ABSTRACT
Respiratory syncytial virus (RSV) and human parainfluenza virus type 3 (HPIV3) are the first and second leading viral agents of severe respiratory tract disease in infants and young children worldwide. Vaccines are not available, and an RSV vaccine is particularly needed. A live attenuated chimeric recombinant bovine/human PIV3 (rB/HPIV3) vector expressing the RSV fusion (F) glycoprotein from an added gene has been under development as a bivalent vaccine against RSV and HPIV3. Previous clinical evaluation of this vaccine candidate suggested that increased genetic stability and immunogenicity of the RSV F insert were needed. This was investigated in the present study. RSV F expression was enhanced 5-fold by codon optimization and by modifying the amino acid sequence to be identical to that of an early passage of the original clinical isolate. This conferred a hypofusogenic phenotype that presumably reflects the original isolate. We then compared vectors expressing stabilized prefusion and postfusion versions of RSV F. In a hamster model, prefusion F induced increased quantity and quality of RSV-neutralizing serum antibodies and increased protection against wild-type (wt) RSV challenge. In contrast, a vector expressing the postfusion F was less immunogenic and protective. The genetic stability of the RSV F insert was high and was not affected by enhanced expression or the prefusion or postfusion conformation of RSV F. These studies provide an improved version of the previously well-tolerated rB/HPIV3-RSV F vaccine candidate that induces a superior RSV-neutralizing serum antibody response.
IMPORTANCE Respiratory syncytial virus (RSV) and human parainfluenza virus type 3 (HPIV3) are two major causes of pediatric pneumonia and bronchiolitis. The rB/HPIV3 vector expressing RSV F protein is a candidate bivalent live vaccine against HPIV3 and RSV. Previous clinical evaluation indicated the need to increase the immunogenicity and genetic stability of the RSV F insert. Here, we increased RSV F expression by codon optimization and by modifying the RSV F amino acid sequence to conform to that of an early passage of the original isolate. This resulted in a hypofusogenic phenotype, which likely represents the original phenotype before adaptation to cell culture. We also included stabilized versions of prefusion and postfusion RSV F protein. Prefusion RSV F induced a larger quantity and higher quality of RSV-neutralizing serum antibodies and was highly protective. This provides an improved candidate for further clinical evaluation.
INTRODUCTION
Human respiratory syncytial virus (RSV) and human parainfluenza virus type 3 (HPIV3) are enveloped, nonsegmented, negative-stranded RNA viruses of the family Paramyxoviridae. They are, respectively, the first and second leading viral causes of severe acute lower respiratory tract infections in infants and children worldwide. RSV alone is responsible for an estimated 34 million annual pediatric cases of lower respiratory tract illness worldwide, with >3.5 million hospitalizations and 66,000 to 199,000 pediatric deaths, which occur predominantly in the developing world (1). Licensed vaccines or effective antiviral drugs are not available for either RSV or HPIV3. Experimental inactivated (RSV and HPIV3) and subunit (RSV) vaccines have been linked to vaccine-induced enhanced disease in young children (inactivated RSV) and experimental animals (subunit RSV and inactivated HPIV3) (2–4). In contrast, disease enhancement is not observed with live attenuated RSV strains or vectors expressing RSV antigens (5–7), indicating that suitably attenuated candidates are safe for immunization of infants and young children.
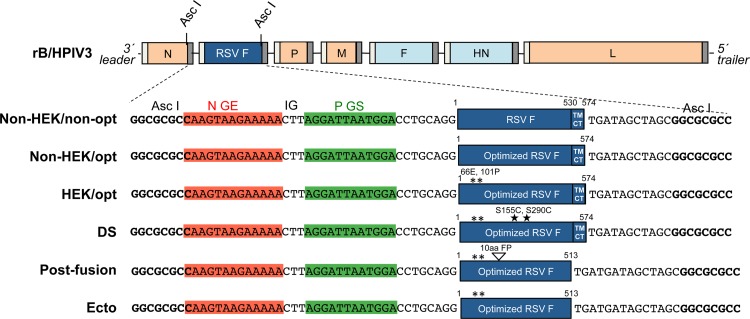
A chimeric recombinant bovine-human PIV3 (rB/HPIV3) (Fig. 1) has been under development as a replication-competent intranasal pediatric vaccine vector. The PIV3 genome is a negative-sense RNA of 15.5 kb that contains six genes in the order 3′-N (nucleoprotein)-P (phosphoprotein)-M (matrix protein)-F (fusion glycoprotein)-HN (hemagglutinin-neuraminidase glycoprotein)-L (polymerase protein)-5′ (Fig. 1). The rB/HPIV3 vector consists of a bovine PIV3 (BPIV3) Kansas strain backbone in which the F and HN genes have been replaced by those of the HPIV3 JS strain (8). This combines the attenuation phenotype of BPIV3 in primates with the HPIV3 F and HN viral neutralization antigens. Inclusion of sequence encoding the RSV F protein, which is the major RSV neutralization and protective antigen, as a supernumerary gene (Fig. 1) provides a bivalent HPIV3/RSV vaccine. In a previous clinical study in children seronegative for both viruses, rB/HPIV3 expressing RSV F from the second (N-P) gene position (MEDI-534) was found to be infectious, attenuated, and well tolerated. However, only 50% of the recipients developed detectable RSV-neutralizing serum antibodies (7), although the sensitivity of the assay would have been reduced because it was done without added complement. Analysis of the shed vaccine virus from vaccinees showed that ∼50% of the specimens contained vaccine virus with mutations predicted to perturb RSV F expression, and 2.5% of the clinical trial material did not express RSV F (9). Our goal was to enhance the immunogenicity of RSV F in the rB/HPIV3 vector and to evaluate its genetic stability during replication in vitro and in vivo.
FIG 1.
rB/HPIV3 vectors expressing different forms of RSV F. The RSV F ORF (strain A2) was inserted into the rB/HPIV3 vector under the control of a set of added BPIV3 N gene end (GE), intergenic (IG), and P gene start (GS) transcription signals so that RSV F would be expressed as a separate mRNA. With the exception of the top construct (Non-HEK/non-opt), each RSV F insert was codon optimized for human expression (GeneArt; Life Technologies). HEK assignments (66E and 101P) are marked by asterisks. The DS mutations (S155C and S290C) are marked with stars. The Ecto form consists of the RSV F ectodomain (amino acids 1 to 513) lacking the transmembrane and cytoplasmic tail domains. The postfusion form is the Ecto form with a further deletion of the first 10 amino acids of the FP. The viruses were recovered in hamster BSR T7/5 cells as described previously (23) and passaged in rhesus monkey LLC-MK2 cells.
Like the F proteins of all paramyxoviruses, the RSV F protein is a type I fusion protein that mediates the fusion of the viral envelope and cellular membranes during entry. It initially assembles into trimers in a metastable (unstable) prefusion conformation on the surfaces of infected cells and virions. Prefusion F can be triggered, such as by contact with an adjacent target cell membrane, to undergo a massive conformational change that mediates membrane fusion, leaving the F protein in a postfusion conformation (10–14). Triggering of RSV F does not require the engagement of attachment protein (G), unlike the triggering of parainfluenza virus F proteins, which is mediated by the binding of attachment protein to a cellular receptor. Prefusion RSV F protein is thought to be susceptible to premature triggering. There also is evidence that much of the RSV F protein expressed during infection is conformationally heterogeneous, which may act as a decoy to reduce the induction of virus-neutralizing antibodies (NAbs) (15). These problems might be obviated by expression of the F protein in a stabilized conformation.
The prefusion form of RSV F appears to be a more effective neutralization antigen than the postfusion form when evaluated as a subunit vaccine in experimental animals (16, 17), although the postfusion form does induce a substantial neutralizing antibody response and has the advantage of being highly stable (13). The prefusion form of RSV F also can be substantially stabilized through structure-based mutations, one of which involves the introduction of a disulfide bond between S155 and S290 (called DS) (17). In the present study, we codon optimized the RSV F open reading frame (ORF), made its amino acid sequence identical to that of an early passage of the original clinical isolate (18), and introduced the stabilized prefusion and postfusion F proteins. The resulting series of rB/HPIV3-RSV F constructs were evaluated for replication, immunogenicity, and protective efficacy in a hamster model and for stability of the RSV F gene in vitro and in vivo. Since the prototype rB/HPIV3-RSV F construct has already been shown to be well tolerated in seronegative children, as noted above, an improved version could advance expeditiously to clinical evaluation.
MATERIALS AND METHODS
Cells and viruses.
Rhesus monkey LLC-MK2 (ATCC CCL-7) cells and African green monkey Vero cells (ATCC CCL-81) were maintained in Opti-MEMI (1×) plus GlutaMax-1 medium (Life Technologies, Grand Island, NY) supplemented with 5% fetal bovine serum (FBS) (HyClone; Thermo Scientific, Atlanta, GA). BSR T7/5 hamster kidney cells, which constitutively express T7 RNA polymerase, were maintained as described previously (19). Recombinant wild-type (wt) RSV and rB/HPIV3 were previously described (8, 20). All rB/HPIV3 vectors were propagated at 32°C in LLC-MK2 cells or Vero cells (21). RSV and its gene sequences were derived from strain A2 (GenBank accession no. M74568); HPIV3 and BPIV3 sequences were from strains JS (Z11575) and Kansas/15626/84 (AF178654), respectively.
Construction of antigenomic cDNA encoding rB/HPIV3 vectors expressing RSV F.
The full-length cDNA of rB/HPIV3 was previously modified to contain a unique AscI restriction site at the 2nd genome position between the N and P genes (22) (Fig. 1). The DNA sequence encoding the RSV F protein with HEK assignments (66E and 101P) (18) was codon optimized (HEK/opt) (Fig. 1) and synthetically derived (GeneArt; Life Technologies). The HEK/opt and Ecto (ectodomain; amino acids [aa] 1 to 513) insert sequences were amplified by PCR with an Advantage HF 2 PCR kit (Clontech, Mountain View, CA) using synthetic HEK/opt RSV F as the template. The shared forward primer for both HEK/opt and Ecto was ATCATGGCGCGCCAAGTAAGAAAAACTTAGGATTAATGGACCTGCAGGATGGAACTGCTGATCCTGAAGGC, the reverse primer for HEK/opt was GAGATGGCGCGCCGCTAGCTATCAGTTGGAGAAGGCGATATTGTTG, and the reverse primer for Ecto was GAGATGGCGCGCCGCTAGCTATCATCACAGCAGCTCGTCGCTCTTTCTGATG (the AscI restriction sites are underlined; the ORF translation initiation and termination codons are in boldface). The PCR products of the HEK/opt and Ecto inserts were cloned into the pCR4-TOPO vector with a TOPO TA cloning kit (Life Technologies), and the sequences were confirmed by automated sequencing. The pCR4-TOPO vectors with non-HEK/opt and DS inserts were generated with a QuikChange Lightning Multi site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) using pCR4-TOPO vector DNA containing a HEK/opt insert as the template. The pCR4-TOPO vector with a postfusion insert was generated by deleting the first 10 amino acids of the fusion peptide (FP) in RSV F (aa 137 to 146) with a QuikChange II site-directed mutagenesis kit (Agilent Technologies) using the pCR4-TOPO vector containing an Ecto insert as the template. To mutate 2 residues in RSV F to non-HEK assignments (P101Q and E66K), two mutagenesis reactions were performed. The following primers were used for mutagenesis: for the P101Q mutation, the primers TGATGCAGTCCACCCAAGCCACCAACAACCGG and CCGGTTGTTGGTGGCTTGGGTGGACTGCATCA were used; for the E66K mutation, the primers TCGAGCTGTCCAACATCAAGAAAAACAAGTGCAACGGCAC and GTGCCGTTGCACTTGTTTTTCTTGATGTTGGACAGCTCGA were used. To stabilize RSV F in the prefusion conformation, two DS mutations (S155C and S290C) were introduced in RSV F by mutagenesis. For the S155C mutation, the primers GGCGTGGCCGTGTGCAAGGTGCTGCACC and GGTGCAGCACCTTGCACACGGCCACGCC were used; for the S290C mutation, the primers CAGAGCTACTCCATCATGTGCATCATCAAAGAAGAGG and CCTCTTCTTTGATGATGCACATGATGGAGTAGCTCTG were used; for the 10-aa FP deletion in the postfusion form of RSV F, the primers GCGGAAGCGGCGGGCCATTGCCTCTG and CAGAGGCAATGGCCCGCCGCTTCCGC were used.
All RSV F sequences in pCR4-TOPO vectors were confirmed by automated sequencing analysis. The RSV F inserts were digested and ligated into full-length cDNA of rB/HPIV3 at the 2nd genome position using the AscI restriction site (Fig. 1), and the sequences were confirmed by automated sequencing. Full-length cDNA of rB/HPIV3 with non-HEK/nonoptimized (non-opt) RSV F in the 2nd genome position was generated previously (23). The genome nucleotide length for each construct was a multiple of 6 (24). The phasing of each gene was maintained as the native phasing (25). The phasing of the inserted RSV F gene at the second gene position was identical to the native phasing of the P gene, which normally is in the second gene position in PIV3 (25).
Recovery of rB/HPIV3-RSV F viruses from cDNA.
The rB/HPIV3-RSV F viruses were recovered by reverse genetics (8) in BSR-T7/5 cells constitutively expressing T7 RNA polymerase (26). The recovered virus was amplified by two passages in LLC-MK2 cells at 32°C. Viral RNA was isolated from the virus stocks using the Qiagen Viral Amp kit (Qiagen, Valencia, CA), followed by DNase I treatment to remove the plasmid DNA used for rescue. Virus sequences were confirmed in their entirety (except for the last 30 and 120 nucleotides at the 3′- and 5′-terminal ends, respectively) by sequence analysis of uncloned reverse transcription (RT)-PCR products amplified from viral RNA. Control RT-PCRs lacking reverse transcriptase indicated that the RT-PCR products were not derived from the input antigenomic cDNAs (data not shown). The percentage of vector virions expressing RSV F in each working stock of inoculum was determined by a fluorescence double-staining plaque assay (described below) to ensure that each stock had 99 to 100% of the virions expressing RSV F and that plaques were homogeneous in size and shape.
Fluorescence double-staining plaque assay.
Vero cell monolayers in 24-well plates were infected with 10-fold serially diluted samples. The infected monolayers were overlaid with culture medium containing 0.8% methylcellulose, incubated at 32°C for 6 days, and fixed by two overnight incubations in 80% methanol at 4°C. The monolayers were washed in 1× phosphate-buffered saline (PBS), followed by incubation in Odyssey blocking buffer (LiCor) for 1 h at room temperature. The monolayers were then incubated with Odyssey blocking buffer containing RSV F-specific monoclonal antibodies (MAbs) diluted at 1:2,000 (19) and HPIV3 hyperimmune serum diluted 1:1,000 for 1 h at room temperature. The HPIV3 hyperimmune serum was generated by immunizing rabbits with sucrose-purified HPIV3 virions, as described previously (23). After washing three times in 1× PBS, the monolayers were incubated for 1 h at room temperature with Odyssey blocking buffer containing IRDye800CW-conjugated goat anti-rabbit antibody and IRDye680LT-conjugated goat anti-mouse antibody (Licor), each diluted at 1:800. The plaques were scanned and visualized with the Odyssey infrared imaging system (LiCor) under 800-nm and 680-nm channels. Fluorescent plaques showing staining for HPIV3 proteins and RSV F were detected as green (800 nm) and red (680 nm), respectively, and were yellow when merged (800 and 680 nm).
Analysis of RSV F expression by Western blotting.
Vero cells (2 × 105) were infected with rB/HPIV3-RSV F viruses at a multiplicity of infection (MOI) of 10 50% tissue culture infective doses (TCID50) or with wt RSV at 10 PFU and incubated at 32°C or 37°C, as indicated. At 48 h postinfection (p.i.), the medium supernatants were removed and the monolayers were washed twice with ice-cold PBS and lysed with 200 μl of ice-cold RIPA buffer containing 1× Complete Cocktail Protease Inhibitor (Roche, Indianapolis, IN). For Western blot analysis under reducing and denaturing conditions, supernatants or lysates were mixed with 1× LDS buffer (Life Technologies) and 1× reducing reagent (Life Technologies) and boiled at 95°C for 5 min. Thirty microliters of reduced, denatured lysate was loaded onto 4 to 12% NuPAGE gels (Novex-Life Technologies) with NuPage antioxidant reagent (Novex-Life Technologies) added to the running buffer in the cathode chamber. For Western blot analysis under nonreducing conditions, the lysates were mixed with 1× LDS buffer without boiling. Thirty microliters of lysate with 1× LDS buffer was loaded onto 4 to 12% NuPAGE gels (Novex-Life Technologies) without adding NuPage antioxidant reagent (Novex-Life Technologies) to the running buffer. The membranes were probed with murine monoclonal anti-RSV F antibody (Ab43812; Abcam, Cambridge, MA), rabbit polyclonal antibodies generated by immunizing rabbits with sucrose-purified wt RSV, goat polyclonal anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (G8795; Sigma, St. Louis, MO), and anti-HPIV3 HN antibodies generated as described previously (23). The following fluorescent-dye-conjugated secondary antibodies were used: donkey anti-rabbit IRDye680 and donkey anti-mouse IRDye 800CW (LiCor, Lincoln, NE). The membranes were scanned, and the fluorescence intensities of the protein bands were quantified using the Odyssey infrared imaging system (Image Studio; LiCor).
Analysis of cell surface RSV F expression by flow cytometry.
Vero cells were infected with rB/HPIV3 vectors at an MOI of 5 TCID50 or with wt RSV at an MOI of 5 PFU. After incubation at 32°C for 48 h, the infected cells were harvested by incubation in PBS containing 1 mM EDTA at 37°C for 5 min and then washed twice in ice-cold fluorescence-activated cell sorter (FACS) buffer (1× PBS, 2% FBS) and incubated with an optimized dilution of Alexa Fluor 488 (AF488)-conjugated RSV F MAb 1129 (27) or unconjugated D25 human monoclonal antibody (11). The AF488-conjugated RSV F MAb was prepared using an Alexa Fluor 488 antibody-labeling kit (Life Technologies) following the kit instructions. Cells stained with D25 were washed twice with ice-cold FACS buffer and stained with Alexa Fluor 488-conjugated goat F(ab)2 anti-human IgG(γ) (Life Technologies). The stained cells were washed twice in ice-cold FACS buffer and incubated in Near-infrared LIVE/DEAD dye (Life Technologies) to discriminate dead cells. The stained cells were analyzed using a BD FACSCanto II flow cytometer.
Hamster studies.
All animal studies were approved by the NIH Institutional Animal Care and Use Committee (IACUC). Six-week-old Golden Syrian hamsters (Harlan Laboratories, Frederick, MD), which were confirmed to be seronegative for HPIV3 and RSV by hemagglutination inhibition (28) assay and RSV-specific virus neutralization assay, respectively (29, 30), were anesthetized and inoculated intranasally (i.n.) with 0.1 ml inoculum containing 105 TCID50 of rB/HPIV3 vectors or 106 PFU of wt RSV (A2 strain). Six hamsters per group were inoculated with each virus. To evaluate vector replication, nasal turbinate and lung tissues were collected separately on days 3 and 5 p.i. for virus quantification by serial dilution on LLC-MK2 cells (rB/HPIV3 vectors) or by plaque assay on Vero cells (wt RSV) (19, 31). To determine vector immunogenicity, sera were collected from hamsters 3 days prior to and 28 days after inoculation. Titers of RSV- and HPIV3-specific NAbs were determined by 60% plaque reduction neutralization assays (PRNT60) on Vero cells and LLC-MK2 cells, respectively, performed in the presence or absence of guinea pig complement (Lonza) (32). (Note that all sera were heated at 56°C for 30 min to destroy endogenous complement prior to assay and the addition of guinea pig complement.) Protective efficacy was determined by challenge infection performed by intranasal infection with 106 PFU of wt RSV in 0.1 ml inoculum at 30 days after immunization. The challenge RSV loads in the nasal turbinates and lungs were determined 3 days after challenge by plaque titration of tissue homogenates on Vero cells, as described previously (33).
RESULTS
Codon optimization and an early-passage form of RSV F protein.
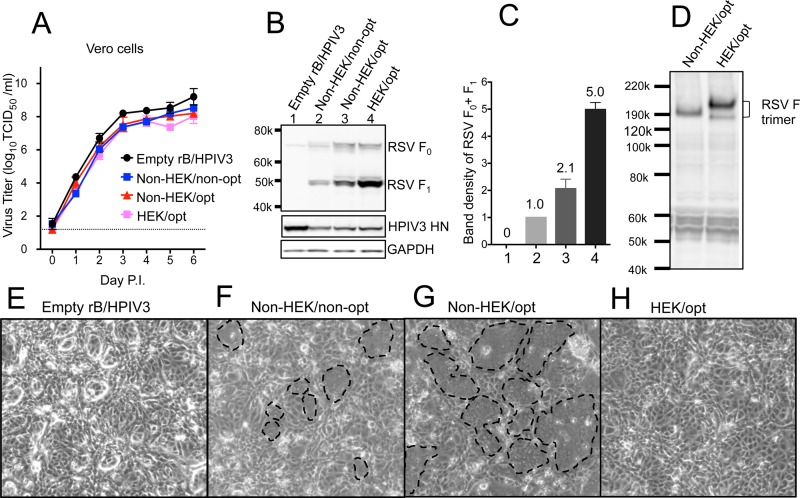
The RSV F sequence (strain A2) was codon optimized (GeneArt; Life Technologies) for human expression and, in addition, was modified by two amino acid substitutions (K66E and Q101P) to be identical in amino acid sequence to an early passage of strain A2 that originally had been propagated in human embryonic kidney cells (HEK F protein) (18). Three versions of the RSV F gene—(i) nonoptimized with non-HEK assignments (non-HEK/non-opt), a construct similar to MEDI-534; (ii) codon optimized with non-HEK assignments (non-HEK/opt); and (iii) codon optimized with HEK assignments (HEK/opt)—were inserted at the 2nd position between the N and P genes of the rB/HPIV3 vector (Fig. 1). Infectious virus was recovered by reverse genetics, as described in Materials and Methods. Multicycle replication growth curves in Vero (Fig. 2A) and LLC-MK2 (not shown) cells showed that all three constructs replicated with similar kinetics and to high peak titers (∼108 TCID50/ml), although they were all slightly more attenuated than the empty vector (Fig. 2A). Virus stocks of these constructs contained only trace amounts of RSV F, indicating that it was not efficiently packaged into the vector particles (not shown).
FIG 2.
Effects of codon optimization and HEK assignments. (A) Multicycle growth kinetics of rB/HPIV3 vectors in African green monkey Vero cells at 32°C. Triplicate Vero cell monolayers were infected with the indicated rB/HPIV3 vectors at a multiplicity of infection (17) of 0.01 TCID50/cell. Samples were collected at 24-h intervals. Virus titers were determined by serial dilution in LLC-MK2 cells (23) and are expressed as means with standard errors of the mean (SEM) (40). (B) Western blot analysis of RSV F expression in Vero cells, with vector HN protein and cellular GAPDH protein analyzed as controls. Vero cells were infected at an MOI of 10 TCID50/cell at 32°C with the indicated rB/HPIV3 vectors. Total lysates were harvested at 48 h p.i. and subjected to gel electrophoresis under reducing and denaturing conditions, followed by Western blotting, as described previously (23). RSV F0 (70 kDa) is the primary translation product of the F ORF, and RSV F1 (47 kDa) is the larger subunit created when RSV F0 is activated by cleavage. (C) Quantification of RSV F expression. The RSV F1 and F0 band densities (from panel B) were quantified and normalized to that of the Non-HEK/non-opt sample shown at a density value of 1. The means of four independent experiments are shown (numbers above the bars) with the SEM. The numbers on the x axis represent the lanes in Fig. 2B. (D) Mobility of RSV F trimers in polyacrylamide gel electrophoresis under nonreducing conditions. RSV F trimers were detected by Western blotting with rabbit polyclonal antibodies generated by immunizing rabbits with sucrose-purified wt RSV particles. (E to H) Formation of syncytia on Vero cell monolayers infected at an MOI of 10 TCID50/cell with the indicated rB/HPIV3 vectors. The cells were incubated at 32°C for 48 h and photographed using a light microscope at a magnification of ×10. Representative syncytia are marked with dashed lines in panels F and G.
Codon optimization and the HEK substitutions conferred 2.1- and 2.4-fold increases in RSV F protein expression, respectively, with an aggregate 5-fold increase (Fig. 2B and C). Also, following the HEK substitutions, the F protein trimer migrated somewhat more slowly than the non-HEK form under nonreducing polyacrylamide gel electrophoresis (Fig. 2D). The non-HEK form of RSV F induced abundant syncytia on Vero monolayers (Fig. 2F), and increasing its expression by codon optimization resulted in more extensive syncytia (Fig. 2G). Interestingly, the HEK form of RSV F conferred a hypofusogenic phenotype (Fig. 2H), despite having the highest level of expression (Fig. 2B and C, lane 4).
Expression of stabilized prefusion and postfusion forms of RSV F.
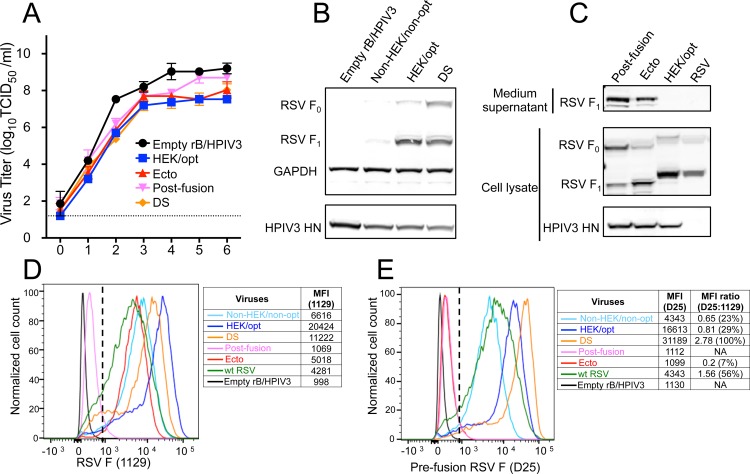
The full-length RSV F protein, (i.e., including the C-terminal transmembrane [TM] and cytoplasmic [CT] domains that anchor it in the membrane) was stabilized in the prefusion form (Fig. 1, DS) by introducing two mutations (S155C and S290C) that generate a new disulfide bond, as previously described (17). Stabilized postfusion RSV F protein was generated as previously described by expressing the ectodomain (aa 1 to 513, lacking the C-terminal TM and CT domains) with further deletion of the first 10 amino acids of the FP (aa 137 to 146) (Fig. 1, Postfusion) (12, 13). Another construct consisting of the entire RSV F ectodomain (aa 1 to 513) was included for comparison (Fig. 1, Ecto). All three forms of RSV F were codon optimized and had HEK assignments.
rB/HPIV3 expressing the prefusion DS, postfusion, and Ecto forms of RSV F protein replicated in Vero cells (Fig. 3A) and LLC-MK2 cells (not shown) at rates and to final titers similar to those of a vector expressing native RSV F protein (HEK/opt). None of these constructs induced visible syncytium formation in vitro (not shown), consistent with the idea that F protein that is not anchored in the membrane or that is stabilized in the prefusion form should not be efficient in directing fusion. The prefusion DS form was expressed at a level similar to that of HEK/opt (Fig. 3B). Cleavage of the prefusion DS F0 precursor was somewhat less efficient than that of HEK/opt. As expected, the postfusion and Ecto constructs were expressed primarily as secreted forms (Fig. 3C), and only the cleaved protein was detected in the medium supernatant. None of the forms was efficiently packaged into vector particles (not shown).
FIG 3.
Expression of prefusion DS, postfusion, and Ecto forms of RSV F. (A) Multicycle replication kinetics of the indicated rB/HPIV3 vectors in Vero cells at 32°C. Titration was carried out as described for Fig. 2A. (B) Expression of the prefusion DS form of RSV F. Vero cells were infected with the indicated rB/HPIV3 vectors at an MOI of 10 TCID50/cell and incubated at 37°C for 48 h. Western blot analysis was performed as described for Fig. 2B. (C) Expression and secretion of postfusion, Ecto, and HEK/opt forms of RSV F. Vero cell monolayers were infected with the indicated vectors at an MOI of 10 TCID50/cell or with wt RSV (A2) at an MOI of 10 PFU/cell and incubated at 32°C for 48 h. Western blot analysis was carried out as described for Fig. 2B. (D and E) Cell surface expression of the total and prefusion RSV F protein. Vero cells were infected with the indicated vectors at an MOI of 5 TCID50/cell or with wt RSV at an MOI of 5 PFU/cell and incubated at 32°C. At 48 h p.i., the unpermeabilized cells were stained with RSV F antibody 1129 (D), which reacts with both prefusion and postfusion F, or antibody D25 (E), which is specific for prefusion F. The x axis shows the intensity of RSV F expression, and the y axis is the percentage of the cell count normalized to the maximum count (100%) in a distribution, with the MFI values shown on the right of each histogram. Gating of live RSV F-positive cells used for analysis is indicated with a dashed line.
Analysis of cell surface expression by flow cytometry using monoclonal antibody 1129 (the murine precursor of palivizumab, which recognizes both prefusion and postfusion F protein) demonstrated very efficient surface expression of the HEK/opt and prefusion DS forms of RSV F (Fig. 3D). Consistent with its lower total expression versus HEK/opt (Fig. 2B and C), the non-HEK/non-opt form had relatively low surface expression (Fig. 3D). The Ecto form was detected at the cell surface at a reduced but still substantial level, whereas the postfusion form was not detected above background (Fig. 3D). The difference in cell surface accumulation between these two secreted forms might reflect the presence of the hydrophobic FP domain in Ecto, which might mediate association with the plasma membrane. Except for the postfusion and Ecto forms, the cell surface expression of RSV F protein by the rB/HPIV3 vectors was more efficient than that by wt RSV (Fig. 3D).
The relative quantity of RSV F protein in the prefusion conformation on the cell surface was measured with human monoclonal antibody D25, which recognizes antigenic site Ø, which is unique to the prefusion conformation and some intermediate forms (11). The greatest cell surface expression of F protein in the prefusion conformation was observed with the DS construct; a smaller amount was observed with HEK/opt (Fig. 3E), even though the latter was more efficiently expressed on the surface, as detected by antibody 1129 (Fig. 3D). This was illustrated by calculating the ratio of the median fluorescence intensity (MFI) value for D25 to that for 1129 (Fig. 3E). This suggested that HEK/opt, i.e., a native form of the RSV F protein, was only partially displayed in the prefusion conformation on the cell surface. The presence of the HEK assignments (i.e., HEK/opt versus non-HEK/non-opt) resulted in a marginal increase in the D25/1129 ratio. Essentially no secretory postfusion form was detected at the cell surface with either antibody 1129 (Fig. 3D) or D25 (Fig. 3E). Also, essentially no Ecto form was detected with antibody D25 (Fig. 3E), suggesting that this form was highly susceptible to triggering. F expressed by wt RSV (strain A2) was also detected in the prefusion conformation (Fig. 3E), albeit in a smaller amount than DS and HEK/opt, likely because the total expression of F by wt RSV was lower (Fig. 3D).
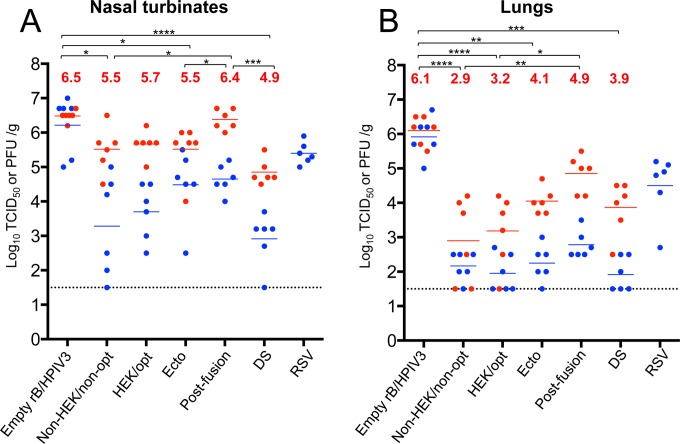
Replication of rB/HPIV3 vectors in the respiratory tract of hamsters.
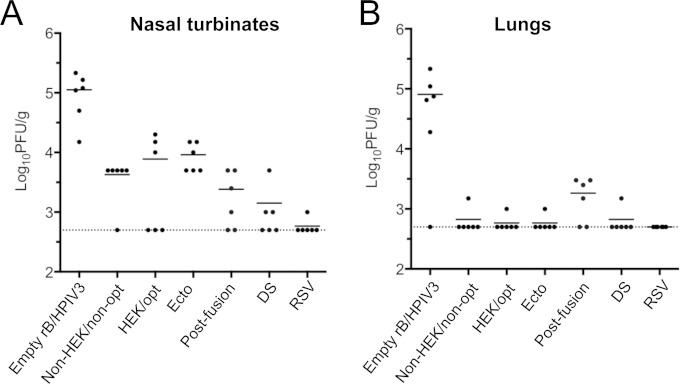
Hamsters were infected i.n. with 105 TCID50 of each vector or with 106 PFU of wt RSV (A2). Replication in the upper and lower respiratory tracts was determined by titrating the viral load in nasal turbinates and lungs on days 3 and 5 (only day 3 for RSV), and samples of the input inocula also were analyzed to confirm the input titer. Compared with the empty vector, vectors bearing an RSV F insert replicated more slowly (i.e., with lower titers on day 3 versus day 5) and were moderately more attenuated in the nasal turbinates (Fig. 4A) and substantially more attenuated in the lungs (Fig. 4B) on both days 3 and 5. The difference between the empty vector and the vector bearing an RSV F insert was the greatest with the DS construct on day 3, where the reduction was more than 3.0 log10 units in the nasal turbinates and 4.0 log10 units in the lungs. There was a single time point at which a vector bearing RSV F was not more attenuated than the empty vector, and that was for the construct expressing postfusion F on day 5 in the nasal turbinates. Among the various vectors, there was some variability in titer, but the differences were significant only in the case of vector expressing postfusion F, which replicated to higher titers in both the nasal turbinates and lungs. It may be that this insert was more easily tolerated by the vector because it was secreted and had minimal association with the infected cells, as shown in Fig. 3. Wt RSV replicated to 100- to 1,000-fold-higher titers than the various vectors expressing RSV F protein, particularly in the lungs, which would be relevant to its relative immunogenicity (see below).
FIG 4.
Replication of rB/HPIV3-RSV-F vectors in hamsters. Golden Syrian hamsters were infected i.n. with 105 TCID50 of the indicated rB/HPIV3 vectors or 106 PFU of wt RSV (A2) in a 0.1-ml inoculum. Hamsters were euthanized (n = 6 per virus per day) on days 3 and 5 postinfection, the nasal turbinates (A) and lungs (B) were collected and homogenized, and the viral titers were determined by limiting dilution on LLC-MK2 (rB/HPIV3 vectors) or Vero (RSV) cells at 32°C. The blue and red dots indicate the titers for individual animals euthanized on days 3 and 5, respectively, and the mean group titer is indicated by a blue or red horizontal line for days 3 and 5, respectively. The mean values of day 5 titers are also shown as red numbers. The limit of detection (LOD) is 1.5 log10 TCID50/g of tissue, indicated with a dotted line. The statistical significance of differences among peak titers on day 5 was determined by one-way analysis of variance (ANOVA) with a Tukey-Kramer test and is indicated by asterisks: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Prefusion RSV F induced highly potent RSV-neutralizing serum antibodies.
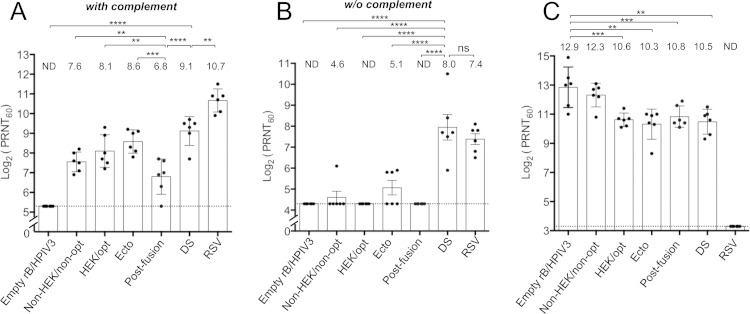
Hamsters were infected with the various vectors or with wt RSV, and sera were collected 28 days later and analyzed for RSV-neutralizing antibodies by a PRNT60 performed in the (i) presence (Fig. 5A) and (ii) absence (Fig. 5B) of added complement. The presence of added complement provides more sensitive detection of virus-specific antibodies, since complement potentially confers viral-lysis and steric-hindrance capabilities (34) to both neutralizing and nonneutralizing antibodies, whereas the complement-independent assay detects only antibodies that directly neutralize the virus.
FIG 5.
RSV- and HPIV3-neutralizing serum antibody titers induced by rB/HPIV3-RSV F vectors. Hamsters (n = 6 per vector) were infected as described for Fig. 4, and serum samples were collected at 28 days postimmunization. (A and B) RSV-neutralizing antibody titers were determined by a PRNT60 performed on Vero cells at 37°C with (A) and without (B) added guinea pig complement. (C) HPIV3-neutralizing antibody titers were determined by a PRNT60 performed on LLC-MK2 cells at 32°C with added guinea pig complement (23). The height of each bar represents the mean titer, which is shown above the bar; the SEM is indicated by the error bars, and the values for individual animals are shown as dots. The statistical significance of differences among groups was determined as described for Fig. 4; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, P > 0.05. The detection limit is indicated with a dotted line. ND, the neutralization titer was below the detection limit.
For the complement-containing assay (Fig. 5A), the most interesting observations were that a vector expressing postfusion F was less immunogenic than the other vectors, even though it replicated to the highest titers in hamsters (Fig. 4), while the vector expressing the prefusion DS form of F induced the highest titer of RSV-neutralizing antibodies among the vectors (Fig. 5A). In the complement-independent assay (Fig. 5B), only two viruses induced similar high titers of potent RSV-neutralizing serum antibodies: vectors expressing prefusion DS RSV F and wt RSV. The observation that the RSV-neutralizing antibodies induced by these two viruses were similar in magnitude is noteworthy, because the vector replicated 100 to 1,000 times less efficiently than wt RSV (Fig. 4), and in addition, the neutralizing activity conferred by wt RSV would have an additional contribution from the RSV G protein. In this regard, it is clear that RSV G is an important independent neutralization and protective antigen in this model, because a previous study comparing rB/HPIV3 expressing RSV F versus rB/HPIV3 expressing RSV G showed that they were statistically indistinguishable with regard to the induction of RSV-neutralizing serum antibodies and protection against RSV challenge (22). Thus, the vector expressing the prefusion DS form of RSV F induced an RSV-specific antibody response that was both quantitatively and qualitatively greater than that of the other vectors and that was very similar to that of wt RSV.
The induction of HPIV3-neutralizing serum antibodies was evaluated by a PRNT60 in the presence of complement. This showed that all of the vectors induced high antibody titers. The titer was greatest for the empty vector and generally was somewhat lower for vectors bearing the RSV F gene (Fig. 5C), likely reflecting the reduced replication of the constructs (Fig. 4).
Protection against RSV challenge.
The immunized hamsters from the experiment shown in Fig. 5 were challenged 30 days postimmunization with 106 PFU of wt RSV administered i.n. The viral loads in the nasal turbinates and lungs on day 3 after challenge were determined by plaque titration of tissue homogenates. In the nasal turbinates (Fig. 6A), the construct expressing the prefusion DS form of RSV F was the most protective of the vectors, although the difference was not significant. In the lungs (Fig. 6B), each of the vectors expressing RSV F completely restricted RSV replication in 5/6 animals in each group, with the exception of the postfusion construct, which was less restrictive. Wt RSV conferred nearly complete resistance in the nasal turbinates and complete resistance in the lungs, but this high level of protection was aided by three factors: (i) wt RSV replicated up to 1,000-fold more efficiently than the attenuated rB/HPIV3-RSV-F vectors; (ii) wt RSV expressed both RSV neutralization antigens, G and F; and (iii) wt RSV expressed all of the other RSV proteins as potential antigens for cellular immunity, which is known to be effective in protection in short-term RSV challenge studies (35).
FIG 6.
Protection of immunized hamsters against wt RSV challenge. The hamsters (n = 6 per vector) that had been immunized as shown in Fig. 5 were challenged i.n. on day 30 postimmunization with 106 PFU of wt RSV (A2) in a 0.1-ml inoculum. On day 3 postchallenge, the hamsters were euthanized, and nasal turbinates (A) and lungs (B) were collected. The RSV titers in tissue homogenates were determined by plaque assay on Vero cells at 37°C. Each symbol represents the RSV titer for an individual animal, and the mean viral titers of the groups are shown as horizontal lines. The detection limit of the assay was 2.7 log10 PFU/g of tissue, indicated by a dotted line.
Stability of RSV F protein expression during replication in vivo.
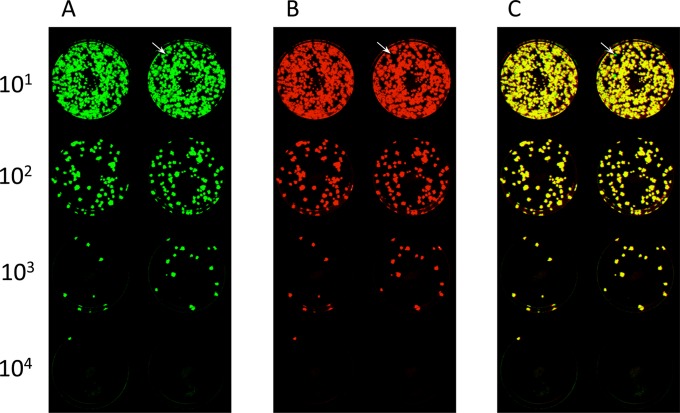
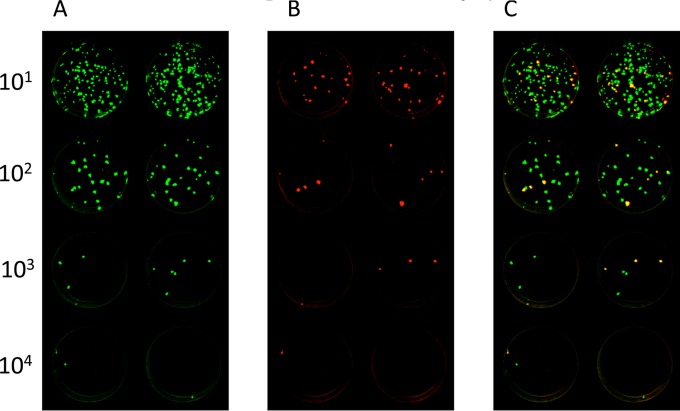
The stability of expression of RSV F protein during replication in vitro and in vivo was evaluated by examining the virus stocks used for immunization, as well as the homogenates of hamster nasal turbinates and lungs from days 3 and 5, from the study shown in Fig. 4. This was done using a fluorescence double-staining plaque assay to simultaneously detect the RSV F and HPIV3 proteins. The results are summarized in Table 1, and fluorographs of plaque staining for two representative in vivo specimens are shown in Fig. 7 and 8. In general, the expression of unmodified RSV F protein (non-HEK/non-opt) was substantially stable, and the stability of RSV F expression was not affected by modifications (DS, postfusion, or Ecto) affecting its structure or level of expression. For the viral stocks, all of the virus plaques present in wells in which individual plaques could be enumerated were positive for expression of RSV F and were scored as 100% (Table 1). The same was true for the majority of tissue homogenate samples (108 out of 120) (Table 1). Note that in some of the lower dilutions that contained too many plaques to clearly distinguish and enumerate, occasional green plaques could be distinguished, suggesting a very low background of virus that did not express RSV F (e.g., Fig. 7). In the other specimens (11 out of 120), there was a greater loss of RSV F expression, usually <10% of all plaques (Table 1). Only a single sample (hamster number 511, nasal turbinate, day 3 [Table 1 and Fig. 8]) had 14% RSV F expression, whereas the lung specimen from the same animal had 100% expression (Table 1). There was no evidence that loss of RSV F expression increased progressively over time, so neither enhanced expression nor structural stabilization of RSV F reduced the genetic stability of the RSV F insert in the vector during replication in vivo.
TABLE 1.
Percentages of vaccine virus plaques that expressed RSV F after replication in vivo
| Virus | Inoculum | Day 3 |
Day 5 |
||||
|---|---|---|---|---|---|---|---|
| Hamster no.a | % of vaccine virus plaques expressing RSV Fb |
Hamster no.a | % of vaccine virus plaques expressing RSV Fb |
||||
| Nasal turbinates | Lungs | Nasal turbinates | Lungs | ||||
| Non-HEK/non-opt | 100 | 469 | NAc | NA | 385 | 100 | 100 (n = 1)d |
| 470 | 100 | 100 (n = 1) | 386 | 100 | 100 (n = 5) | ||
| 471 | 100 | 100 (n = 1) | 387 | 99 | 100 | ||
| 472 | 100 (n = 1) | 100 (n = 1) | 388 | 97 | 100 | ||
| 473 | NA | 100 (n = 1) | 389 | 100 | 100 (n = 1) | ||
| 474 | 100 | 100 (n = 1) | 390 | 100 | NA | ||
| Non-HEK/opt | 100 | 475 | 100 | NA | 391 | 100 | 100 (n = 2) |
| 476 | 100 (n = 5) | 100 (n = 5) | 392 | 100 | 100 (n = 2) | ||
| 477 | 100 (n = 5) | NA | 393 | 100 | 100 | ||
| 478 | 98 | NA | 394 | 98 | 100 | ||
| 479 | 100 (n = 5) | NA | 395 | 97 | 100 | ||
| 480 | 100 (n = 5) | NA | 396 | 100 | 100 | ||
| HEK/opt | 100 | 481 | 100 | NA | 397 | 100 | NA |
| 482 | 100 (n = 5) | NA | 398 | 100 | NA | ||
| 483 | 100 | NA | 399 | 100 | 100 | ||
| 484 | 100 | NA | 400 | 100 | 100 (n = 4) | ||
| 485 | 100 (n = 2) | 100 (n = 1) | 401 | 100 | 100 | ||
| 486 | 100 (n = 7) | 100 (n = 3) | 402 | 100 | 100 | ||
| Ecto | 100 | 511 | 14 | 100 (n = 4) | 427 | 100 (n = 4) | 100 (n = 8) |
| 512 | 100 | NA | 428 | 100 | 92 | ||
| 513 | 100 | NA | 429 | 100 | 100 | ||
| 514 | 100 | 100 (n = 1) | 430 | 100 | 100 | ||
| 515 | 100 (n = 3) | NA | 431 | 98 | 100 | ||
| 516 | 100 | 100 (n = 5) | 432 | 98 | 100 | ||
| Postfusion | 100 | 517 | 100 | NA | 433 | 100 | 100 |
| 518 | 100 | 100 (n = 8) | 434 | 100 | 100 | ||
| 519 | 100 (n = 6) | 100 (n = 6) | 435 | 100 | 100 | ||
| 520 | 100 | 100 (n = 3) | 436 | 100 | 100 | ||
| 521 | 98 | 100 (n = 8) | 437 | 100 | 100 | ||
| 522 | 100 | 100 | 438 | 100 | 100 | ||
| DS | 100 | 523 | NA | 100 (n = 1) | 439 | 98 | NA |
| 524 | 100 | NA | 440 | 100 | 100 | ||
| 525 | 100 (n = 2) | 100 (n = 1) | 441 | 100 | 100 | ||
| 526 | 100 | NA | 442 | 100 | 100 (n = 1) | ||
| 527 | 100 | NA | 443 | 100 | 100 | ||
| 528 | 100 (n = 1) | 100 (n = 1) | 444 | 95 | 100 (n = 3) | ||
Hamster numbers were assigned consecutively to six hamsters in each group.
The percentage of RSV F-expressing viruses was determined by double-staining plaque assay. It was calculated using the wells on which distinct plaques could be enumerated (i.e., <50 plaques per well [Fig. 7]).
NA, no plaque was obtained from the sample due to low titer.
Samples with fewer than 10 plaques are marked “(n = x),” where x is their total number of plaques.
FIG 7.
Example of fluorescence double-staining plaque titration of a hamster nasal turbinate homogenate. Vero cells were inoculated with a 10-fold serially diluted homogenate of nasal turbinates of hamster number 428 (Table 1), which had been infected with rB/HPIV3 expressing the Ecto form of RSV F. The cells were incubated under a methylcellulose overlay and subjected to double staining for RSV F and HPIV3 antigens. The dilution factors are shown on the left. Shown are staining of duplicate wells for HPIV3 antigens (green) (A) and RSV F (red) (B) (40) and a merged image (plaques expressing RSV F are yellow; plaques that express only HPIV3 antigens are green) (C). The arrows point to a plaque that did not express RSV F in a lower-dilution well. In panel C, there were no green plaques at the dilutions for which individual plaques could be counted (102, 103, and 104), and hence, this sample was scored “100%” for RSV F expression (Table 1), even though sporadic green plaques (e.g., the arrow in panel C) were detected against the yellow background in the 101 dilution.
FIG 8.
Example of fluorescence double staining of plaques from a hamster nasal turbinate homogenate with only 14% of recovered vaccine viruses expressing RSV F. Homogenates of nasal turbinates from hamster number 511 (Table 1), which had been infected with rB/HPIV3 expressing the Ecto form of RSV F, were prepared as a 10-fold dilution series, inoculated onto LLC-MK2 cells, incubated under a methylcellulose overlay, and subjected to double staining for RSV F and HPIV3 antigens as described for Fig. 7. The dilution factors are shown on the left. Shown are staining of duplicate wells for HPIV3 antigens (green) (A) and RSV F (red) (40) (B) and a merged image (plaques expressing RSV F are yellow; plaques that express only HPIV3 antigens are green) (C).
DISCUSSION
RSV is a major pediatric respiratory pathogen causing a significant disease burden worldwide, particularly in developing countries. Despite decades of effort, a licensed RSV vaccine is still elusive. Live attenuated RSV strains and live attenuated viral vectors expressing RSV protective antigens (especially the F protein) are currently being developed as intranasal vaccines for infants and children (reviewed in references 36, 37, and 38). The B/HPIV3 vector expressing the RSV F protein, as evaluated in the present study, has a number of potential advantages compared to a live-attenuated RSV vaccine strain. As noted (see the introduction), the vector provides a bivalent vaccine against RSV and HPIV3, the latter virus being second in importance only to RSV as a worldwide viral agent of severe pediatric respiratory tract diseases. Additionally, the use of a PIV vector avoids the poor replication, large filaments, and poor stability characteristic of RSV, which would greatly facilitate vaccine manufacture and distribution and would greatly facilitate extending RSV vaccines to developing countries. A vectored RSV vaccine approach enables immunization with a stabilized form of the optimal RSV antigen, the highly immunogenic prefusion form of RSV F. Stabilized prefusion F cannot be expressed as part of a live attenuated RSV strain because it is nonfunctional in fusion. Stabilized prefusion F protein may provide better-than-nature priming during the first exposure to RSV antigen in life, in contrast to the metastable native RSV F protein, which induces a substantial content of suboptimal antibodies (15).
Also, in experimental animals, immunization with PIV-vectored F protein delivered as a boost following primary immunization with a live attenuated RSV strain was more immunogenic than a second dose of the live attenuated vaccine (unpublished observations). Conversely, one disadvantage of a PIV vector expressing the RSV F protein is that it provides only one of the two RSV neutralization and major protective antigens and lacks all of the other RSV proteins as potential antigens for cellular immunity.
Recent clinical evaluation of a version of the rB/HPIV3 vector expressing an unmodified RSV F insert (MEDI-534) indicated that it was infectious, well tolerated, and safe in seronegative children but displayed poor immunogenicity for RSV F (evaluated in the absence of complement) and exhibited evidence of substantial instability of RSV F expression (7). Therefore, current efforts are focused on improving the immunogenicity and enhancing the expression and stability of RSV F in this vector.
When the RSV F amino acid sequence was modified by the HEK assignments to be identical to that of an early passage of the same strain, the F protein had a slight decrease in F trimer gel mobility and acquired a hypofusogenic phenotype. It also exhibited an ∼2-fold increase in accumulation that might reflect increased stability. A similar effect on fusion activity involving one of the HEK assignments (position 66) was observed previously in one version of the live attenuated RSV vaccine candidate ΔM2-2 (39). However, that mutation also caused a substantial reduction in RSV growth, and it was unclear whether the reduced syncytium formation was due directly to the mutation or was a consequence of reduced growth. The present study demonstrates a direct effect on syncytium formation. Position 66 in RSV F is part of the site Ø epitope in the prefusion structure, and position 101 is inside the trimer cavity and just at the end of an alpha helix at the C terminus of the F2 chain that dips into the opening of the cavity (11). Mutation at either site might affect the efficiency of conformational change during fusion. A GenBank search revealed that most RSV clinical isolates have the HEK assignments (data not shown), indicating they are representative of circulating RSV. We suggest that the presence of the HEK assignments in early-passage RSV strain A2, and their substitution in later-passage strain A2, reflects an adaptation to cell culture that resulted in a hyperfusogenic phenotype and increased growth. While the HEK version of RSV F may be a closer match to circulating strains, its effect of reducing RSV replication probably would make it unsuitable for inclusion in a live attenuated RSV strain, since impaired growth in vitro would complicate vaccine manufacture. However, HEK F protein can be readily accommodated as a passenger in a heterologous vector, such as rB/HPIV3.
Codon optimization provided a modest increase in expression that was additive to that of the HEK mutations, resulting in an aggregate 5-fold increase in expression. However, this did not result in significant increases in RSV-neutralizing serum titers or protective efficacy in the hamster model. Increased expression of viral antigen typically provides increased immunogenicity (40, 41), and we previously observed that a 30- to 69-fold increase in the expression of RSV F from position 1 or 2 versus 6 in the rB/HPIV3 vector induced significant increases in protective efficacy in hamsters (23). It may be that any increase in immunogenicity due to the modest increase in antigen expression in the present study was simply too small to be reliably detected. It also may be that codon optimization for human expression might not be effective in hamsters. Whether this 5-fold increase in expression and the presence of the HEK assignments might confer increased immunogenicity and protection in a more permissive primate host, or against circulating virus bearing HEK assignments, remains to be seen.
As a subunit vaccine evaluated in experimental animals, prefusion DS RSV F protein induced higher titers of neutralizing antibodies than the postfusion form (17). However, it was not clear if in vivo expression of a stabilized prefusion form would be advantageous, since unmodified F protein expressed in vivo presumably is primarily in the prefusion form. Also, whether these novel forms could be efficiently expressed by a live viral vector, and what effects they might have on vector attenuation, insert stability, immunogenicity, and other factors, were unknown. We found that all of the inserts were well tolerated and efficiently expressed by the rB/HPIV3 vector and that prefusion DS F was the most immunogenic and protective among the vectored forms of F. Surprisingly, prefusion DS RSV F was the only vectored form that induced a high titer of RSV-neutralizing antibodies detected in the complement-independent assay, which also was observed with wt RSV. The titers of these “high-quality” antibodies were similar for prefusion DS F and wt RSV, which was remarkable, given that the vector replicated 100- to 1,000-fold less efficiently than wt RSV in vivo and expressed only one of the two RSV neutralization antigens.
In contrast, the postfusion form of F was very inefficient in inducing neutralizing antibodies detected by the complement-independent assay and also was the least immunogenic form in the complement-containing assay. The postfusion form also allowed the greatest challenge virus replication in the lower respiratory tract. These findings indicate that, despite its high level of stability (42), the postfusion form is poorly immunogenic, particularly for high-quality RSV-neutralizing antibodies. The possibility that this might be due to its secreted expression seems unlikely, because the Ecto form, which also is secreted and thus serves as a control, did not have similar reductions in immunogenicity and protection. Therefore, the postfusion conformation appears to have reduced immunogenicity and efficacy when expressed from a vector.
It was somewhat surprising that all of the vectored forms of F other than prefusion DS were very inefficient in inducing neutralizing antibodies detected by the complement-independent assay, in contrast to wt RSV. This could not be attributed solely to their inefficient expression of prefusion F protein. For example, the HEK/opt construct expressed a substantial level of cell surface prefusion RSV F protein yet did not induce RSV-neutralizing serum antibodies detected by the complement-independent assay. One possible explanation is that, because vector-expressed F protein is not actively packaged into the vector particle and is not secreted, it presumably depends on necrosis or apoptosis to gain exposure to the immune system, and it may be that the F protein becomes substantially denatured or triggered during the time needed for this to occur. This would be countered by stabilization of the prefusion conformation by the DS mutation. In the case of wt RSV, the F protein may be somewhat stabilized by the presence of other viral structural proteins (such as G or M), and it may be that packaging of the F protein into the RSV virion provides a more rapid and efficient means of antigen presentation in a native state. It also is noteworthy that the non-HEK/non-opt construct is very similar to the MEDI-534 construct that was previously evaluated in clinical studies and induced low titers of RSV-neutralizing antibodies in young children when quantified by a complement-independent assay (7). The present results suggest that (i) the immunogenicity of MEDI-534 might appear substantially greater if the assays were performed in the presence of complement (as is commonly done in pediatric RSV vaccine trials [36]) and (ii) the prefusion DS construct provides a much more immunogenic vaccine candidate.
Despite inducing serum RSV-neutralizing antibodies at levels similar to that of wt RSV, the prefusion DS construct provided marginally lower protection against wt RSV challenge in the upper respiratory tract than the wt RSV control. The marginally greater protection induced by wt RSV may reflect a contribution from the RSV G protein, which is the other major RSV neutralization and protective antigen. For example, in an earlier study, rB/HPIV3 vectors that expressed RSV G or RSV F were indistinguishable in the levels of RSV-neutralizing serum antibodies and protection against RSV challenge induced in the hamster model (22), confirming that G is an important neutralization and protective antigen in this system. Another factor is that wt RSV expresses all of the other RSV proteins, providing additional antigens for stimulating cellular responses that can be highly protective in short-term studies in rodents. Nonetheless, expression of the DS form of the F protein by the rB/HPIV3 vector was almost as protective as wt RSV expressing all of the viral antigens, even though (as noted) the latter virus was administered at a 10-fold-higher dose and replicated to up to a 1,000-fold-higher titer. The ability of the DS construct to induce levels of neutralizing antibodies and protection comparable to those of wt RSV despite all the factors mentioned above clearly indicates the superior performance of the DS form of RSV F as a neutralization and protective antigen.
As noted (see the introduction), the previously evaluated MEDI-534 vaccine had evidence of substantial instability of the RSV F insert during replication in young children that likely reduced its immunogenicity. Analysis of shed vaccine virus demonstrated (i) mutations in the transcription termination sequence preceding the RSV F insert expected to promote read-through transcription and downregulate translation of RSV F and (ii) mutations introducing stop codons within the RSV F ORF, also expected to reduce F expression. In addition, 2.5% of the MEDI-534 clinical trial material had lost expression of RSV F during preparation in vitro. Our experience in this study and a previous study (23) is that some vector preparations indeed can have a substantial background of virus that fails to express RSV F but that this appears to be a sporadic occurrence, and most preparations have a very low background, as observed in the present study. Similarly, there occasionally was evidence of loss of expression in vivo in a small subset of specimens. There may be selective pressure favoring amplification of vector that has lost the expression of RSV F, but this appears to be a minor effect, and there was not a progressive accumulation. None of the manipulations in the present or previous study appeared to increase the frequency of inactivation of expression of the RSV F insert. Thus, while loss of expression of RSV F occurs (and certainly would be expected given the high error rate of this type of virus), it is at a relatively low level. Instability probably can be controlled by placing RSV F at the first position before the BPIV3 N gene so that no gene end signal precedes it. Also, careful monitoring can detect and exclude the sporadic loss of expression in virus preparations.
In summary, we modified the rB/HPIV3-RSV F vaccine—a candidate that has previously been in clinical trials—by codon optimization, by changing the RSV F amino acid sequence to be identical to an early passage of strain A2, and by expressing stabilized prefusion and postfusion forms. The prefusion DS construct was unique among the vectors in inducing a remarkably high titer of RSV-neutralizing antibodies detected by a complement-independent assay, in sharp contrast to a construct that is very similar to the previously evaluated MEDI-534 vaccine candidate. The prefusion construct should be further developed as a pediatric RSV/HPIV3 vaccine candidate.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIAID, NIH.
We thank Ursula J. Buchholz for advice and for kindly providing wt RSV for hamster studies and Fatemeh Davoodi for technical assistance.
REFERENCES
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottolini MG, Porter DD, Hemming VG, Prince GA. 2000. Enhanced pulmonary pathology in cotton rats upon challenge after immunization with inactivated parainfluenza virus 3 vaccines. Viral Immunol 13:231–236. doi: 10.1089/vim.2000.13.231. [DOI] [PubMed] [Google Scholar]
- 3.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. [DOI] [PubMed] [Google Scholar]
- 4.Connors M, Collins PL, Firestone CY, Sotnikov AV, Waitze A, Davis AR, Hung PP, Chanock RM, Murphy BR. 1992. Cotton rats previously immunized with a chimeric RSV FG glycoprotein develop enhanced pulmonary pathology when infected with RSV, a phenomenon not encountered following immunization with vaccinia-RSV recombinants or RSV. Vaccine 10:475–484. doi: 10.1016/0264-410X(92)90397-3. [DOI] [PubMed] [Google Scholar]
- 5.Belshe RB, Newman FK, Tsai TF, Karron RA, Reisinger K, Roberton D, Marshall H, Schwartz R, King J, Henderson FW, Rodriguez W, Severs JM, Wright PF, Keyserling H, Weinberg GA, Bromberg K, Loh R, Sly P, McIntyre P, Ziegler JB, Hackell J, Deatly A, Georgiu A, Paschalis M, Wu SL, Tatem JM, Murphy B, Anderson E. 2004. Phase 2 evaluation of parainfluenza type 3 cold passage mutant 45 live attenuated vaccine in healthy children 6–18 months old. J Infect Dis 189:462–470. doi: 10.1086/381184. [DOI] [PubMed] [Google Scholar]
- 6.Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O'Shea AF, Gruber WC, Murphy BR. 2007. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein D, Malkin E, Abughali N, Falloon J, Yi T, Dubovsky F. 2012. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr Infect Dis J 31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt AC, McAuliffe JM, Huang A, Surman SR, Bailly JE, Elkins WR, Collins PL, Murphy BR, Skiadopoulos MH. 2000. Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates. J Virol 74:8922–8929. doi: 10.1128/JVI.74.19.8922-8929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C-F, Wang CK, Malkin E, Schickli JH, Shambaugh C, Zuo F, Galinski MS, Dubovsky F, Tang RS. 2013. Implication of respiratory syncytial virus (RSV) F transgene sequence heterogeneity observed in phase 1 evaluation of MEDI-534, a live attenuated parainfluenza type 3 vectored RSV vaccine. Vaccine 31:2822–2827. doi: 10.1016/j.vaccine.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Calder LJ, Gonzalez-Reyes L, Garcia-Barreno B, Wharton SA, Skehel JJ, Wiley DC, Melero JA. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 11.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A 108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakurai H, Williamson RA, Crowe JE, Beeler JA, Poignard P, Bastidas RB, Chanock RM, Burton DR. 1999. Human antibody responses to mature and immature forms of viral envelope in respiratory syncytial virus infection: significance for subunit vaccines. J Virol 73:2956–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A 109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR. 1998. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol 72:4467–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luongo C, Winter CC, Collins PL, Buchholz UJ. 2013. Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature. J Virol 87:1985–1996. doi: 10.1128/JVI.02769-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci U S A 92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall SL, Stokes A, Tierney EL, London WT, Belshe RB, Newman FC, Murphy BR. 1992. Cold-passaged human parainfluenza type 3 viruses contain ts and non-ts mutations leading to attenuation in rhesus monkeys. Virus Res 22:173–184. doi: 10.1016/0168-1702(92)90049-F. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt AC, McAuliffe JM, Murphy BR, Collins PL. 2001. Recombinant bovine/human parainfluenza virus type 3 (B/HPIV3) expressing the respiratory syncytial virus (RSV) G and F proteins can be used to achieve simultaneous mucosal immunization against RSV and HPIV3. J Virol 75:4594–4603. doi: 10.1128/JVI.75.10.4594-4603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang B, Munir S, Amaro-Carambot E, Surman S, Mackow N, Yang L, Buchholz UJ, Collins PL, Schaap-Nutt A. 2014. Chimeric bovine/human parainfluenza virus type 3 expressing respiratory syncytial virus (RSV) F glycoprotein: effect of insert position on expression, replication, immunogenicity, stability, and protection against RSV infection. J Virol 88:4237–4250. doi: 10.1128/JVI.03481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calain P, Roux L. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol 67:4822–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol 72:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchholz UJ, Finke S, Conzelmann K-K. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beeler JA, van Wyke Coelingh K. 1989. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol 63:2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang RS, MacPhail M, Schickli JH, Kaur J, Robinson CL, Lawlor HA, Guzzetta JM, Spaete RR, Haller AA. 2004. Parainfluenza virus type 3 expressing the native or soluble fusion (F) protein of respiratory syncytial virus (RSV) confers protection from RSV infection in African green monkeys. J Virol 78:11198–11207. doi: 10.1128/JVI.78.20.11198-11207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Wyke Coelingh KL, Winter CC, Tierney EL, London WT, Murphy BR. 1988. Attenuation of bovine parainfluenza virus type 3 in nonhuman primates and its ability to confer immunity to human parainfluenza virus type 3 challenge. J Infect Dis 157:655–662. doi: 10.1093/infdis/157.4.655. [DOI] [PubMed] [Google Scholar]
- 30.Coates HV, Alling DW, Chanock RM. 1966. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol 83:299–313. [DOI] [PubMed] [Google Scholar]
- 31.Durbin AP, Cho CJ, Elkins WR, Wyatt LS, Moss B, Murphy BR. 1999. Comparison of the immunogenicity and efficacy of a replication-defective vaccinia virus expressing antigens of human parainfluenza virus type 3 (HPIV3) with those of a live attenuated HPIV3 vaccine candidate in rhesus monkeys passively immunized with PIV3 antibodies. J Infect Dis 179:1345–1351. doi: 10.1086/314769. [DOI] [PubMed] [Google Scholar]
- 32.Coates HV, Forsyth BR, Chanock RM. 1966. Biophysical studies of respiratory syncytial virus. I. Density of respiratory syncytial virus and associated complement-fixing antigens in cesium chloride density gradient. J Bacteriol 91:1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luongo C, Winter CC, Collins PL, Buchholz UJ. 2012. Increased genetic and phenotypic stability of a promising live-attenuated respiratory syncytial virus vaccine candidate by reverse genetics. J Virol 86:10792–10804. doi: 10.1128/JVI.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoder SM, Zhu Y, Ikizler MR, Wright PF. 2004. Role of complement in neutralization of respiratory syncytial virus. J Med Virol 72:688–694. doi: 10.1002/jmv.20046. [DOI] [PubMed] [Google Scholar]
- 35.Connors M, Kulkarni AB, Collins PL, Firestone CY, Holmes KL, Morse HC III, Murphy BR. 1992. Resistance to respiratory syncytial virus (RSV) challenge induced by infection with a vaccinia virus recombinant expressing the RSV M2 protein (Vac-M2) is mediated by CD8+ T cells, while that induced by Vac-F or Vac-G recombinants is mediated by antibodies. J Virol 66:1277–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karron RA, Buchholz UJ, Collins PL. 2013. Live-attenuated respiratory syncytial virus vaccines. Curr Top Microbiol Immunol 372:259–284. doi: 10.1007/978-3-642-38919-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: Still crazy after all these years. Virus Res 162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loomis RJ, Johnson PR. 2013. Gene-based vaccine approaches for respiratory syncytial virus. Curr Top Microbiol Immunol 372:307–324. doi: 10.1007/978-3-642-38919-1_15. [DOI] [PubMed] [Google Scholar]
- 39.Lawlor HA, Schickli JH, Tang RS. 2013. A single amino acid in the F2 subunit of respiratory syncytial virus fusion protein alters growth and fusogenicity. J Gen Virol 94:2627–2635. doi: 10.1099/vir.0.055368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao F, Li Y, Decker J, Peyerl F, Bibollet Ruche F, Rodenburg C, Chen Y, Shaw D, Allen S, Musonda R, Shaw G, Zajac A, Letvin N, Hahn B. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res Hum Retroviruses 19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- 41.Carnero E, Li W, Borderia AV, Moltedo B, Moran T, Garcia-Sastre A. 2009. Optimization of human immunodeficiency virus gag expression by Newcastle disease virus vectors for the induction of potent immune responses. J Virol 83:584–597. doi: 10.1128/JVI.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Arguello MB, Martin D, Wharton SA, Calder LJ, Martin SR, Cano O, Calero M, Garcia-Barreno B, Skehel JJ, Melero JA. 2004. Thermostability of the human respiratory syncytial virus fusion protein before and after activation: implications for the membrane-fusion mechanism. J Gen Virol 85:3677–3687. doi: 10.1099/vir.0.80318-0. [DOI] [PubMed] [Google Scholar]