ABSTRACT
The propensity for transspecies prion transmission is related to the structural characteristics of the enciphering and new host PrP, although the exact mechanism remains incompletely understood. The effects of variability in prion protein on cross-species prion transmission have been studied with animal bioassays, but the influence of prion protein structure versus that of host cofactors (e.g., cellular constituents, trafficking, and innate immune interactions) remains difficult to dissect. To isolate the effects of protein-protein interactions on transspecies conversion, we used recombinant PrPC and real-time quaking-induced conversion (RT-QuIC) and compared chronic wasting disease (CWD) and classical bovine spongiform encephalopathy (cBSE) prions. To assess the impact of transmission to a new species, we studied feline CWD (fCWD) and feline BSE (i.e., feline spongiform encephalopathy [FSE]). We cross-seeded fCWD and FSE into each species' full-length, recombinant PrPC and measured the time required for conversion to the amyloid (PrPRes) form, which we describe here as the rate of amyloid conversion. These studies revealed the following: (i) CWD and BSE seeded their homologous species' PrP best; (ii) fCWD was a more efficient seed for feline rPrP than for white-tailed deer rPrP; (iii) conversely, FSE more efficiently converted bovine than feline rPrP; (iv) and CWD, fCWD, BSE, and FSE all converted human rPrP, although not as efficiently as homologous sCJD prions. These results suggest that (i) at the level of protein-protein interactions, CWD adapts to a new species more readily than does BSE and (ii) the barrier preventing transmission of CWD to humans may be less robust than estimated.
IMPORTANCE We demonstrate that bovine spongiform encephalopathy prions maintain their transspecies conversion characteristics upon passage to cats but that chronic wasting disease prions adapt to the cat and are distinguishable from the original prion. Additionally, we showed that chronic wasting disease prions are effective at seeding the conversion of normal human prion protein to an amyloid conformation, perhaps the first step in crossing the species barrier.
INTRODUCTION
Prion diseases are characterized by the seeded misfolding and aggregation of the cellular prion protein, PrPC, to an amyloid fibrillar state, PrPRes. All mammals express PrPC, with only minor polymorphisms within and between species (1). Prion disease has been identified in humans (Creutzfeldt-Jakob disease [CJD]), cattle (bovine spongiform encephalopathy [BSE]), cervids (chronic wasting disease [CWD]), felines (feline spongiform encephalopathy [FSE]), and sheep and goats (scrapie), as well as other mammals (2). The transmissibility of prion diseases makes them unique among other protein misfolding diseases, including Alzheimer's and Parkinson's diseases. Though prion diseases vary in their proclivity to transmit to new species, the classical BSE (cBSE) epidemic and subsequent transmission to humans (variant CJD [vCJD]), felines, and several ungulate species illustrate the importance of understanding zoonotic transmission (2–8).
The transspecies transmissibility of cBSE and CWD has been studied in natural hosts, conventional and transgenic rodents, and in vitro models. CWD has been transmitted experimentally to sheep, felines, cattle, and squirrel monkeys (9–14). BSE has been transmitted experimentally to sheep, European red deer, and macaques (15–17). Transgenic mice have been developed to explore the potential transmissibility of cBSE and CWD to humans. In four studies with mice expressing the human prion protein (TgHu), cBSE inoculation produced mixed results, with attack rates ranging from 0 to ∼50% (18–21). Inoculation of TgHu mice with CWD has yielded no infections (21–24). Additionally, three distinct in vitro amyloid amplification methods have also shown that BSE is better able to convert human PrP than is CWD (25–31).
That prion characteristics are not solely defined by primary structure can be inferred by the existence of prion strains (25). Indeed, several studies have noted the similarity that transspecies-passaged cBSE prions (as represented by FSE, vCJD, and sheep BSE) have to the original cBSE agent, despite the variable primary structure of the passaged prions (19, 26, 32, 33). Therefore, it is logical to conclude that variability in prion conformation and the interactions of PrPRes and PrPC are essential to prion species barriers. We employ real-time quaking-induced conversion (RT-QuIC) to assess the complementarity of seed-substrate pairing and, by extension, the possibility of transspecies conversion, of two prions pertinent to human health, i.e., the agents causing CWD and BSE.
In the studies to follow, we hypothesized that in RT-QuIC, cBSE and passaged forms of cBSE would have similar transspecies seeding characteristics and would readily convert human rPrP, whereas passaged forms of CWD would maintain fewer CWD seeding tendencies and would convert human rPrP poorly.
MATERIALS AND METHODS
rPrP production.
The coding regions for full-length (amino acids [aa] 23 to 231) recombinant prion protein (rPrP) from each species (bank vole, bovine, feline, human M129, and white-tailed deer) were kindly provided by Glenn Telling. Full-length constructs (bovine, feline, human M129, and white-tailed deer) were cloned into the pET100D expression system (Life Technologies). The truncated Syrian hamster construct in Escherichia coli BL21 was kindly provided by Byron Caughey. The bank vole PRNP coding region was truncated using specific primers to isolate the sequence for amino acids 90 to 231 and was cloned into the vector that contained the Syrian hamster construct. E. coli BL21 Star cells (Life Technologies) were used to express rPrP. Briefly, BL21 cells from a glycerol stock were spiked into 5 ml LB medium and 5 μg/ml ampicillin and grown overnight with shaking at 37°C. LB medium (1 liter) was inoculated with the 5-ml culture plus 5 μg/ml ampicillin and autoinduction reagents [final concentrations, 0.5 M (NH4)2SO4, 1 M KH2PO4, 1 M Na2HPO4, 0.5% glycerol, 0.05% glucose, 0.2% α-lactose, 0.001 M MgSO4]. Bacteria were harvested when the optical density at 600 nm (OD600) reached approximately 3.0; cells were lysed and inclusion bodies isolated according to the manufacturer's protocol with Bug Buster and Lysonase (EMD-Millipore). Inclusion bodies were solubilized and rotated in 8 M guanidine hydrochloride (GdnHCl) and 100 mM Na2HPO4 for at least 1 h at room temperature. The denatured rPrP was bound to Superflow nickel resin (Qiagen) and refolded at a rate of 0.75 ml/min on the column with a gradient from 6.0 M GdnHCl–100 mM Na2HPO4–10 mM Tris (pH 8.0) to the same buffer with no GdnHCl. A gradient to 0.5 M imidazole–100 mM NaH2PO4–10 mM Tris (pH 5.5) with a flow rate of 2.0 ml/min resulted in rPrP elution. The eluted rPrP was filtered and dialyzed at a concentration of ∼0.4 mg/ml in two changes of 4.0 liters 20 mM NaH2PO4 (pH 5.5). After dialysis, the rPrP was filtered and stored at 4°C, its concentration having been determined by A280. Purity of the rPrP samples was tested by performing gel electrophoresis (12% bis-Tris gels with 1× MOPS [morpholinepropanesulfonic acid; Bio-Rad], 190 V, 55 min) and staining with Coomassie brilliant blue (Bio-Rad).
RT-QuIC.
rPrP (0.1 mg/ml) was combined with premixed reaction buffer (final concentrations, 400 mM NaCl, 1 mM EDTA, 20 mM NaH2PO4) and freshly made thioflavin T (ThT; final concentration, 10 μM) for a final volume of 95 μl per well in a 96-well plate. Brain material (Table 1) was homogenized in 1× phosphate-buffered saline (PBS) as a 10% solution with a bead beater (Next Advance). Brain homogenate was aliquoted in single-use tubes and frozen at −80°C. Homogenate aliquots were thawed and diluted into 1× PBS plus 0.05% SDS (first set of conditions, here termed “condition 1”) or 0.1% SDS (“condition 2”). Two microliters of diluted brain seed was added to 95 μl protein substrate mix in the well of an optical bottom 96-well plate. A real-time quaking-induced conversion (RT-QuIC) experiment consisted of 400 cycles (100 h) of shaking and incubation at 45°C; specifically, plates were shaken for 1 min (700 rpm, double orbital) followed by 1 min of rest. Fluorescence (450-nm excitation and 480-nm emission, 20 flashes/well) was recorded every 15 min using a gain of 1,700 (96-well plate).
TABLE 1.
Sources of the prion seeds used in the study experimentsa
| Prion type | Species (n) | Prion disease status | Tissue | Inoculum | Inoculation | Sample ID |
|---|---|---|---|---|---|---|
| CWD | WTD (1) | + | Caudal brain sections | Colorado Department of Wildlife positive deer LA01 | Oral | 104 |
| Exper. CWD | WTD (6) | + | Whole brain | CSU CWD-positive deer 700, 800 series | Oral or aerosol | CBP6 |
| Negative deer | WTD (1) | − | Caudal brain sections | CSU CWD-negative deer UGA 1/2 | Oral | 123 |
| Feline CWD | Feline (2) | + | Multiple brain sections | CSU CWD-positive deer | Intracranial | 4137/4152 |
| Negative cat | Feline (1) | − | Obex | CSU CWD-negative deer UGA 1/2 | Oral | 4141 |
| FSEb | Feline (1) | + | Medulla | Natural infection | Assumed oral | FSE |
| BSE | Bovine (1) | + | Obex | Natural infection | Assumed oral | BSE |
| Negative cow | Bovine (1) | − | Brainstem | Not inoculated | Negative bovine | |
| Field CWD 1c | WTD | + | Obex | Field isolate | H92 | |
| Field CWD 2c | WTD | + | Obex | Field isolate | 98933968 |
In order to ensure that the results observed in our RT-QuIC experiments were not a result of specific experimental conditions, all the experiments in this study were completely repeated under two sets of conditions. For condition 1, one or two batches of each rPrP species (bovine, feline, white-tailed deer, human) were used and the concentration of SDS used to dilute the brain material was 0.05%. For condition 2, new batches (one or two) of each species of rPrP were used, with an SDS concentration of 0.1% in the sample dilution step. For both condition 1 and condition 2, there were 6 to 8 replicates (on 3 or 4 plates) of every reaction. Because the reaction conditions resulted in different kinetics (kinetics using 0.1% SDS were faster than 0.05% SDS), rates of amyloid conversion could not be compared between experiments. The data shown here are from condition 2, but these patterns were maintained in condition 1.
Artificial seeds.
In order to assess each species' inherent propensity to convert from rPrPC to rPrPRes (amyloidogenicity), we created synthetic rPrPThT+ seeds from truncated Syrian hamster rPrPC (SH; amino acids 90 to 231) or truncated bank vole rPrPC (BV; amino acids 90 to 231). For the SH artificial seeds, 0.5 mg/ml SH rPrPC with 2 M GdnHCl was shaken at 37°C overnight. BV artificial seeds were made by shaking 0.3 mg/ml BV rPrPC at 37°C without GdnHCl. In both cases, 2 μl artificial seed (or 10-fold dilutions thereof) was added to the RT-QuIC reaction mixture in place of a brain-derived seed.
Western blotting and quantification.
A volume of 5 μl, 9 μl, or 13 μl of a 5% homogenate of BSE or CWD (sample 104) was added to 1 μl of proteinase K (PK; 100 μg/ml; Invitrogen) and 1 μl of 2% SDS and brought to a volume of 15 μl with water. These samples were incubated at 37°C for 30 min and then at 45°C for 10 min. Five microliters of a 4× sample loading buffer/reducing agent (Life Technologies) was added to the samples, and they were incubated at 93°C for 3 min. Eighteen microliters was added to each well of a 12-well, 12% bis-Tris gel. The samples were electrophoresed for 10 min at 115 V and then for 1 h at 150 V. Transfers were performed in cold transfer buffer (20% MeOH, 192 mM glycine, 25 mM Tris base) for 1.5 h at 115 V, and the membranes were blocked overnight at 4°C in Tris-buffered saline plus 0.1% Tween (TBST) and dry milk. 6H4 was bound to the membrane in TBST plus milk for 1 h at a concentration of 1:5,000, and then the membrane was washed with TBST for 1 h. The secondary antibody, a Licor-tagged goat anti-mouse antibody, was bound to the membrane in TBST plus milk for 1 h at a concentration of 1:20,000. The membrane was again washed with TSBT for 1 h. Images were collected on the Odyssey CLx Infrared Imager (Li-Cor Biosciences, Inc.) and analyzed using Image Studio Lite Version 4.0 (Li-Cor Biosciences, Inc.). Briefly, the intensity was calculated for each lane of the Western blot and then divided by the number of microliters of 5% brain homogenate added to the well. Finally, the intensity per microliter for each PK+ lane was divided by the intensity of the PK− lane to determine the relative quantity of PK-resistant material.
Quantitative analysis.
The lag phases of positive and negative samples were determined using MARS software (BMG Labtech) by calculating the time required to meet a threshold for positive samples (34). The threshold was defined as the average baseline fluorescence plus 5 standard deviations for each experiment. The rate is the inverse of the lag phase for each sample (1/lag phase), with units of hours−1. We use the rate of amyloid conversion here instead of lag phase in order to account for the samples that never cross the threshold (lag phase for these samples is undefined, but the rate can be conservatively assigned a value of 0 h−1). Despite their similar purity, conformation, and functionality, the rPrP substrates were not equally sensitive. In order to compare one seed's behavior across multiple substrates without confounding by the inherent amyloidogenicity differences between the substrates, relative rates were calculated and used for analysis. Specifically, all rates for a given substrate were divided by the highest rate in that substrate; the fastest sample has a rate of 1.0. Average relative rates of amyloid conversion from 6 to 12 replicates are displayed as the means, with error bars indicating the standard errors of the means. The difference between rates at each dilution was tested with the nonparametric Mann-Whitney U (MWU) test (when both samples had a nonzero median) in Prism 5.0 (Graphpad) or the nonparametric one-sample Wilcoxon signed-rank (WSR) test (when one of the samples had a median of zero) in R. A statistically significant difference was defined by a P value of <0.05 from the appropriate test. Significant differences by the MWU test are indicated in the figures by an asterisk (*), while significant differences by the WSR test are indicated by a pound sign (#).
RESULTS
Assessment of recombinant bovine, feline, human, and white-tailed deer PrP and quality.
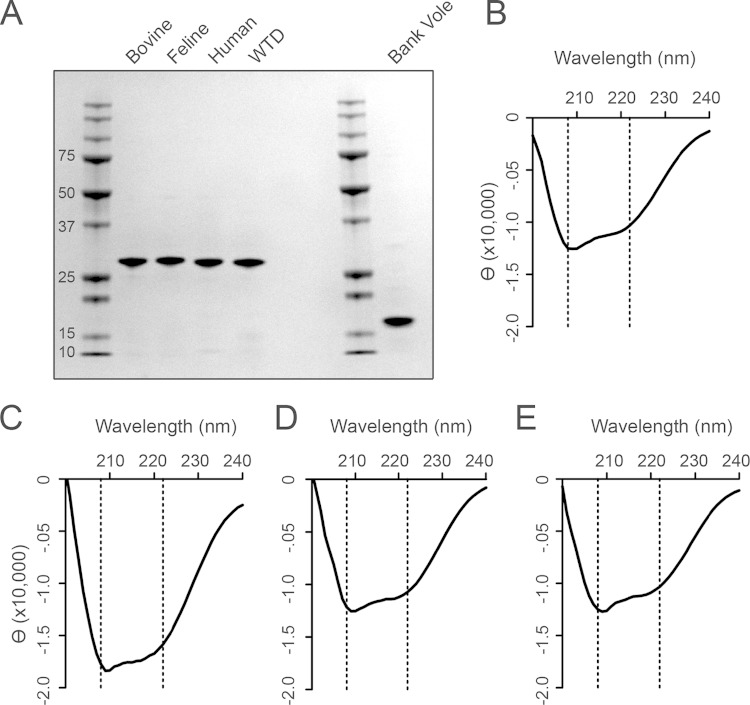
In order to compare the behavior of a given prion seed across multiple rPrP substrates, it was necessary to ensure that every batch of every substrate had similar purity, structure, and functionality. Each rPrP preparation was analyzed by gel electrophoresis and Coomassie blue staining to show that the substrates were of comparable purity (Fig. 1A). To ensure that the secondary structures from each preparation were comparable, we used circular dichroism (CD) spectroscopy (Fig. 1B to E). All substrates had minima at 208 nm and 222 nm, indicating a predominance of α-helical secondary structure, as expected. Each preparation of recombinant protein was analyzed with the homologous species' prion seed in RT-QuIC to verify its ability to form amyloid upon addition of seed. Homologous seeds (e.g., CWD brain samples in full-length white-tailed deer substrate) were considered positive controls, and negative brain samples (deer, bovine, and feline) were negative controls. A batch was considered functional if the wells containing positive-control seeds exhibited ThT fluorescence emission signals that crossed the threshold within 20 h (rate, ≥1/20, 0.05 h−1) and if the negative controls had an average rate less than 0.015 h−1. By these metrics, we determined that all batches of rPrP were comparable and that seed behavior could consequently be compared among substrates.
FIG 1.
rPrP substrates are of comparable quality. (A) Coomassie blue visualization of 1.5 μg rPrP substrate indicates the purity of each rPrP. Bands represent, from left to right, ladder, bovine (amino acids 23 to 231), feline (aa 23 to 231), human M129 (aa 23 to 231), and white-tailed deer (aa 23 to 231) rPrP, ladder, and bank vole (aa 90 to 231) rPrP. (B) CD spectrum for bovine rPrP. (C) CD spectrum for feline rPrP. (D) CD spectrum for human M129 rPrP. (E) CD spectrum for white-tailed deer rPrP. The dotted lines indicate the anticipated minima for α-helical structure (B to F).
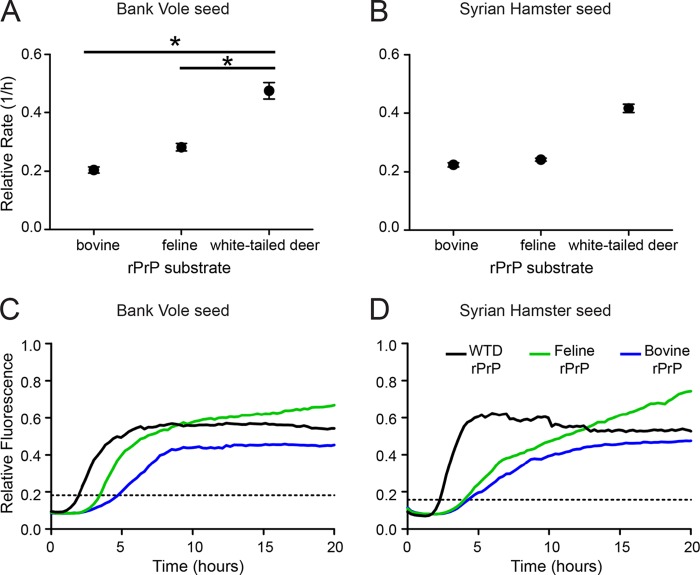
rPrP species differ in inherent propensity for conversion, as reflected by the rate of amyloid conversion.
Despite their similar purity, conformation, and functionality, the rPrP substrates did not have an equal propensity to convert to an amyloid conformation (amyloidogenicity), which may reflect inherent features from each species, as multiple purified batches of rPrP had the same behavior. With a variety of prion seeds (CWD, BSE, fCWD, FSE), the highest rate of amyloid conversion for full-length white-tailed deer rPrP (0.27 h−1) was nearly twice the highest rate for full-length bovine (0.13 h−1) or feline (0.17 h−1) rPrP. We assessed the inverse of the lag phase, the rate of amyloid conversion, in order to include samples that never become positive (which were conservatively assigned a rate of 0).
To verify the observed differences among substrates, we used artificial seeds (rPrPThT+ created by shaking rPrPC with guanidine hydrochloride at 37°C) and compared the rates of amyloid conversion. Mimicking our observation with brain-derived prion seeds, white-tailed deer rPrP converted the fastest with the addition of truncated bank vole rPrPThT+, with a rate approximately twice the rate of bovine and feline rPrP conversion (Fig. 2A). We observed the same pattern when the seed was prepared from truncated Syrian hamster rPrP (Fig. 2B). The same trends were visualized with representative raw data (Fig. 2C and D). These experiments with artificial seeds confirmed observations in experiments with natural seeds, demonstrating the inherently different propensities for amyloid conversion in different species. In order to compare the behavior of a given seed among species with various levels of amyloidogenicity, we normalized the rates by considering the maximum rate in each substrate to be 1.0 h−1.
FIG 2.
rPrP substrates have different inherent amyloidogenicity. (A) Points indicate the average rate of amyloid conversion for each substrate (bovine, feline, and white-tailed deer) upon addition of artificial truncated bank vole rPrPThT+. (B) Points indicate the rate of amyloid conversion for each substrate upon the addition of artificial truncated Syrian hamster rPrPThT+. Error bars represent the standard errors of the means (SEM) (A and B). (C and D) Each line represents the average ThT fluorescence for 4 replicates for each seed-substrate combination, and the dotted line indicates the threshold for determination of the lag phase.
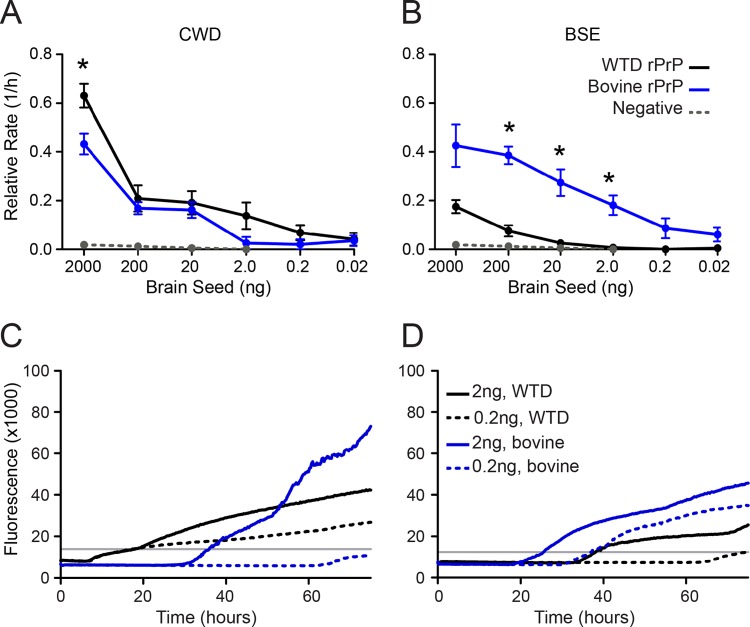
RT-QuIC using full-length rPrP recapitulates BSE and CWD species barriers in vitro.
We hypothesized that real-time conversion would reflect in vivo species barriers, as defined by preferential seeding of the native host species rPrP. By using quantitative RT-QuIC analysis, we found the native prion of a given species to be most compatible (the fastest conversion to amyloid) with its host substrate. To demonstrate an in vitro species barrier, we added cBSE or CWD PrPRes to the nonhomologous rPrP substrate. As anticipated, CWD converted full-length white-tailed deer rPrP faster than it did full-length bovine rPrP (Fig. 3A). Likewise, cBSE seeded full-length bovine rPrP relatively faster than full-length white-tailed deer rPrP (Fig. 3B). Thus, RT-QuIC recapitulated the in vivo species-seeding proclivities of CWD and cBSE prions.
FIG 3.
Preference for intraspecies conversion by BSE and CWD prions is recapitulated in RT-QuIC. (A) CWD was seeded into white-tailed deer rPrP (black line) or bovine rPrP (blue line). (B) BSE is seeded into white-tailed deer (WTD) rPrP (black line) or bovine rPrP (blue line). Each point represents the relative rate of amyloid conversion for each seed concentration (in nanograms), and the error bars represent SEM. The rate of spontaneous amyloid conversion is designated by the dotted gray line in panels A and B. Significant differences between substrates were tested by the MWU test (*, P < 0.05). (C and D) Each line represents the average ThT trace for 6 replicates of 3 dilutions of each seed-substrate combination, and the gray line indicates the threshold for determination of the lag phase.
Enciphering characteristics of cBSE and cBSE-derived prions are conserved after transspecies transmission.
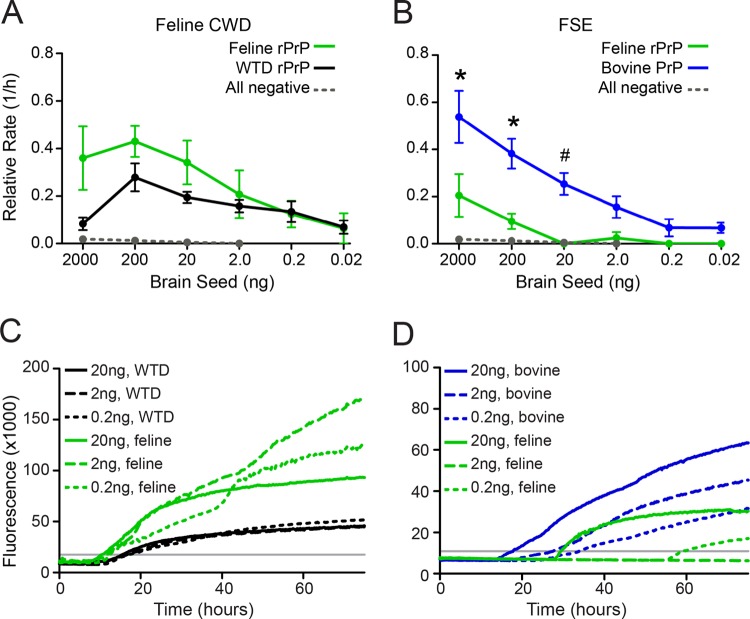
cBSE and CWD are prion diseases that have been naturally passaged in their respective species (cattle and deer), whereas feline spongiform encephalopathy (FSE) and feline chronic wasting disease (fCWD) are first-passage infections in a new host species (cat). To investigate the biochemical properties of cBSE and CWD after transspecies transmission to felines, we compared the amyloidogenicity of fCWD and FSE in the original host and in feline substrate. We found fCWD to be a more efficient seed for its new (feline) host, suggesting that adaptation to the new host had occurred (Fig. 4A). In contrast, FSE remained a more efficient seed for its enciphering (bovine) host, despite its derivation from feline brain PrPC (Fig. 4B). Thereby, these cross-species seeding experiments in RT-QuIC indicated that the characteristics of cBSE were maintained upon passage to a new species whereas CWD had adapted to its new host. These findings in felids suggest that cBSE may retain its ability to cross species barriers even after transmission to a new host species and that CWD may change substantially upon transspecies transmission.
FIG 4.
BSE and CWD prions passed into felines demonstrate either (i) maintenance of original species characteristics (BSE) or (ii) adaptation to the new species (CWD). (A) Points indicate the average rate of amyloid conversion for white-tailed deer rPrP (black line) or feline rPrP (green line) upon seeding with an fCWD seed. (B) Points indicate the rate of amyloid conversion for bovine rPrP (blue line) or feline rPrP (green line) upon addition of an FSE seed. The rate of spontaneous amyloid conversion is designated by the dotted gray line, and error bars represent SEM. Significant differences between substrates were tested by the MWU test (*, P < 0.05) or the WSR test (#, P < 0.05). (C, D) Each line represents the average ThT trace for 6 replicates of 3 dilutions of each seed-substrate combination, and the gray line indicates the threshold for determination of the lag phase.
Human rPrPC can be converted by bovine, feline, and cervid prions.
The threat of zoonotic transmission of prion disease is evident and well documented, yet such transmission is uncommonly observed and incompletely understood. We thereby explored the propensity of heterologous prions to convert human rPrP. In these human rPrPC experiments, we used sporadic CJD brain as a positive control and normal bovine, white-tailed deer, and feline brain as negative controls. sCJD, as expected, seeded human rPrPC most efficiently, so all other seeds were normalized to the rate of conversion of sCJD. We found human rPrPC to be a competent substrate in RT-QuIC for CWD, fCWD, cBSE, and FSE (Fig. 5A). Interestingly, CWD and fCWD converted human rPrPC more efficiently than did cBSE and FSE. These data suggest that at the level of PrPC-PrP seed interaction, CWD has the ability to template the conversion of human rPrPC to ThT-positive amyloid. In order to assess whether CWD was faster than cBSE due to an increased concentration of prion seed, we performed Western blotting on the seed inocula. Western blots indicated that the cBSE sample had a higher concentration of PrPRes than the CWD sample, indicating that CWD was not a better seed than cBSE due to PrPRes content (Fig. 5B). Finally, we assessed the behavior of 8 CWD field isolates, brain samples from white-tailed deer infected naturally and verified to be positive using full-length white-tailed deer RT-QuIC (Fig. 5C). All 8 of these isolates converted human rPrPC, confirming that our observations were not due to the use of experimentally CWD (Fig. 5D). In all, these experiments suggest that the CWD prions naturally circulating in the western United States have the capacity to convert human rPrPC in this assay of protein-protein interactions.
FIG 5.
CWD is capable of efficiently seeding the conversion of human rPrPC. (A) Points represent the average rate of conversion of full-length human M129 rPrP by sporadic CJD (black line), CWD (red line), fCWD (pink line), BSE (dark blue line), and FSE (light blue line). Error bars indicate SEM. The rate of spontaneous amyloid conversion is indicated by the gray line. (B) Western blot of BSE and CWD. PK-digested lanes indicate the presence of more PrPRes in the BSE sample than in the CWD sample. Densitometry indicates that BSE has more PrPRes/microliter relative to total undigested PrP/microliter than does CWD. (C) Points indicate the average rate of amyloid conversion for multiple field isolates of CWD in full-length human M129 rPrPC. The gray line indicates the rate of spontaneous amyloid conversion. (D) Points indicate the average rate of amyloid conversion for multiple field isolates of CWD in full-length white-tailed deer rPrPC. The gray line indicates the rate of spontaneous amyloid conversion.
DISCUSSION
Despite decades of investigation, a complete characterization of barriers to transspecies transmission of prion diseases remains elusive. Many animals, including multiple lines of PrP transgenic mice, have been inoculated with various prions to define prion disease species barriers and understand the effects of passage into a new host. Likewise, in vitro assays have been used to model propensities for transspecies PrPC conversion. We have employed RT-QuIC since it permits observation of amyloid conversion in real time and, consequently, comparison of the conversion efficiency of seed (PrPSc)-substrate (rPrPC) combinations. Though RT-QuIC, as performed here, likely produced noninfectious amyloid products, we believe that RT-QuIC offers valuable insight regarding the efficiency of the initial amyloid conversion of a substrate by a seed.
We report that cBSE, after passage to felids in the form of FSE, remained a more efficient seed for the prion protein of the enciphering (bovine) host than for the new (feline) host (Fig. 4). This pattern has been observed in other contexts as well: cBSE-derived prions maintain many of their characteristics upon experimental or natural transmission to a new species (32, 33). We show here that these features are maintained as a result of the conformation of cBSE prions and not based solely on cellular cofactors. Conversely, fCWD was a more efficient seed for the new (feline) host than for the enciphering (white-tailed deer) host (Fig. 4). This suggests that when felids are infected with CWD, the resulting feline CWD has adapted to the new host. This appears to be an example of the difference between prions that adapt to new hosts upon passage and amplification that occurs without adaptation (J. Bian, V. Khaychuck, K. Bowling, K. Angers, N. Fernandez-Borges, E. Vidal, C. Meyerett-Reid, S. Kim, C. Calvi, J. Bartz, E. Hoover, U. Agrimi, J. Richt, J. Castilla, and G. C. Telling, submitted for publication) It would be interesting to test additional passaged BSE and CWD samples, particularly in light of the evidence for prion strains, but these samples are rare. It is important to note that the behavior of the fCWD and FSE in this paper may be dependent on the CWD and BSE that infected the cats.
We also assessed the seeded conversion of human rPrPC by BSE, CWD, FSE, and fCWD. Previous in vitro work using protein misfolding cyclic amplification (PMCA) and two seeded fibrillization assays found human (polymorphism M129) PrPC to be a weakly competent substrate for conversion by CWD and cBSE (25, 28, 29). We demonstrate that both FSE and fCWD have the ability to seed human rPrPC as well. Our finding that cBSE was a poor, if not ineffective, seed for human rPrPC in RT-QuIC was also observed by Orrú et al. (35). In contrast, our finding that CWD is an efficient seed for human rPrPC (albeit not as efficient as human sCJD) differs from previous results using PMCA or other seeded fibrillization assays (25, 28, 29). Perhaps the disparities between these in vitro assays reflect that RT-QuIC measures the rate of amyloid conversion (indicating the initial transspecies seeding) versus total PrPRes after conversion (25, 26, 28–30). We understand the rate of amyloid conversion to depend on both the quantity of prions in the seed and the competence of the seed to convert the substrate (34). Because our cBSE brain seed had a higher concentration of PrPRes relative to the total PrP than did our CWD brain sample, we interpreted the increased rate of amyloid conversion in human rPrPC to reflect the relative compatibility of seed with substrate. Indeed, our analysis supports the notion that human rPrPC is a competent substrate for other several nonhuman prions. Of course, we also understand that any in vitro estimation of prion species barriers carries the innate caveats of a reductionist model of complex in vivo processes.
In summary, real-time conversion demonstrates that CWD and BSE prions differ in their enciphering rigidity and plasticity across species barriers. One illustration is the conservation versus adaptation of enciphering prion characteristics upon passage to cats. These experiments also demonstrate that human rPrP can be converted to amyloid by both cBSE and CWD prions. These data point to the importance of deciphering the mechanisms by which prions infect and adapt to a new species and of prompt continued vigilance regarding indirect pathways that may facilitate transspecies prion transmission.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01-NS-061902.
We thank Geoff Pearson (University of Edinburgh, Edinburgh, Scotland) for providing the FSE samples and Maurice Bardsley and James Hope (Animal and Plant Health Agency, Surrey, United Kingdom) for providing the classical BSE samples. We thank Nicholas Haley and Clare Hoover for their reviews of the manuscript. Finally, we thank Byron Caughey, who was critical to the initiation of the RT-QuIC technology in our laboratory.
REFERENCES
- 1.van Rheede T, Smolenaars MMW, Madsen O, de Jong WW. 2003. Molecular evolution of the mammalian prion protein. Mol Biol Evol 20:111–121. doi: 10.1093/molbev/msg014. [DOI] [PubMed] [Google Scholar]
- 2.Sigurdson C, Miller MW. 2003. Other animal prion diseases. Br Med Bull 66:199–212. doi: 10.1093/bmb/66.1.199. [DOI] [PubMed] [Google Scholar]
- 3.Bruce ME, Will R, Ironside J, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C. 1997. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature 389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 4.Kirkwood J, Cunningham A. 1994. Epidemiological observations on spongiform encephalopathies in captive wild animals in the British Isles. Vet Rec 135:296–303. doi: 10.1136/vr.135.13.296. [DOI] [PubMed] [Google Scholar]
- 5.Lezmi S, Bencsik A, Monks E, Petit T, Baron T. 2003. First case of feline spongiform encephalopathy in a captive cheetah born in France: PrPsc analysis in various tissues revealed unexpected targeting of kidney and adrenal gland. Histochem Cell Biol 119:415–422. [DOI] [PubMed] [Google Scholar]
- 6.Will RG, Ironside JW, Zeidler M, Estibeiro K, Cousens SN, Smith PG, Alperovitch A, Poser S, Pocchiari M, Hofman A. 1996. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet 347:921–925. [DOI] [PubMed] [Google Scholar]
- 7.Wyatt J, Pearson G, Smerdon T, Gruffydd-Jones T, Wells G. 1990. Spongiform encephalopathy in a cat. Vet Rec 126:513. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt J, Pearson G, Smerdon T, Gruffydd-Jones T, Wells G, Wilesmith J. 1991. Naturally occurring scrapie-like spongiform encephalopathy in five domestic cats. Vet Rec 129:233–236. doi: 10.1136/vr.129.11.233. [DOI] [PubMed] [Google Scholar]
- 9.Hamir AN, Kunkle RA, Cutlip RC, Miller JM, O'Rourke KI, Williams ES, Miller MW, Stack MJ, Chaplin MJ, Richt JA. 2005. Experimental transmission of chronic wasting disease agent from mule deer to cattle by the intracerebral route. J Vet Diagn Investig 17:276–281. doi: 10.1177/104063870501700313. [DOI] [PubMed] [Google Scholar]
- 10.Hamir AN, Kunkle RA, Cutlip RC, Miller JM, Williams ES, Richt JA. 2006. Transmission of chronic wasting disease of mule deer to Suffolk sheep following intracerebral inoculation. J Vet Diagn Investig 18:558–565. doi: 10.1177/104063870601800606. [DOI] [PubMed] [Google Scholar]
- 11.Hamir AN, Miller JM, Kunkle RA, Hall SM, Richt JA. 2007. Susceptibility of cattle to first-passage intracerebral inoculation with chronic wasting disease agent from white-tailed deer. Vet Pathol 44:487–493. doi: 10.1354/vp.44-4-487. [DOI] [PubMed] [Google Scholar]
- 12.Marsh RF, Kincaid AE, Bessen RA, Bartz JC. 2005. Interspecies transmission of chronic wasting disease prions to squirrel monkeys (Saimiri sciureus). J Virol 79:13794–13796. doi: 10.1128/JVI.79.21.13794-13796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathiason CK, Nalls AV, Seelig DM, Kraft SL, Carnes K, Anderson KR, Hayes-Klug J, Hoover EA. 2013. Susceptibility of domestic cats to chronic wasting disease. J Virol 87:1947–1956. doi: 10.1128/JVI.02592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Race B, Meade-White KD, Miller MW, Barbian KD, Rubenstein R, LaFauci G, Cervenakova L, Favara C, Gardner D, Long D, Parnell M, Striebel J, Priola SA, Ward A, Williams ES, Race R, Chesebro B. 2009. Susceptibilities of nonhuman primates to chronic wasting disease. Emerg Infect Dis 15:1366–1376. doi: 10.3201/eid1509.090253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagleish MP, Martin S, Steele P, Finlayson J, Sisó S, Hamilton S, Chianini F, Reid HW, González L, Jeffrey M. 2008. Experimental transmission of bovine spongiform encephalopathy to European red deer (Cervus elaphus elaphus). BMC Vet Res 4:17. doi: 10.1186/1746-6148-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasmézas CI, Comoy E, Hawkins S, Herzog C, Mouthon F, Konold T, Auvré F, Correia E, Lescoutra-Etchegaray N, Salès N. 2005. Risk of oral infection with bovine spongiform encephalopathy agent in primates. Lancet 365:781–783. [DOI] [PubMed] [Google Scholar]
- 17.Lasmezas CI, Deslys JP, Demaimay R, Adjou KT, Lamoury F, Dormont D, Robain O, Ironside J, Hauw JJ. 1996. BSE transmission to macaques. Nature 381:743–744. doi: 10.1038/381743a0. [DOI] [PubMed] [Google Scholar]
- 18.Asante EA, Linehan JM, Desbruslais M, Joiner S, Gowland I, Wood AL, Welch J, Hill AF, Lloyd SE, Wadsworth JDF, Collinge J. 2002. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J 21:6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asante EA, Linehan JM, Gowland I, Joiner S, Fox K, Cooper S, Osiguwa O, Gorry M, Welch J, Houghton R, Desbruslais M, Brandner S, Wadsworth JDF, Collinge J. 2006. Dissociation of pathological and molecular phenotype of variant Creutzfeldt–Jakob disease in transgenic human prion protein 129 heterozygous mice. Proc Natl Acad Sci U S A 103:10759–10764. doi: 10.1073/pnas.0604292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadsworth JDF, Asante EA, Desbruslais M, Linehan JM, Joiner S, Gowland I, Welch J, Stone L, Lloyd SE, Hill AF, Brandner S, Collinge J. 2004. Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science 306:1793–1796. doi: 10.1126/science.1103932. [DOI] [PubMed] [Google Scholar]
- 21.Wilson R, Plinston C, Hunter N, Casalone C, Corona C, Tagliavini F, Suardi S, Ruggerone M, Moda F, Graziano S, Sbriccoli M, Cardone F, Pocchiari M, Ingrosso L, Baron T, Richt J, Andreoletti O, Simmons M, Lockey R, Manson JC, Barron RM. 2012. Chronic wasting disease and atypical forms of bovine spongiform encephalopathy and scrapie are not transmissible to mice expressing wild-type levels of human prion protein. J Gen Virol 93:1624–1629. doi: 10.1099/vir.0.042507-0. [DOI] [PubMed] [Google Scholar]
- 22.Kong Q, Huang S, Zou W, Vanegas D, Wang M, Wu D, Yuan J, Zheng M, Bai H, Deng H, Chen K, Jenny AL, O'Rourke K, Belay ED, Schonberger LB, Petersen RB, Sy MS, Chen SG, Gambetti P. 2005. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J Neurosci 25:7944–7949. doi: 10.1523/JNEUROSCI.2467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandberg MK, Al-Doujaily H, Sigurdson CJ, Glatzel M, O'Malley C, Powell C, Asante EA, Linehan JM, Brandner S, Wadsworth JD, Collinge J. 2010. Chronic wasting disease prions are not transmissible to transgenic mice overexpressing human prion protein. J Gen Virol 91:2651–2657. doi: 10.1099/vir.0.024380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamguney G, Giles K, Bouzamondo-Bernstein E, Bosque PJ, Miller MW, Safar J, DeArmond SJ, Prusiner SB. 2006. Transmission of elk and deer prions to transgenic mice. J Virol 80:9104–9114. doi: 10.1128/JVI.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barria MA, Balachandran A, Morita M, Kitamoto T, Barron R, Manson J, Knight R, Ironside JW, Head MW. 2014. Molecular barriers to zoonotic transmission of prions. Emerg Infect Dis 20:88. doi: 10.3201/eid2001.130858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones M, Wight D, Barron R, Jeffrey M, Manson J, Prowse C, Ironside JW, Head MW. 2009. Molecular model of prion transmission to humans. Emerg Infect Dis 15:2013–2016. doi: 10.3201/eid1512.090194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurt TD, Jiang L, Fernández-Borges N, Bett C, Liu J, Yang T, Spraker TR, Castilla J, Eisenberg D, Kong Q, Sigurdson CJ. 2015. Human prion protein sequence elements impede cross-species chronic wasting disease transmission. J Clin Invest 125:1485–1496. doi: 10.1172/JCI79408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luers L, Bannach O, Stöhr J, Wördehoff MM, Wolff M, Nagel-Steger L, Riesner D, Willbold D, Birkmann E. 2013. Seeded fibrillation as molecular basis of the species barrier in human prion diseases. PLoS One 8:e72623. doi: 10.1371/journal.pone.0072623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond GJ, Bossers A, Raymond LD, O'Rourke KI, McHolland LE, Bryant PK, Miller MW, Williams ES, Smits M, Caughey B. 2000. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J 19:4425–4430. doi: 10.1093/emboj/19.17.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymond GJ, Hope J, Kocisko DA, Priola SA, Raymond LD, Bossers A, Ironside J, Will RG, Chen SG, Petersen RB, Gambetti P, Rubenstein R, Smits MA, Lansbury PT, Caughey B. 1997. Molecular assessment of the potential transmissibilities of BSE and scrapie to humans. Nature 388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 31.Tamgüney G, Miller MW, Giles K, Lemus A, Glidden DV, DeArmond SJ, Prusiner SB. 2009. Transmission of scrapie and sheep-passaged bovine spongiform encephalopathy prions to transgenic mice expressing elk prion protein. J Gen Virol 90:1035–1047. doi: 10.1099/vir.0.007500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill AF, Desbruslais M, Joiner S, Sidle KCL, Gowland I, Collinge J, Doey LJ, Lantos P. 1997. The same prion strain causes vCJD and BSE. Nature 389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 33.Torres J-M, Espinosa J-C, Aguilar-Calvo P, Herva M-E, Relaño-Ginés A, Villa-Diaz A, Morales M, Parra B, Alamillo E, Brun A, Castilla J, Molina S, Hawkins SAC, Andreoletti O. 2014. Elements modulating the prion species barrier and its passage consequences. PLoS One 9:e89722. doi: 10.1371/journal.pone.0089722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson DM, Davenport KA, Haley NJ, Denkers ND, Mathiason CK, Hoover EA. 2015. Quantitative assessment of prion infectivity in tissues and body fluids by real-time quaking-induced conversion. J Gen Virol 96:210–219. doi: 10.1099/vir.0.069906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orrú CD, Favole A, Corona C, Mazza M, Manca M, Groveman BR, Hughson AG, Acutis PL, Caramelli M, Zanusso G, Casalone C, Caughey B. 2015. Detection and discrimination of classical and atypical L-type bovine spongiform encephalopathy by real-time quaking-induced conversion. J Clin Microbiol 53:1115–1120. doi: 10.1128/JCM.02906-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angers RC, Browning SR, Seward TS, Sigurdson CJ, Miller MW, Hoover EA, Telling GC. 2006. Prions in skeletal muscles of deer with chronic wasting disease. Science 311:1117. doi: 10.1126/science.1122864. [DOI] [PubMed] [Google Scholar]