baker SC, Lessard JL. 1986. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc Natl Acad Sci U S A 83:5405–5409. doi: 10.1073/pnas.83.15.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill VM, Harmon SA, Summers DF. 1986. Stimulation of vesicular stomatitis virus in vitro RNA synthesis by microtubule-associated proteins. Proc Natl Acad Sci U S A 83:5410–5413. doi: 10.1073/pnas.83.15.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnell MJ, Mebatsion T, Conzelmann KK. 1994. Infectious rabies viruses from cloned cDNA. EMBO J 13:4195–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finke S, Granzow H, Hurst J, Pollin R, Mettenleiter TC. 2010. Intergenotypic replacement of lyssavirus matrix proteins demonstrates the role of lyssavirus M proteins in intracellular virus accumulation. J Virol 84:1816–1827. doi: 10.1128/JVI.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orbanz J, Finke S. 2010. Generation of recombinant European bat lyssavirus type 1 and inter-genotypic compatibility of lyssavirus genotype 1 and 5 antigenome promoters. Arch Virol 155:1631–1641. doi: 10.1007/s00705-010-0743-8. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann B, Freuling CM, Wakeley PR, Rasmussen TB, Leech S, Fooks AR, Beer M, Muller T. 2010. Improved safety for molecular diagnosis of classical rabies viruses by use of a TaqMan real-time reverse transcription-PCR “double check” strategy. J Clin Microbiol 48:3970–3978. doi: 10.1128/JCM.00612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Kruger D, Grebnev G, Kuban M, Strumillo M, Uyar B, Budd A, Altenberg B, Seiler M, Chemes LB, Glavina J, Sanchez IE, Diella F, Gibson TJ. 2014. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 42:D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marston DA, McElhinney LM, Ellis RJ, Horton DL, Wise EL, Leech SL, David D, de Lamballerie X, Fooks AR. 2013. Next generation sequencing of viral RNA genomes. BMC Genomics 14:444. doi: 10.1186/1471-2164-14-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marston DA, McElhinney LM, Johnson N, Muller T, Conzelmann KK, Tordo N, Fooks AR. 2007. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G-L 3′ nontranslated region. J Gen Virol 88:1302–1314. doi: 10.1099/vir.0.82692-0. [DOI] [PubMed] [Google Scholar]
- 24.Gluska S, Zahavi EE, Chein M, Gradus T, Bauer A, Finke S, Perlson E. 2014. Rabies virus hijacks and accelerates the p75NTR retrograde axonal transport machinery. PLoS Pathog 10:e1004348. doi: 10.1371/journal.ppat.1004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapali P, Szenes A, Radnai L, Bakos A, Pal G, Nyitray L. 2011. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J 278:2980–2996. doi: 10.1111/j.1742-4658.2011.08254.x. [DOI] [PubMed] [Google Scholar]
- 26.Davis PL, Holmes EC, Larrous F, Van der Poel WH, Tjornehoj K, Alonso WJ, Bourhy H. 2005. Phylogeography, population dynamics, and molecular evolution of European bat lyssaviruses. J Virol 79:10487–10497. doi: 10.1128/JVI.79.16.10487-10497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourhy H, Kissi B, Lafon M, Sacramento D, Tordo N. 1992. Antigenic and molecular characterization of bat rabies virus in Europe. J Clin Microbiol 30:2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohan R, Panda D. 2008. Kinetic stabilization of microtubule dynamics by estramustine is associated with tubulin acetylation, spindle abnormalities, and mitotic arrest. Cancer Res 68:6181–6189. doi: 10.1158/0008-5472.CAN-08-0584. [DOI] [PubMed] [Google Scholar]
- 29.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruthel G, Demmin GL, Kallstrom G, Javid MP, Badie SS, Will AB, Nelle T, Schokman R, Nguyen TL, Carra JH, Bavari S, Aman MJ. 2005. Association of Ebola virus matrix protein VP40 with microtubules. J Virol 79:4709–4719. doi: 10.1128/JVI.79.8.4709-4719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canter DM, Perrault J. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376–386. doi: 10.1006/viro.1996.0263. [DOI] [PubMed] [Google Scholar]
- 32.Bauer A, Nolden T, Schroter J, Romer-Oberdorfer A, Gluska S, Perlson E, Finke S. 2014. Anterograde glycoprotein-dependent transport of newly generated rabies virus in dorsal root ganglion neurons. J Virol 88:14172–14183. doi: 10.1128/JVI.02254-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finke S, Brzozka K, Conzelmann KK. 2004. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J Virol 78:12333–12343. doi: 10.1128/JVI.78.22.12333-12343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asthana J, Kuchibhatla A, Jana SC, Ray K, Panda D. 2012. Dynein light chain 1 (LC8) association enhances microtubule stability and promotes microtubule bundling. J Biol Chem 287:40793–40805. doi: 10.1074/jbc.M112.394353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finke S, Cox JH, Conzelmann KK. 2000. Differential transcription attenuation of rabies virus genes by intergenic regions: generation of recombinant viruses overexpressing the polymerase gene. J Virol 74:7261–7269. doi: 10.1128/JVI.74.16.7261-7269.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jurado S, Conlan LA, Baker EK, Ng JL, Tenis N, Hoch NC, Gleeson K, Smeets M, Izon D, Heierhorst J. 2012. ATM substrate Chk2-interacting Zn2+ finger (ASCIZ) is a bi-functional transcriptional activator and feedback sensor in the regulation of dynein light chain (DYNLL1) expression. J Biol Chem 287:3156–3164. doi: 10.1074/jbc.M111.306019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luthra P, Jordan DS, Leung DW, Amarasinghe GK, Basler CF. 2015. Ebola virus VP35 interaction with dynein light chain 8 regulates viral RNA synthesis. J Virol 89:5148–5153. doi: 10.1128/JVI.03652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]



