ABSTRACT
Lipid-enveloped viruses replicate and bud from the host cell where they acquire their lipid coat. Ebola virus, which buds from the plasma membrane of the host cell, causes viral hemorrhagic fever and has a high fatality rate. To date, little has been known about how budding and egress of Ebola virus are mediated at the plasma membrane. We have found that the lipid phosphatidylserine (PS) regulates the assembly of Ebola virus matrix protein VP40. VP40 binds PS-containing membranes with nanomolar affinity, and binding of PS regulates VP40 localization and oligomerization on the plasma membrane inner leaflet. Further, alteration of PS levels in mammalian cells inhibits assembly and egress of VP40. Notably, interactions of VP40 with the plasma membrane induced exposure of PS on the outer leaflet of the plasma membrane at sites of egress, whereas PS is typically found only on the inner leaflet. Taking the data together, we present a model accounting for the role of plasma membrane PS in assembly of Ebola virus-like particles.
IMPORTANCE The lipid-enveloped Ebola virus causes severe infection with a high mortality rate and currently lacks FDA-approved therapeutics or vaccines. Ebola virus harbors just seven genes in its genome, and there is a critical requirement for acquisition of its lipid envelope from the plasma membrane of the human cell that it infects during the replication process. There is, however, a dearth of information available on the required contents of this envelope for egress and subsequent attachment and entry. Here we demonstrate that plasma membrane phosphatidylserine is critical for Ebola virus budding from the host cell plasma membrane. This report, to our knowledge, is the first to highlight the role of lipids in human cell membranes in the Ebola virus replication cycle and draws a clear link between selective binding and transport of a lipid across the membrane of the human cell and use of that lipid for subsequent viral entry.
INTRODUCTION
Lipid-enveloped viruses harbor a lipid membrane bilayer derived from their host cell during the budding process. This envelope provides the virus stability, protection of its genetic contents, and a reservoir for its transmembrane glycoprotein, which mediates entry into cells (1, 2). The viral lipid envelope may be a viable target for drug development, as particular alterations in the lipid coat or receptor-lipid interaction can inhibit viral entry (3–6). The lipid-dependent budding and egress of some lipid-enveloped viruses have been investigated. For example, it is well established that HIV-1 binds and utilizes 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate) [PI(4,5)P2] enriched in the plasma membrane (PM) inner leaflet for assembly and egress from the cell (7, 8). Enteroviruses and flaviviruses use a phosphatidylinositol-4-phosphate [PI(4)P]-enriched organelle to replicate (9), and enteroviruses are packaged into phosphatidylserine (PS)-enriched vesicles, thereby enhancing the efficiency of viral transmission (10). The budding and egress of filoviruses (11, 12), however, are poorly understood.
Ebola virus (EBOV), a member of the Filoviridae family, is a negative-sense single-stranded RNA virus that assembles and buds from the inner leaflet of the plasma membrane (13). EBOV contains seven proteins in its genome, which in concert with host machinery coordinate the entry, viral replication, and budding required to sustain and spread the infection. VP40, a matrix protein, is one of the seven genes that the virus encodes, and it coats the inner leaflet of the viral lipid envelope (14–16). In mammalian cells, VP40 expression in the absence of other EBOV proteins is sufficient for assembly and formation of virus-like particles (VLPs) that are similar in size and shape to and nearly indistinguishable from the authentic virions (17–20). To this end, VP40 has served as an excellent model to investigate Ebola virus budding in vitro. VP40 has been shown to form a high-affinity dimer (21), which is able to transform into hexameric filaments that can concentrate at the inner leaflet of the plasma membrane (21–24). While VP40 has been shown to associate with lipid vesicles or model membrane systems harboring phosphatidylserine (PS) in vitro (24–28), little information is available on how VP40 assembles and buds from the plasma membrane of human cells and on what the targets in these processes might be for antiviral intervention.
The inner leaflet of the mammalian cell plasma membrane contains ∼20 mol% anionic lipid. This anionic charge creates a negative electric field that can contribute to cationic peripheral protein recruitment (29). The electronegativity of the plasma membrane is attributed in part to the enrichment of polyvalent phosphoinositides, including PI(4)P, PI(4,5)P2, and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate] [PI(3,4,5)P3] (30), which make up a small fraction of the phospholipids on the inner leaflet of the plasma membrane. Phosphatidylserine (PS) is the most abundant anionic lipid in the plasma membrane cytosolic leaflet (31) and can represent ∼15% to ∼20% of the inner leaflet. PS contributes to the recruitment of proteins with cationic clusters (32) as well as lipid-binding domains containing a motif for PS recognition (33). The studies presented here aimed to determine how EBOV matrix protein VP40 associates with the plasma membrane. We have used several human and mammalian cell lines to investigate the mechanism of VP40 plasma membrane binding and oligomerization, which we demonstrate to be dependent upon the plasma membrane PS content. Additionally, we demonstrate that interaction of VP40 with the plasma membrane is sufficient to induce specific exposure of PS at sites of budding. This translocation of PS from inner to outer leaflets and subsequent decoration of the budded viral surface with PS provide the mechanism by which filoviruses may acquire the surface PS needed for capture by cellular TIM-1 (3–5). A unique feature of filovirus entry is that its specific GP-binding receptor, Niemann Pick C1, is encountered only after the virus has entered the endosome (34, 35). Initial capture of the virion on cells is mediated through lectin capture of glycoprotein and TIM-1 capture of viral lipid (3–5, 36). Taken together, our results provide new insight into the role of cellular membrane PS in inducing VP40 oligomerization and assembly to form a new viral particle.
MATERIALS AND METHODS
Materials.
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphol-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate) [PI(4,5)P2], 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate] [PI(3,4,5)P3], and d-erythro-sphingosine (sphingosine) were from Avanti Polar Lipids (Alabaster, AL) and used without further purification.
Glutathione S-transferase GST resin (Novagen, San Diego, CA), thrombin (GE Healthcare, Pittsburgh, PA), HisTag resin (Qiagen), L1 sensor chips (GE Healthcare), VP40 antibody (IBT Bioservices, Gaithersburg, MD), secondary goat anti-rabbit horseradish peroxidase (HRP) conjugate (Bio-Rad, Hercules, CA), anti-phosphatidylserine [4B6] antibody (Abcam, Cambridge, MA), and l-amino acid oxidase (LAAO) (from Crotalus adamanteus venom; Worthington, Lakewood, NJ) were obtained from the indicated sources. Phospholipase D (PLD) from Streptomyces chromofuscus, docosahexaenoic acid (DHA), antimycin, 2-deoxy-glucose, and a BioPorter QuikEase protein delivery kit were from Sigma (St. Louis, MO). Ionomycin was from EMD Chemicals (Philadelphia, PA), anti-calnexin was from BioVision (Milpitas, CA), and Complete Mini-EDTA-free protease inhibitor cocktail was from Roche (Indianapolis, IN). A bicinchoninic acid (BCA) assay kit was from Thermo Fisher Scientific (Rockford, IL). Neutravidin beads, Lipofectamine 2000, Lipofectamine LTX and Plus reagent, Amplex Red reagent, and Amplex red stop reagents were from Life Technologies (Grand Island, NY). Peroxidase from horseradish roots was obtained from Oriental Yeast (Tokyo, Japan). An enhanced green fluorescent protein (EGFP) enzyme-linked immunosorbent assay (ELISA) kit was from Cell Bio Labs, Inc. (San Diego, CA).
Plasmids.
pGex4T-1 VP40 was made by cloning VP40 from the pcDNA3.1 (EGFP-VP40) plasmid. The lactadherin C2 (Lact C2) EGFP, Lact C2 mCherry, Lact C2 pet28a, and Rpre-mRFP plasmids were kind gifts from Sergio Grinstein (University of Toronto). PLCδ PH GFP was a kind gift from Tamas Balla (NIH), Akt1 PH GFP and Inp54 were from Tobias Meyer (Stanford University), and HIV-1 gag EGFP was a kind gift from Tom Hope (Northwestern University). The W95A/E160 mutation of EGFP-VP40 was prepared as previously described (22).
Protein purification.
GST-VP40 (24) and the lactadherin C2 domain (37) were expressed in Escherichia coli and purified as previously described in detail.
SPR.
All surface plasmon resonance (SPR) measurements were performed at 25°C. A detailed protocol for coating the L1 sensor chip has been described elsewhere (27, 38). Lipid vesicles containing either POPC:POPE (80:20) or POPC:POPE:POPS (60:20:20) were injected at 5 μl/min to give a response of 3,000 response units (RU) for the control channel or the active surface channel, respectively. Each lipid layer was stabilized by injecting 10 μl of 50 mM NaOH three times at 100 μl/min following lipid coating. SPR measurements were done at the flow rate of 5 μl/min, and 80 to 90 μl of protein–10 mM HEPES (pH 7.4)–0.16 M KCl was injected to give the association time required to reach saturation of binding signal (Req). The lipid surface was regenerated using two pulses of 10 μl of 50 mM NaOH at 30 μl/min. After sensorgrams were obtained for five or more different concentrations of each protein within a 10-fold range of dissociation constant (Kd) values, each of the sensorgrams was corrected for the refractive index change by subtracting the control surface response value. Req values were then plotted versus protein concentrations (C), and the Kd value was determined by a nonlinear least-squares analysis of the binding isotherm using the equation Req = Rmax/(1 + Kd/C) (27, 38). Analysis of each data set was repeated three or more times to calculate a standard deviation.
Cell culture maintenance.
HEK293 cells were cultured and maintained at 37°C in a 5% CO2 humidified incubator supplemented with Dulbecco's modified Eagle's medium (DMEM) (low glucose) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Pen/Strep). After trypsinization, cells were transferred from a T-25 tissue culture flask to an 8-well plate for confocal imaging. HEK293 cells were then grown to 50 to 80% confluence and transfected with 1 μg DNA/dish using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. A549 cells were cultured and maintained at 37°C in a 5% CO2 humidified incubator supplemented with DMEM-RPMI medium (50:50) containing 10% FBS and 1% Pen/Strep. For imaging, A549 cells were subjected to trypsinization and then transferred to the respective plates for imaging. After cells reached ∼70% to ∼80% confluence, transfections were performed with Lipofectamine LTX and Plus reagent according to the manufacturer's protocol. HUH7.5 cells were maintained in DMEM (low glucose) containing 10% FBS and 1% Pen/Strep at 37°C in a 5% CO2 humidified incubator. Cells were grown to 70 to 80% confluence prior to passaging or transfection. Transfections were performed using Lipofectamine LTX and Plus reagent according to the manufacturer's protocol.
CHO-K1 cells were maintained in DMEM–F-12 (50:50) (Invitrogen) containing 10% FBS and 1% Pen/Strep at 37°C in a 5% CO2 humidified incubator. Cells were grown to 70 to 80% confluence before passaging or transfection. Transfections were performed using Lipofectamine LTX and Plus reagent according to the manufacturer's protocol. PSA-3 cells (a kind gift from O. Kuge, Kyushu University) were grown to 70 to 80% confluence before passaging or transfection. PSA-3 cells were maintained in DMEM–F-12 (50:50) containing 10% FBS and 1% Pen/Strep at 37°C in a 5% CO2 humidified incubator. Transfections were performed using Lipofectamine LTX and Plus reagent according to the manufacturer's protocol. In the experiments in which PS was added as a supplement, 30 μM POPS–chloroform was dried under nitrogen and resuspended in 1 ml of sterile phosphate-buffered saline (PBS) (pH 7.4). The solution was then sonicated for 15 min at 4°C and added to the media as previously reported (39). For experiments using PSA-3 plus DHA, 40 μM DHA was added to the media after preparation as described above to stimulate PS synthesis.
Pharmacological treatments.
Pharmacological treatments were performed at 37°C unless otherwise noted. A sphingosine stock solution was made by evaporation of chloroform under nitrogen gas. The resulting film was then resuspended in ethanol. Sphingosine was then added to cells in culture to achieve a final concentration of 75 μM.
Confocal imaging and analysis.
Routine imaging of all cell lines was performed on a Zeiss LSM 710 inverted microscope using a Plan Apochromat 63× 1.4-numerical-aperture (NA) oil objective and the 488-nm-wavelength laser line or the 561-nm-wavelength laser line or both laser lines for colocalization and dual-visualization experiments. Laser power was maintained at a constant level (i.e., at a setting of 1.0) during the experiment as were the gain and offset settings. EGFP was imaged using the 488-nm-wavelength line or the Ar ion laser with the emission collected through a 493-to-550-nm-wavelength filter. mCherry or monomeric red fluorescent protein (mRFP) was collected with the laser line from the 561-nm-wavelength laser with an emission filter collection of wavelengths from 600 to 700 nm. For assessment of VP40 localization in the presence of anti-PS antibody 4B6, a BioPorter QuikEase Protein delivery kit was used according to the manufacturer's instructions. In experiments where cells were counted for localization, at least 100 cells were counted in each experiment (unless otherwise noted) for three separate experiments and counts were averaged to calculate the standard deviation (or standard error of the mean if indicated) and P values. The Zeiss software package was used to assess the membrane/(membrane plus cytosol) ratio for quantification of membrane and cytosolic distribution and was also used for assessment of localization.
RICS data acquisition.
Raster image correlation spectroscopy (RICS) data were acquired on a commercial laser scanning confocal microscope (Zeiss LSM710 inverted microscope) using a Plan Apochromat 63× 1.4-numerical-aperture oil objective. The 488-nm-wavelength line of the Ar ion laser was used for excitation of EGFP. The laser power was maintained at 1% throughout the experiment with the emission collected through a 493-to-550-nm-wavelength filter. The data were collected as images of 256-by-256 pixels with a pixel dwell time of 12.6 μs. RICS analysis was done with SimFCS software using 100 frames of the image series.
For number and brightness (N&B) analysis, cells expressing monomeric EGFP were used as a brightness standard and were imaged under the same conditions as EGFP-VP40 and respective mutations. The brightness of the EGFP was used as the brightness of the monomer. The selection window for analysis in the brightness-versus-intensity plot was based on the average brightness of a monomer (0.104), which allowed selection of oligomeric size based upon the multiple of the monomer. Thus, the selection window for each species is based upon the average brightness, which yields an average population of each species in the respective area of analysis.
N&B analysis is based upon moment analysis and allows measurement of the average number of molecules as well as of the brightness in each pixel of a fluorescence microscopy image (22–24, 40–43). Here, the average brightness of a particle is determined from the ratio (of variance to intensity) at each pixel. Fluctuating particles can be determined by dividing the average intensity value by the brightness value at each pixel. In live cells, intensity may be attributed to the presence of autofluorescence, scattering, bright immobile particles, or fast moving particles that are dim. To control for these parameters, the N&B analysis accounts for these limitations by calculating the total variance, which also incorporates the detector noise. For particles fluctuating in the focal volume, the variance is proportional to the particle brightness; however, the variance of the immobile particles, scattering, autofluorescence, and detector noise is proportional to the intensity of those components. Thus, only fluorescent fluctuations that are dependent upon the mobile particles (square of the brightness) have a ratio of variance to intensity of >1. Brightness maps then allow pixel-level resolution of the clustering of fluorescently labeled proteins.
The following equations were used to compute the number and brightness values:
| (1) |
| (2) |
Here, N and B are the apparent number and the brightness of the molecule, < I > is the average intensity, σ2 is the variance, and offset and σ02 are the intensity and noise variance due to the detector. With these parameters properly calibrated using EGFP, we obtained the distribution of the brightness values of each individual pixel in the image of the cell under investigation. More details of the N&B analysis of VP40 in live cells can be found in references 22, 23, 24, 27, and 44.
VLP purification and quantification and Western blot analyses.
VLPs were isolated from cellular media as previously described (22, 24, 27, 40). Cell lysates and VLP samples were mixed with SDS loading buffer and were boiled before loading on an 8% SDS-PAGE gel. The gel was then transferred to a nitrocellulose membrane that was later incubated with primary rabbit polyclonal anti-EGFP antibody (Thermo Scientific) (1:1,000 dilutions) or rabbit polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Santa Cruz Biotechnology) followed by goat anti-rabbit HRP (Bio-Rad) conjugate as the secondary antibody according to the protocol of the manufacturer (Thermo Scientific). Blots were exposed using ImageQuant LAS 4000 (GE Healthcare). This method allows a comparison of VP40 in VLPs versus cell lysate using GAPDH as a loading control.
In order to collect cell lysates, the respective cell lines were washed once with PBS and then trypsinized and collected via centrifugation for 6 min at 900 rpm. The resulting pellet was washed via resuspension of the pellet in 5 ml of PBS. The resuspended pellet was centrifuged at 900 rpm for 6 min, resuspended in 5 ml of PBS (pH 7.4), and spun down one more time for collection with a final resuspension in 150 μl of PBS. Cells were lysed via addition of PBS containing 1.6 μl of protease inhibitor and 10 μl of 0.5% Triton X-100 for quick sonication (at probe setting 5 with 5 quick pulses on ice). Incubation was performed for 5 min on ice followed by centrifugation (13,200 rpm for 15 min at 4°C). The supernatant was then collected for analysis.
The samples were run on an SDS-PAGE gel (8%) and transferred onto a nitrocellulose membrane at 60 V for 3 h at 25°C or 35 V overnight at 4°C in transfer buffer (1 liter final volume; 14.4 g glycine, 3.02 g Tris, 100 ml methanol, 2 ml 10% SDS). The gels were stained to check the efficiency of the transfer. The membrane blots were placed in blocking buffer (2.5 g dry milk–50 ml Tris-buffered saline [TBS]) for 1 h at 25°C. The membrane blots were sealed in a plastic bag with dilute primary antibody (anti-calnexin-rabbit, 1:1,000 ratio)–1% dry milk–TBS-Tween 20 (TBS-T) and mixed for 2 h at 25°C. The membranes were then washed multiple times (for 5, 5, 10, 5, and 5 min) and sealed in a plastic bag with dilute secondary antibody (anti-rabbit HRP conjugate; 1:15,000 ratio)–1% dry milk–TBS-T and allowed to mix for 1 h at 25°C. The membrane blots were washed again multiple times with TBS-T (for 5, 5, 10, 5, and 5 min) and TBS (for 10 min) before incubation with substrate/enhancer (6 ml/6 ml; 1:1 [vol/vol]), covered, and exposed for imaging with an ImageQuant LAS 4000 imager (GE Healthcare).
ELISA.
At 48 h posttransfection, media and cells were separately collected and lysed as described above to measure the egress of VP40 or HIV-1 gag. An EGFP ELISA kit (Cell Bio Labs, Inc., San Diego, CA) was used to quantify VLP release. VLPs were isolated from cellular media as previously described (22, 24, 27, 40). Cell lysates or VLP lysates were incubated at 37°C for 2 h in the concentration range of 10 to 220 ng/μl and diluted to a total volume of 100 μl in each microwell of a 96-well plate. GFP standards were incubated in a similar fashion to generate a standard curve. After 2 h, each well was washed four times with 250 μl of wash buffer. The plate was tapped dry over a Kimwipe, and 100 μl of biotinylated anti-EGFP antibody at a 1:1,000 dilution was added to each well and incubated at 25°C on an orbital shaker at 50 rpm for 2 h. Microwells were washed again four times each with 250 μl of wash buffer and tapped dry, and 100 μl of streptavidin-enzyme conjugate was added to each well. The plate was incubated for 1 h at 25°C on an orbital shaker rotating at 50 rpm. The wells were then once again washed four times with 250 μl of wash buffer and tapped dry, and 100 μl of substrate solution was added to each well. The plate was incubated at 25°C for between 2 and 15 min while the color change of the standards was observed. The reaction was stopped by adding 100 μl of stop solution, and the results were scanned immediately for absorbance at 450 nm on a Molecular Devices Spectramax M5e reader. Individual sample experiments were performed in duplicate, with separate experiments from different cell plates done in triplicate. The results are expressed as the ratio of EGFP-VP40 or EGFP–HIV-1 gag in VLP versus cell lysates. Cells transfected with EGFP were used as a control.
Plasma membrane isolation and purification.
To purify plasma membrane from A549, CHOK-1, PSA-3, or PSA-3 cells supplemented with PS or DHA, the following protocol was used. Cells of interest were seeded at 2 × 105 in five T150 flasks (150 cm2) and were allowed to grow for 2 to 3 days to reach 80 to 90% confluence. To begin the plasma membrane purification, culture flasks were moved to 4°C and allowed to cool for 10 min prior to biotinylation. Cells were then washed twice with 10 ml PBS. A 1 mg/ml concentration of N-hydroxysuccinimide (NHS)-sulfo-long chain (LC)-biotin–PBS (pH 8) (0.02 ml/cm2) was then added to each flask to initiate the biotinylation reaction. Cells were incubated for 20 min with gentle rocking at 4°C. After 20 min, cells were washed twice with PBS (10 ml per flask). Subsequently, a 0.1 M glycine wash was used to quench unreactive biotinylation reagents (10 ml per flask) followed by another wash with PBS (10 ml each flask). Cells were then scraped into 20 mM Tris (pH 8.35) containing 200 mM sucrose and 0.2 mM MgCl2 (0.01 ml/cm2 or 1.5 ml per flask) and transferred to a 5-ml test tube. Streptavidin-conjugated M-280 Dynabeads (in PBS [pH 7.4] containing 0.1% bovine serum albumin [BSA] and 0.02% NaN3) were then added to the cells using 0.5 μl of beads/cm2 at the concentration of 4 × 108 beads per ml. The suspension was carefully mixed to allow binding to proceed at 4°C for 20 to 30 min. A light microscope is used to monitor binding of the cells to the beads. After 30 min, an EasySep magnet was used to remove bound cells from the solution. This was performed quickly to prevent sedimentation of unbound cells. The supernatant was poured off, and the collected cells were washed twice with 1 ml of 20 mM Tris (pH 8.35) containing 200 mM sucrose and 0.2 mM MgCl2. A 3-ml volume of freshly prepared hypoosmotic buffer (20 mM Tris [pH 9.1] containing 25 mM sucrose and 0.2 mM MgCl2) was then added, and the reaction mixture was incubated for 10 min at 4°C with mixing. The hypoosmotic buffer was removed by using an EasySep magnet and pipetting off the supernatant. Buffer exchange was then performed by resuspending the plasma membrane coated beads with 3 ml of 250 nM latrunculin A and incubation at room temperature with mixing for 5 min. Using this procedure, there should not have been many if any whole cells under the microscope at that juncture. The buffer was then removed as described above. Beads were then washed with 50 mM Tris (pH 6.8) containing 0.2 M NaBr, 0.2 M KCl, and 250 nM latrunculin A and transferred to a new tube. Beads were then washed twice with 0.1 M Na2CO3 (pH 11) and once with 25 mM Tris (pH 6.8) containing 0.1 M KCl and 0.1 M NaBr. A final wash with freshly prepared 150 mM NH4HCO3 (pH 8) was then performed. Each wash step described used an EasySep magnet for supernatant removal as described earlier. The resulting plasma membrane was resuspended in 200 liters of 150 mM NH4HCO3 for respective analysis. The purity of the plasma membrane was assessed for contamination by probing for calnexin, an endoplasmic reticulum (ER) resident protein. The ER comprises about 80% of the membrane of the cell and represents the primary lipid contaminant throughout the purification procedure.
Phosphatidylserine quantification.
Measurement of PS content of the plasma membrane was performed using a three-reagent system as recently reported (45). Reagent S1 contained 600 U/ml PLD, 20 U/ml LAAO, 50 mM NaCl, and 50 mM Tris-HCl (pH 7.4). Reagent S2 contained 6.25 U/ml peroxidase, 187.5 μM Amplex Red, 0.125% Triton X-100, 50 mM NaCl, and 50 mM Tris-HCl (pH 7.4). Reagent S3 was the Amplex Red Stop reagent and was obtained from Molecular Probes. PS (POPS) standard solutions were dissolved in 1% Triton X-100 aqueous solution before use. To assess PS content, a purified plasma membrane sample (5 μl) was added to reagent S1 (10 μl) and the reaction mixture was incubated at 25°C for 4 h. After the incubation, reagent S2 (80 μl) was added and the reaction mixture was incubated at 25°C for 15 min. Next, Amplex Red stop reagent (20 μl) was added, the reaction mixture was incubated at 25°C for 10 min, and the fluorescence intensity was measured using a fluorescence microplate reader (Spectramax M5e; Molecular Devices, CA). The excitation and emission wavelength filters were set at 544 and 590 nm, respectively. PS concentrations in the analyzed samples were determined from the standard curve, with the procedure repeated in triplicate. Thin-layer chromatography was used to spot and assess total plasma membrane lipid amounts. Image J was used to quantify the density of all visible spots.
Exoplasmic leaflet phosphatidylserine detection with flow cytometry.
HEK293 cells were plated at 5 × 105 cells per plate in 60-mm-diameter plates and incubated at 37°C for 24 h. Cells were then washed twice with 1× phosphate-buffered saline (PBS) (pH 7.4) and given 3 ml DMEM before transfection. Plasmid DNA (8.5 μg) was complexed with 8.5 μl Plus reagent–250 μl DMEM for 15 min at room temperature, mixed with 3.3 μl Lipofectamine LTX diluted in 250 μl DMEM, and incubated at room temperature for 30 min. This mixture was then added dropwise to the 60-mm-diameter plates. Cells were then harvested, stained, and processed for 24 to 26 h posttransfection. Camptothecin (Sigma catalog no. C9911) was added to cells 12 h before harvesting to achieve a final concentration of 10 μM. Ionomycin (MP Biomedicals, Santa Ana, CA) was added with a treatment buffer (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 20 mM HEPES, 100 μM EGTA [pH 7.4], 0.7 mM CaCl2) to achieve a final concentration of 10 μM 30 min before harvesting.
Cells were washed once with 1× PBS (pH 7.4) and treated with 0.5 ml trypsin for 3 to 5 min at 37°C. Cells were then washed twice with PBS (pH 7.4) and resuspended in 1 ml of 1× binding buffer (diluted from 10× binding buffer; BD Biosciences catalog no. 556454). Cells were divided into 300-μl aliquots and stained with either allophycocyanin (APC) annexin V (BD Biosciences catalog no. 556454) or phycoerythrin (PE) annexin V (BD Biosciences catalog no. 559763) and 7-aminoactinomycin D (7-AAD) (BD Biosciences catalog no. 559925) according to the annexin V staining protocol from BD Biosciences. Cells were collected using a Beckman FC500 flow cytometer. EGFP and GFP were excited at 488 nm and collected at between 515 and 535 nm. APC annexin V was excited at 633 nm and collected at between 660 and 690 nm. PE annexin V was excited at 488 nm and collected at between 567 and 583 nm. 7-AAD was excited at 488 nm and collected at between 660 and 690 nm.
Exoplasmic leaflet phosphatidylserine detection with confocal microscopy.
HEK293 cells were grown in an 8-well plate to 60 to 80% confluence, at which time they were transfected with 0.3 μg total EGFP construct with 0.5 μl Lipofectamine LTX–DMEM (without FBS or Pen/Strep) for 18 to 24 h. One hour prior to imaging, the medium was removed from each well and replaced with 100 μl 1× annexin V binding buffer. Four microliters of annexin V-APC was added to each sample 15 to 20 min prior to imaging. To prevent imaging of apoptotic cells, CellMetrix Green LIVE/DEAD Stain was added as instructed by the manufacturer (ImmunoChemistry Technologies, LLC, Bloomington, MN). Images were collected on a Zeiss 710 LSM confocal microscope, with a 63× 1.4-NA oil immersion objective. EGFP and annexin V-APC were excited at 488 nm and 561 nm and collected at between 485 and 550 nm and between 650 and 800 nm, respectively. Laser power and gain were held constant for annexin V-APC data collection. The EGFP gain was adjusted to saturated pixels. A total of 8 to 10 cells were imaged per condition over four independent experiments. Imaging data were analyzed for percent intensity overlap and percent area overlap in MATLAB (The MathWorks, Natick, MA) with a custom script (available upon request). The data (averaged ± standard errors of the means [SEM]) were plotted and significance was determined with a confidence level of a P value of <0.05 by Student's t test. The percentage of annexin V-APC that did not overlap EGFP was also determined. To discount any data with a false annexin V-APC signal, images with 25% or more false binding were removed from the data analysis.
RESULTS
Determining the role of PS in VP40 plasma membrane assembly.
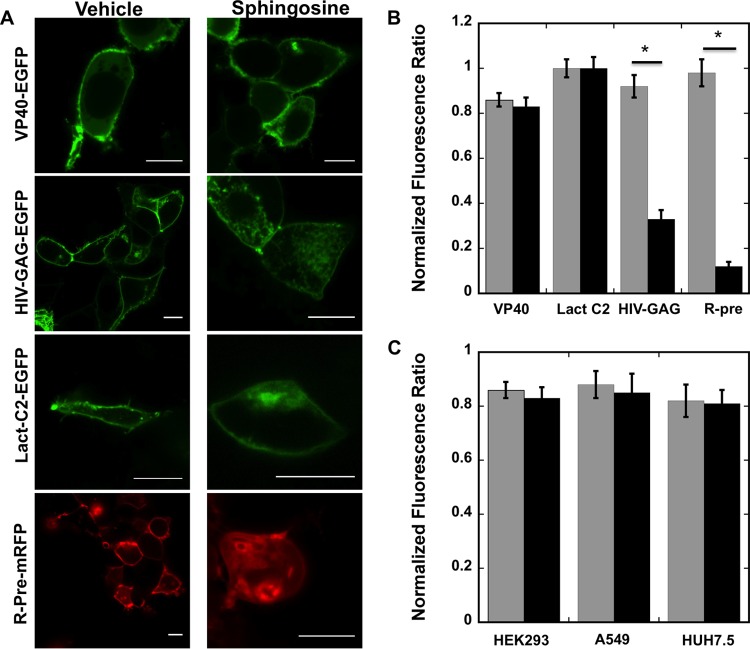
To assess the role of PS in the cellular localization of VP40 and egress of EBOV, we used several cell lines, including HEK293, A549, CHOK-1, and HUH7.5, a natural target cell for EBOV infections (Fig. 1 and 2). It has been well established that VP40 localizes at the plasma membrane inner leaflet (14, 16, 17, 19, 21–24, 27, 40, 46); however, the nature of this interaction has not been well understood. First, the negativity of the plasma membrane inner leaflet was neutralized with the membrane permeant base sphingosine (47) or vehicle control (Fig. 1). Previous studies have demonstrated that nonspecific electrostatic interactions at the plasma membrane can be greatly reduced when sphingosine is added to a cell culture (47). The redistribution of phosphoinositide sensors (HIV-1–gag) (Fig. 1B) and anionic charge reporters (Rpre) (Fig. 1D) but not VP40 or Lact C2 (Fig. 1A or C) suggested that the interaction of VP40 with the membrane is not dependent solely on the anionic charge of the plasma membrane (47).
FIG 1.
Cellular imaging and distribution change with and without sphingosine treatment. (A) HEK293 cells expressing fluorescent fusion constructs of VP40, HIV-1 gag, Lact C2, or Rpre are shown after 1 h of treatment with vehicle (dimethyl sulfoxide [DMSO]) or 75 μM sphingosine. Bars, 10 μm. (B) HEK293 cells displaying detectable PM localization were quantified for the respective constructs following treatment with vehicle (gray bars) or 75 μM sphingosine (black bars). Three independent experiments were performed to determine the standard deviation (SD) for each measurement. *, P < 0.002. (C) HEK293, A549, or HUH7.5 cells displaying detectable plasma membrane localization were quantified with vehicle (gray bars) or 75 μM sphingosine (black bars). Three independent experiments were performed to determine the SD for each measurement.
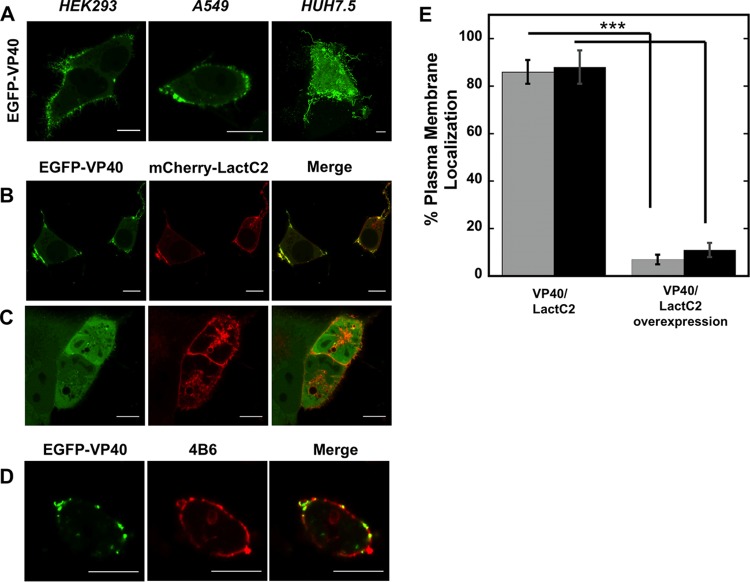
FIG 2.
Cellular localization of VP40. (A) EGFP-VP40 robustly localizes and induces filamentous plasma membrane assembly in HEK293, A549, and HUH7.5 cells. Bars, 10 μm. (B) EGFP-VP40 cotransfected with an equimolar concentration of mCherry-Lact C2 displays colocalization signal with Lact C2 at regions of VP40 plasma membrane localization in HEK293 cells. One limitation of the colocalization analysis is local folding of the plasma membrane, which cannot be ruled out for the colocalization signals as shown. Bars, 10 μm. (C) A 5-fold difference in the mCherry-Lact C2/EGFP-VP40 transfection ratio significantly reduces VP40 plasma membrane localization in A549 cells. Bars, 10 μm. (D) EGFP-VP40 exhibits colocalization signal with the anti-phosphatidylserine antibody 4B6. CHOK-1 cells are shown. Bars, 10 μm. (E) Quantification of plasma membrane localization of VP40 plasma membrane localization in HEK293 cells (gray bars) and A549 cells (black bars) at an equimolar transfection ratio (left column) and a 5-fold increase in the Lact C2/VP40 transfection ratio. n = 3 independent experiments, with 500 cells assessed per experiment to determine SD. ***, P < 0.001.
Next, the PS biosensor Lact C2 was coexpressed in cells with VP40. We observed overlap of VP40 staining in membrane locations in which the PS sensor was detected (Fig. 2B). Similarly, an anti-PS antibody (4B6) which detected PS (Fig. 2D) also showed a VP40 signal overlap at the plasma membrane. While the measurements imply a role for cellular PS in VP40 assembly, they still have their limitations. First, colocalization of VP40 and Lact C2 may be enhanced due to local folding of the plasma membrane. Second, cellular delivery of the anti-PS antibody may have affected some of the plasma membrane localization of VP40. While Lact C2 can also detect PS on endomembranes (47, 48), it is predominantly found at the plasma membrane and thus can compete for PS at the PM. When Lact C2 was overexpressed with VP40, Lact C2 could competitively displace VP40 plasma membrane localization (Fig. 2C and E), further suggesting an important role for PS in VP40 localization. Again, there are some limitations to this interpretation, as VP40 seems to require a threshold level of expression to achieve detectable plasma membrane localization (data not shown).
Membrane affinity of VP40 for lipid vesicles containing phosphatidylserine.
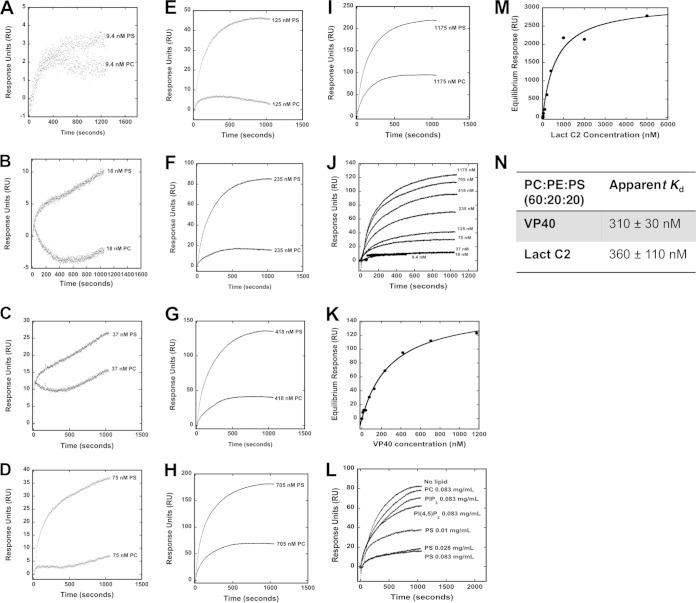
VP40 has previously been shown to associate with membranes containing the anionic lipid PS (24–28). To assess the membrane binding affinity of VP40, we employed a surface plasmon resonance (SPR) assay as shown in Fig. 3A to I. Here, VP40 binding was measured for POPC:POPE:POPS (60:20:20) vesicles versus control POPC:POPE (80:20) vesicles at increasing concentrations of VP40 (Fig. 3A to I). VP40 displayed high affinity for lipid vesicles containing 20 mol% PS compared to control vesicles containing zwitterionic lipids. VP40-bound vesicles contained 20 mol% PS, a concentration representative of the PS content of the cytoplasmic leaflet of the plasma membrane, with an apparent Kd of 310 ± 30 nM affinity (Fig. 3K, L, and N). To gauge how VP40 apparent affinity compares to that of a consensus PS binding protein, we tested the binding of the lactadherin C2 domain (Lact C2) to the same lipid vesicle system. Lact C2 is a high-affinity binding protein with specificity for PS and has been utilized extensively for detection of PS localization in cellular studies (47, 49, 50). Lact C2 bound these PS-containing vesicles with comparable affinity, at an apparent Kd of 360 ± 110 nM (Fig. 3M and N). These values for Lact C2 also agree with a recent affinity estimate for PS-containing vesicles using a different assay (37). Thus, VP40 and the PS sensor Lact C2 bind PS-containing vesicles with similar apparent Kd values.
FIG 3.
SPR binding measurements of VP40 and Lact C2 with PS-containing vesicles. (A to I) Different concentrations of VP40 were injected into the Biacore X instrument, where flow cell 1 contained POPC:POPE (80:20) vesicles and flow cell 2 contained POPC:POPE:POPS (60:20:20). A flow rate of 5 μl/min was used for each injection. Flow cell 1 and flow cell 2 response unit (RU) values before VP40 injections were normalized to zero to present VP40 binding sensorgrams as shown. (J) Increasing concentrations (9.4 to 1,175 nM) of VP40 were assessed for binding to POPC:POPE:POPS (60:20:20) vesicles to determine the saturation response at each concentration to determine the apparent Kd. POPC:POPE (80:20) vesicles were used on a different flow channel for the control and subtracted to yield the respective curves (as derived from panels A to I). (K) The saturation response values determined from the data in panel J were plotted against the VP40 concentration to determine the apparent Kd value for PS-containing vesicles. (L) A SPR competition assay indicates that PS-containing vesicles but not PC-, PI(4,5)P2-, or PI(3,4,5)P3-containing vesicles preincubated with VP40 are able to compete for VP40 binding to a sensor surface coated with PS-containing vesicles. (M) The saturation response values determined for Lact C2 at increasing protein concentrations were plotted to determine the apparent Kd value for PS-containing vesicles. (N) Apparent Kd values determined for VP40 and Lact C2 binding to POPS-containing vesicles. n = 3 for VP40 and n = 4 for Lact C2 to determine the standard deviation (SD).
To demonstrate the specificity of VP40 for PS, we performed a competition assay with lipid vesicles of different anionic lipids (Fig. 3L). When VP40 was preincubated with PS-containing vesicles prior to injection over the SPR sensor surface, a marked reduction in VP40 association with PS vesicles was detected. This specificity was further demonstrated, as PC-, PIP2-, or PIP3-containing vesicles preincubated with VP40 did not display as much of a significant reduction in VP40 binding to PS-containing vesicles (Fig. 3L). The reduction in binding to the PS vesicles in the SPR system correlated with increasing concentrations of PS-containing vesicles incubated with VP40 prior to injection into the SPR system.
Plasma membrane PS levels are critical for VP40 assembly.
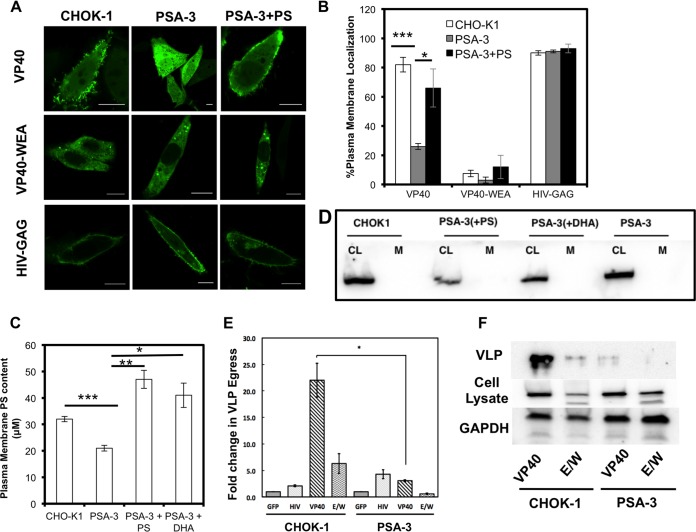
The level of PS in the plasma membrane may regulate VP40 assembly and egress. To test this hypothesis, we employed a mutant CHO-K1 cell line (PSA-3) (51) where the PS content is constitutively reduced. We found that localization of both VP40 and Lact C2 to the plasma membrane was significantly reduced in PSA-3 cells compared to wild-type (WT) CHO-K1 cells (Fig. 4A and B). Localization of charge and phosphoinositide reporters and localization of HIV-1 gag, which is dependent upon the presence of PI(4,5)P2 (7, 8), were unchanged (Fig. 4A and B). HIV-1 gag has also been shown to harbor an important interaction with PS (52, 53). However, the molecular details of HIV-1 gag interactions with PS in cellular membranes are not as well understood as the in vitro interactions. Thus, perhaps the plasma membrane of PSA-3 cells, despite an ∼30% reduction in the PS level, still harbored enough PS to promote HIV-1 gag plasma membrane localization. A double mutation of VP40 (W95A/E160A) which has previously been shown to greatly reduce VP40 plasma membrane localization, oligomerization, and budding was used as a negative control (Fig. 3). VP40 VLP release from the PSA-3 cells was also dramatically reduced (Fig. 4E and F). In order to determine if PS supplementation could rescue the defect in VP40 localization and assembly in PSA-3 cells, we supplemented the PSA-3 cells with either PS or docosahexaenoic acid (DHA), as described in Materials and Methods (Fig. 4C), which raises the PS content (39, 54). The PS enzymatic assay demonstrated that PS or DHA supplementation restored plasma membrane content and actually increased PS levels compared to the results seen with the parent CHOK-1 cell line (Fig. 4C). In PS- and DHA-supplemented PSA-3 cells, VP40 localization and Lact C2 PM localization were restored (Fig. 4A and B). The purification of the plasma membrane was probed for contamination by other cellular membranes using an antibody against the ER resident protein calnexin. The absence of calnexin in the plasma membrane purification was used to demonstrate the specificity of the plasma membrane PS assessment (Fig. 4D).
FIG 4.
Plasma membrane PS content regulates VP40 localization, assembly, and viral egress. (A) VP40 lacked significant plasma membrane localization and evidence of egress from the PS-deficient PSA-3 cell line compared to the parent CHOK-1 cell line. VP40 plasma membrane localization and evidence of egress could be restored with the supplementation of POPS in the media. VP40 WEA and HIV-GAG images of CHOK-1 cells, PSA cells, and PSA-plus-PS cells are also shown. Bars, 10 μm. (B) Cells were counted for detectable plasma membrane localization. Experiments were repeated in triplicate to calculate the standard errors of the means (SEM) as indicated. n = 3 independent experiments to determine the SD. Plasma membrane localization of WT VP40 was significantly increased when PSA-3 cells were supplemented with PS compared to PSA-3 cells without PS. (C) The PM PS content was assessed using the enzymatic assay described in Materials and Methods. PSA-3 cells display a nearly 30% reduction in PM PS content, whereas supplementing the cellular media with either PS or DHA restores and increases the PM PS content. (D) Cell lysates (CL) and purified plasma membranes (M) from CHOK-1, PSA-3, and PSA-3 cells treated with either PS or DHA were analyzed via Western blotting using an anticalnexin antibody to assess for ER contamination of the purified plasma membrane. CL, cell lysate; M, plasma membrane. (E) An ELISA demonstrates that PSA-3 cells have a 10-fold reduction in VP40 egress. n = 3 independent experiments to determine the standard errors of the means (SEM). (F) Western blot analysis was performed in triplicate to detect VLPs from CHOK-1 and PSA-3 cells using an anti-EGFP antibody. Cell lysate and GAPDH were also monitored for VP40 expression and a loading control, respectively. *, P < 0.01; **, P < 0.001; ***, P < 0.0001.
Defining the role of PS in VP40 oligomerization in live cells.
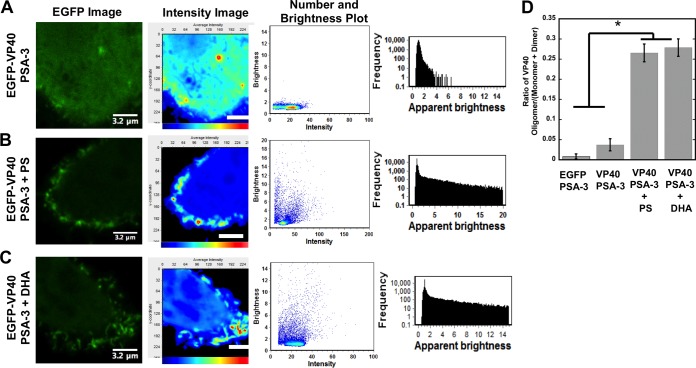
Oligomerization of VP40 serves as a marker of EBOV assembly, egress, and infectivity (21, 55). Plasma membrane lipid binding by VP40 has been hypothesized to induce the structural arrangements needed for VP40 hexamerization and filament formation (16, 21, 24). We assessed the ability of VP40 to oligomerize on the plasma membrane inner leaflet with a combination of confocal microscopy and number and brightness (N&B) analysis (22, 41–43). Here we also employed a double mutant of VP40 (W95A/E160A), which has previously been shown to significantly reduce VP40 plasma membrane localization (22, 55), by encouraging formation of an alternate, perinuclear ring structure instead (21). As previously reported, VP40 oligomers are enriched in membrane assembly sites extending from the plasma membrane, consistent with sites of viral egress (Fig. 5) (22–24). The PSA-3 cell line displayed a low level of VP40 oligomerization (Fig. 5A) which could be restored with supplementation of PS or DHA in the media (Fig. 5B and C).
FIG 5.
Plasma membrane PS content regulates VP40 plasma membrane oligomerization. (A to C) VP40 oligomerization was assessed using confocal microscopy with N&B analysis in PSA-3 cells without supplementation (A), PSA-3 cells supplemented with PS (B), or PSA-3 cells supplemented with DHA (C). The EGFP image acquired is shown in the first column; the cellular average-intensity map (second column; scale bars are 3.2 μm) demonstrates the lack of EGFP-VP40 plasma membrane signal in PSA-3 cells and lack of enriched intensity characteristic of oligomerization. The second column of each panel shows a brightness-versus-intensity plot demonstrating the lack of VP40 oligomerization in PSA-3 cells. A frequency-versus-apparent-brightness plot (third column of each) demonstrates significant oligomerization of VP40 at or near the PM of PSA-3 cells upon PS supplementation. n = 4 independent experiments for each condition. y-coordinate, pixels. (D) Histogram plot of the VP40 oligomer/VP40 (monomer-plus-dimer) ratio to demonstrate the reduction in VP40 oligomerization in PSA-3 cells without supplementation compared to PSA-3 cells supplemented with PS or DHA. n = 4 independent experiments. One-way analysis of variance (ANOVA) was used to calculate the SEM and P values. *, P < 0.001.
To quantify these effects, we calculated the VP40 oligomer/VP40 dimer ratio. Specifically, we measured the increase in VP40 oligomerization relative to that of the VP40 dimer (23). The calibration performed with monomeric EGFP allowed the selection of oligomers based upon the brightness of the monomer. Thus, we chose to analyze monomers and dimers together, as the separation of monomers from dimers in the brightness-versus-intensity plot can be difficult to interpret accurately. This provided a method (23) of analyzing VP40 monomers/dimers and VP40 assemblies larger than dimers that may assemble into hexamers or hexamers that concatenate into larger filaments (21–23). This allows assessment of a statistically significant reduction in the oligomerization of VP40. VP40 exhibited an approximately 6-fold increase in oligomerization when PS levels were restored to PSA-3 cells (Fig. 5D). In addition, overexpression of Lact C2 also abrogated VP40 oligomer formation as well as detectable protrusion sites (data not shown), presumably by competing with VP40 for available PS.
VP40 plasma membrane binding is sufficient to induce PS exposure on the exoplasmic plasma membrane leaflet of human cells.
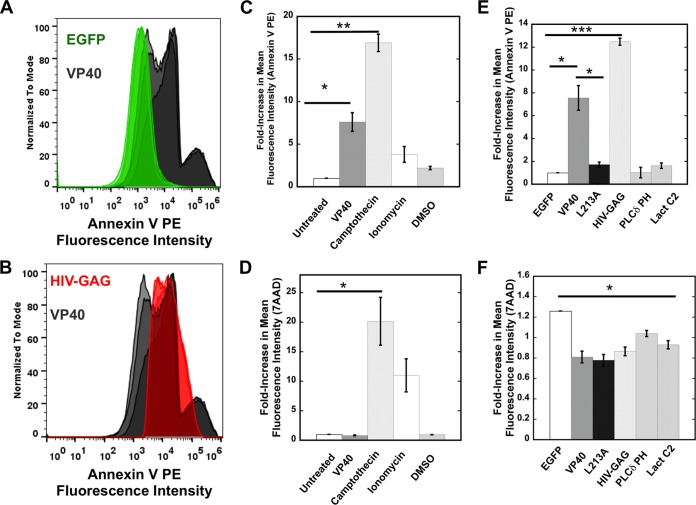
Viruses and virus-like particles of Ebola virus (56) and several other lipid-enveloped viruses require PS to be on their outer membrane in order for viral entry to occur (5). PS, however, is typically localized to the inner leaflet of the plasma membrane from which virions bud, suggesting that the topology of PS should be internal following scission of a virion or VLP. We asked if expression of VP40 was sufficient to induce PS exposure to the outer leaflet at sites of VLP formation, using flow cytometry and confocal imaging with the robust PS sensor annexin V (Fig. 6 and 7). Annexin V is an ideal tool to detect PS exposure on the outer leaflet of cell membranes (47) as it permeates only disrupted membranes of necrotic cells, which can be identified with cell viability stains (see Materials and Methods). The results seen with 7-AAD dye, which is used to stain dead cells, indicated that VP40 expression did not induce a significant change in cell viability compared to the results seen with untreated cells or vehicle-treated cells (Fig. 6D). However, treatment of cells with camptothecin or the calcium ionophore ionomycin, both of which induce PS exposure, did cause significant changes in cell viability (Fig. 6D).
FIG 6.
Flow cytometry analysis of PS exposure. (A) HEK293 cells expressing EGFP or EGFP-VP40 were stained with annexin V PE according to the protocol detailed in Materials and Methods. The fluorescence intensity was normalized to the mode; data represent the results of independent experiments performed in triplicate. (B) HEK293 cells expressing EGFP–HIV-1 gag or EGFP-VP40 were stained with annexin V PE according to the protocol detailed in Materials and Methods. Note that the VP40 signals shown in panels A and B are the same to facilitate comparisons. (C) Annexin V staining was performed for HEK293 cells with or without VP40 expression. The median value of fluorescent intensity was measured for each sample to calculate the fold increase in mean fluorescence intensity for each sample normalized to untreated HEK293 cells. (D) 7-AAD staining was used to determine cell viability under the conditions indicated in panel A and to determine the median value of fluorescent intensity for each sample as described for panel A. This was used to calculate the fold increase in mean fluorescence intensity for each sample normalized to untreated HEK293 cells. (E) Annexin V staining, performed as described for panel A, was used to compare HEK293 cells expressing EGFP-VP40, EGFP, HIV-1 gag, and other fluorescent constructs. Note that the VP40 signals shown in panels C and E are the same to facilitate comparisons. (F) 7-AAD staining, performed as described for panel B, was used to determine the viability of HEK293 cells expressing EGFP-VP40, EGFP, HIV-1 gag, and other fluorescent constructs. Note that the VP40 signals shown in panels D and F are the same to facilitate comparisons. n = 3 for each condition; results were used to determine the SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
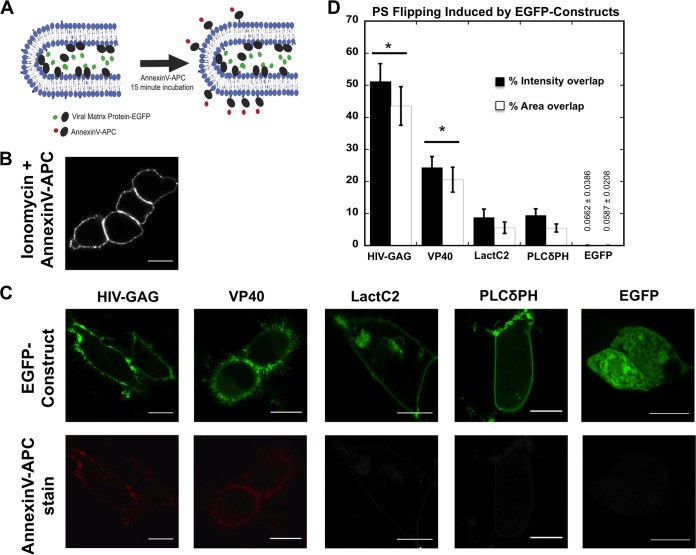
FIG 7.
Imaging of PS exposure on live cells. (A) Cartoon representation of PS exposure imaging experiment where annexin V-APC was bound to extracellular PS exposed on a virus-like particle induced by an EGFP-viral matrix protein transfected into HEK293 cells. (B) Representative cells treated with ionomycin as directed by the manufacturer and with annexin V-APC stain in 1× annexin V binding buffer for 15 to 20 min. (C) Representative images of HEK293 cells transfected for 18 to 24 h with HIV-1 gag-EGFP, VP40-WT-EGFP, Lact C2-EGFP, PLCδPH-EGFP, and EGFP. Transfected cells were treated with annexin V-APC stain in 1× annexin V binding buffer for 15 to 20 min. All scale bars are 10 μM. (D) Quantification of PS exposure detected by annexin V-APC stain with EGFP constructs. A total of 25 to 40 cells per construct were imaged for each experiment, and four or more independent experiments were performed. Percent intensity and percent area overlap of annexin V-APC and EGFP at the plasma membrane were determined in MATLAB for each cell (see Materials and Methods). The results are displayed ± SEM, and significant (P < 0.05) increases in PS exposure from controls Lact C2, PLCδPH, and EGFP are marked with a star (*).
To gauge the specificity of PS exposure from HEK293 cells, we also tested expression of EGFP (Fig. 6A and D), the PS sensor Lact C2 (Fig. 6B and E), the PI(4,5)P2 sensor PLCδ PH (Fig. 6E), and the HIV-1 gag protein (Fig. 6B and E). HIV-1 gag (Fig. 6B), like that of VP40 (Fig. 6A and B), induces a significant amount of PS exposure. This is not unexpected, however, as previous studies have demonstrated that HIV particles have PS exposed on the outer leaflet (6). In contrast, EGFP, Lact C2, and PLCδ PH do not induce a statistically significant amount of PS exposure on the exoplasmic leaflet of human cells. To determine if VP40 plasma membrane localization, and not just VP40 cellular expression, is necessary for PS exposure, we employed the L213A mutation of VP40, which has previously been shown to drastically reduce plasma membrane localization (24, 46) (data not shown). The PS levels detected on cells expressing L213A were reduced ∼7-fold compared to the WT level of VP40 expression (Fig. 6E).
We next used confocal microscopy to image and quantify VP40 expression and PS exposure on single cells. In consonance with the flow cytometry data, VP40-expressing cells displayed a significant increase in the number of cells with PS exposed as detected by the annexin V assay (Fig. 7A to C). Additionally, annexin V intensity analysis indicated that VP40-expressing cells display a statistically significant increase in PS on the cells' outer leaflets (Fig. 7). MATLAB was then used to quantify the percentage of intensity overlap between EGFP-VP40 and annexin V signals as well as area overlap of the signals (Fig. 7D). As expected, expression of HIV-1 gag induced a significant amount of PS exposure, expression of Lact C2 and PLCδ PH induced a small amount of PS exposure, and expression of EGFP did not induce any statistically significant exposure of PS (Fig. 7D). Enhanced sensitivity of the confocal detection system likely leads to detection of exposed PS for Lact C2 and PLCδ PH in the confocal assay but not in the flow cytometry assay. Together, these studies demonstrate that VP40 PM localization is sufficient to induce significant exposure of PS at sites of egress.
DISCUSSION
Despite its initial discovery having occurred almost 4 decades ago (57), little is known about how EBOV assembles and buds from the inner leaflet of the plasma membrane of human cells. Specifically, there is a lack of information on how VP40 interacts with lipid membranes in human cells to form the filamentous particles that emanate from the plasma membrane or how VP40 may orchestrate or control these processes in its favor. VP40 was first shown to bind PS-containing vesicles more than a decade ago (25, 26). Here, we show that the VP40-PS membrane interaction occurs with nanomolar affinity. Recently, VP40 was shown to dimerize and align its C-terminal domains so that both present their large cationic patch on the same filament surface (21). Lysine residues in this patch seem to be important for plasma membrane localization and budding and may thus interact with the anionic plasma membrane. Additionally, mutation of residues in the N-terminal domain dimeric interface of VP40 also greatly reduced budding and plasma membrane localization (21). This suggested that the dimer may be necessary for transport to the plasma membrane or that the dimer has higher affinity than the monomer for the plasma membrane interface. Our data indicate that VP40 association with plasma membrane PS is essential for its plasma membrane association and formation of Ebola virus VLPs.
Reductions in levels of plasma membrane PS may serve to inhibit or reduce assembly of VP40 and egress of the Ebola virus. VP40 plasma membrane localization and oligomerization were sensitive to the PS content of the PSA-3 cell line versus the CHOK-1 cell line. The PSA-3 cell line, which has an ∼30% decrease in plasma membrane PS content, did not support significant plasma membrane localization of VP40, and VLP formation was barely detectable. VP40 may rely heavily on electrostatic interactions for plasma membrane association and thus may require a threshold amount of PS to achieve binding under physiological conditions. This would help explain why moderate decreases in the plasma membrane PS content or competition assays were able to significantly reduce VP40 plasma membrane binding and assembly. Additionally, VP40 seems to require a higher moles percent of PS in the membrane than Lact C2 to achieve detectable binding, where we found it difficult to detect VP40 binding below 8 mol% PS in the vesicles (28; S. Soni and R. Stahelin, unpublished results). In contrast, HIV-1 gag behaved similarly in PSA-3 and CHOK-1 cells. The PS interaction of HIV-1 gag is well established (52, 53), but the cellular amount of PS required for HIV-1 gag assembly and budding is not known. Thus, VP40 may require higher PS content than HIV-1 gag to support assembly and budding. Admittedly, the current study was just the first step in understanding the complex set of interactions that VP40 likely undergoes with the membrane interface and host proteins to generate infectious virions. Nonetheless, VP40 appears to have a high sensitivity to the PS content of the plasma membrane.
The experiments performed here with sphingosine, which neutralizes the plasma membrane anionic charge, likely demonstrate that VP40 assembly is not simply dependent on anionic charge sensing. Additionally, the oligomerization of VP40, which serves as a marker of Ebola virus budding, was drastically reduced when plasma membrane PS levels decreased or PS binding was specifically blocked by Lact C2. Clearly, the interaction of VP40 with PS is essential to assemble VP40 on the plasma membrane and facilitate egress in human cell lines but other interactions with lipids or proteins must not be neglected. To date, many lipid-binding proteins identified have recognized two specific lipids or a specific lipid and membrane physical property such as curvature (58). For instance, the HIV-1 matrix domain has recently been shown to have additional binding sites for plasma membrane lipids (53, 59) in addition to consensus PI(4,5)P2 binding (60). The HIV-1 matrix domain is also sensitive to the acyl chain composition and cholesterol content of membranes (52). More-systematic analysis of VP40 lipid selectivity is needed to discover the optimal lipid composition with which VP40 interacts or if VP40 lipid binding is able to induce changes in lipid packing or membrane order. This is especially important considering the complex chemical landscape of the plasma membrane, including enrichment of cholesterol, sphingomyelin, differences in acyl chain length and saturation, and the presence of liquid ordered and disordered domains. The role of PS in the assembly and egress of other viruses, particularly those that cause viral hemorrhagic fevers, should also be evaluated.
A number of viruses, including EBOV, have been shown to utilize PS on their outer membrane envelope to enhance entry into target cells (6, 10, 56, 61). PS interacts with the TIM-1 receptor on human cells, and inhibition of this interaction may be of therapeutic value in a number of viral infections (5, 56, 61). Additionally, VP40 VLPs lacking the transmembrane glycoprotein are able to enter target cells in a PS-dependent manner (56). This suggested to us that the driving force behind PS exposure on the outer surface of VLPs may be VP40 alone. Indeed, VP40 and HIV-1 gag are sufficient to induce significant exposure of PS at sites of egress. A mutation that greatly reduced plasma membrane localization (L213A) (24, 46, and data not shown) abrogated PS detection on the exoplasmic leaflet. The idea of PS being flipped may seem counterintuitive, since it seems to be an essential component of VP40 assembly on the inner leaflet; however, some PS redistribution across the plasma membrane may also be advantageous for generating changes in membrane curvature. For example, a lipid such as PE, which can induce negative membrane curvature, could become more concentrated in the inner leaflet subsequent to PS flipping. Additionally, the VP40 oligomers may be less sensitive to the PS content than the dimers that are continuously needed for initial assembly. Thus, VP40 oligomers may be stabilized by other components of the membrane bilayer or by interactions with each other. The mechanisms by which viral matrix proteins are able to provoke changes in PS distribution are still unknown and will require more mechanistic investigations. Research focused on how viruses and viral matrix proteins induce PS redistribution across the plasma membrane interface will also help us to understand how these viruses facilitate entry into the host cell and provide potential avenues for development of pan-viral therapy.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (AI081077) and the Notre Dame Boler-Parseghian Center for Rare and Neglected Diseases to R.V.S. R.V.S. also acknowledges support from the Indiana University School of Medicine—South Bend Imaging and Flow Cytometry CORE as well as the Notre Dame Integrated Imaging Facility (NDIIF). E.A.-G. was supported by an Eck Institute for Global Health predoctoral fellowship. J.L.S. was supported by an AHA predoctoral fellowship (12PRE11960030) and a NIH CBBI predoctoral fellowship (T32GM075762). E.G. and M.A.D. acknowledge support of NIH grants P41-RRO3155 and P50-GM076516 and Keck's Foundation grant 44769549507.
R.V.S. and E.A.-G. thank Yi Xue for excellent technical assistance. R.V.S. thanks Erica Ollmann Saphire for critical readings of the manuscript.
R.V.S. and E.A.-G. conceived and designed the studies. E.A.-G., K.A.J., M.E.F., J.L.S., S.P.S., R.V.S., and K.R.J. performed the experiments. E.A.-G., K.A.J., M.E.F., J.L.S., S.P.S., R.V.S., and K.R.J. analyzed and interpreted the data. C.R.T., M.A.D., and E.G. provided technical consultation for experimentation and data analysis. R.V.S. wrote the paper with input from all authors.
REFERENCES
- 1.Bale S, Liu T, Li S, Wang Y, Abelson D, Fusco M, Woods VL Jr, Saphire EO. 2011. Ebola virus glycoprotein needs an additional trigger, beyond proteolytic priming for membrane fusion. PLoS Negl Trop Dis 5:e1395. doi: 10.1371/journal.pntd.0001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moller-Tank S, Albritton LM, Rennert PD, Maury W. 2014. Characterizing functional domains for TIM-mediated enveloped virus entry. J Virol 88:6702–6713. doi: 10.1128/JVI.00300-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moller-Tank S, Kondratowicz AS, Davey RA, Rennert PD, Maury W. 2013. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol 87:8327–8341. doi: 10.1128/JVI.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moller-Tank S, Maury W. 2014. Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology 468–470:565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soares MM, King SW, Thorpe PE. 2008. Targeting inside-out phosphatidylserine as a therapeutic strategy for viral diseases. Nat Med 14:1357–1362. doi: 10.1038/nm.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monde K, Chukkapalli V, Ono A. 2011. Assembly and replication of HIV-1 in T cells with low levels of phosphatidylinositol-(4,5)-bisphosphate. J Virol 85:3584–3595. doi: 10.1128/JVI.02266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A 101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolnik O, Kolesnikova L, Becker S. 2008. Filoviruses: interactions with the host cell. Cell Mol Life Sci 65:756–776. doi: 10.1007/s00018-007-7406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittler E, Kolesnikova L, Herwig A, Dolnik O, Becker S. 2013. Assembly of the Marburg virus envelope. Cell Microbiol 15:270–284. doi: 10.1111/cmi.12076. [DOI] [PubMed] [Google Scholar]
- 13.Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet 377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harty RN. 2009. No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res 81:189–197. doi: 10.1016/j.antiviral.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radzimanowski J, Effantin G, Weissenhorn W. 2014. Conformational plasticity of the Ebola virus matrix protein. Protein Sci 23:1519–1527. doi: 10.1002/pro.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahelin RV. 2014. Membrane binding and bending in Ebola VP40 assembly and egress. Front Microbiol 5:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J Virol 75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licata JM, Johnson RF, Han Z, Harty RN. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol 78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol 76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. 2001. Vesicular release of Ebola virus matrix protein VP40. Virology 283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- 21.Bornholdt ZA, Noda T, Abelson DM, Halfmann P, Wood MR, Kawaoka Y, Saphire EO. 2013. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. 2012. Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys J 102:2517–2525. doi: 10.1016/j.bpj.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adu-Gyamfi E, Soni SP, Jee CS, Digman MA, Gratton E, Stahelin RV. 2014. A loop region in the N-terminal domain of Ebola virus VP40 is important in viral assembly, budding, and egress. Viruses 6:3837–3854. doi: 10.3390/v6103837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adu-Gyamfi E, Soni SP, Xue Y, Digman MA, Gratton E, Stahelin RV. 2013. The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J Biol Chem 288:5779–5789. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruigrok RW, Schoehn G, Dessen A, Forest E, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. 2000. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J Mol Biol 300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 26.Scianimanico S, Schoehn G, Timmins J, Ruigrok RH, Klenk HD, Weissenhorn W. 2000. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J 19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soni SP, Adu-Gyamfi E, Yong SS, Jee CS, Stahelin RV. 2013. The Ebola virus matrix protein deeply penetrates the plasma membrane: an important step in viral egress. Biophys J 104:1940–1949. doi: 10.1016/j.bpj.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soni SP, Stahelin RV. 2014. The Ebola virus matrix protein VP40 selectively induces vesiculation from phosphatidylserine-enriched membranes. J Biol Chem 289:33590–33597. doi: 10.1074/jbc.M114.586396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olivotto M, Arcangeli A, Carla M, Wanke E. 1996. Electric fields at the plasma membrane level: a neglected element in the mechanisms of cell signalling. Bioessays 18:495–504. doi: 10.1002/bies.950180612. [DOI] [PubMed] [Google Scholar]
- 30.van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vance JE, Steenbergen R. 2005. Metabolism and functions of phosphatidylserine. Prog in Lipid Res 44:207–234. doi: 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin S, Murray D. 2005. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 33.Cho W, Stahelin RV. 2006. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta 1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brudner M, Karpel M, Lear C, Chen L, Yantosca LM, Scully C, Sarraju A, Sokolovska A, Zariffard MR, Eisen DP, Mungall BA, Kotton DN, Omari A, Huang IC, Farzan M, Takahashi K, Stuart L, Stahl GL, Ezekowitz AB, Spear GT, Olinger GG, Schmidt EV, Michelow IC. 2013. Lectin-dependent enhancement of Ebola virus infection via soluble and transmembrane C-type lectin receptors. PLoS One 8:e60838. doi: 10.1371/journal.pone.0060838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. 2012. Phosphatidylserine dynamics in cellular membranes. Mol Biol Cell 23:2198–2212. doi: 10.1091/mbc.E11-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. 2002. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem 277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 39.Huang BX, Akbar M, Kevala K, Kim HY. 2011. Phosphatidylserine is a critical modulator for Akt activation. J Cell Biol 192:979–992. doi: 10.1083/jcb.201005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. 2012. Single-particle tracking demonstrates that actin coordinates the movement of the Ebola virus matrix protein. Biophys J 103:L41–L43. doi: 10.1016/j.bpj.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalal RB, Digman MA, Horwitz AF, Vetri V, Gratton E. 2008. Determination of particle number and brightness using a laser scanning confocal microscope operating in the analog mode. Microsc Res Tech 71:69–81. doi: 10.1002/jemt.20526. [DOI] [PubMed] [Google Scholar]
- 42.Digman MA, Brown CM, Horwitz AR, Mantulin WW, Gratton E. 2008. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophys J 94:2819–2831. doi: 10.1529/biophysj.107.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Digman MA, Dalal R, Horwitz AF, Gratton E. 2008. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys J 94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahelin RV. 2013. Monitoring peripheral protein oligomerization on biological membranes. Methods Cell Biol 117:359–371. doi: 10.1016/B978-0-12-408143-7.00019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita SY, Shirakawa S, Kobayashi Y, Nakamura K, Teraoka R, Kitagawa S, Terada T. 2012. Enzymatic measurement of phosphatidylserine in cultured cells. J Lipid Res 53:325–330. doi: 10.1194/jlr.D021808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy SE, Johnson RF, Zhang YA, Sunyer JO, Harty RN. 2007. Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J Virol 81:11452–11460. doi: 10.1128/JVI.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 48.Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, Iemura S, Natsume T, Kuwahara R, Nakagawa T, Nishikawa K, Mukai K, Miyoshi E, Taniguchi N, Sheff D, Lencer WI, Taguchi T, Arai H. 2011. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc Natl Acad Sci U S A 108:15846–15851. doi: 10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fairn GD, Hermansson M, Somerharju P, Grinstein S. 2011. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat Cell Biol 13:1424–1430. doi: 10.1038/ncb2351. [DOI] [PubMed] [Google Scholar]
- 50.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. 2011. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol 194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuge O, Nishijima M, Akamatsu Y. 1986. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. II. Isolation and characterization of phosphatidylserine auxotrophs. J Biol Chem 261:5790–5794. [PubMed] [Google Scholar]
- 52.Dick RA, Goh SL, Feigenson GW, Vogt VM. 2012. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A 109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vlach J, Saad JS. 2013. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc Natl Acad Sci U S A 110:3525–3530. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glomset JA. 2006. Role of docosahexaenoic acid in neuronal plasma membranes. Sci STKE 2006:pe6. doi: 10.1126/stke.3212006pe6. [DOI] [PubMed] [Google Scholar]
- 55.Hoenen T, Biedenkopf N, Zielecki F, Jung S, Groseth A, Feldmann H, Becker S. 2010. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol 84:7053–7063. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, Bu X, Farzan M, Freeman GJ, Umetsu DT, Dekruyff RH, Choe H. 2013. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog 9:e1003232. doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emond RT, Evans B, Bowen ET, Lloyd G. 1977. A case of Ebola virus infection. Br Med J 2:541–544. doi: 10.1136/bmj.2.6086.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moravcevic K, Oxley CL, Lemmon MA. 2012. Conditional peripheral membrane proteins: facing up to limited specificity. Structure 20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olety B, Ono A. 2014. Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res 193:108–115. doi: 10.1016/j.virusres.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morizono K, Chen IS. 2014. Role of phosphatidylserine receptors in enveloped virus infection. J Virol 88:4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]