ABSTRACT
Early biochemical studies of viral replication suggested that most viruses produce double-stranded RNA (dsRNA), which is essential for the induction of the host immune response. However, it was reported in 2006 that dsRNA could be detected by immunofluorescence antibody staining in double-stranded DNA and positive-strand RNA virus infections but not in negative-strand RNA virus infections. Other reports in the literature seemed to support these observations. This suggested that negative-strand RNA viruses produce little, if any, dsRNA or that more efficient viral countermeasures to mask dsRNA are mounted. Because of our interest in the use of dsRNA antibodies for virus discovery, particularly in pathological specimens, we wanted to determine how universal immunostaining for dsRNA might be in animal virus infections. We have detected the in situ formation of dsRNA in cells infected with vesicular stomatitis virus, measles virus, influenza A virus, and Nyamanini virus, which represent viruses from different negative-strand RNA virus families. dsRNA was also detected in cells infected with lymphocytic choriomeningitis virus, an ambisense RNA virus, and minute virus of mice (MVM), a single-stranded DNA (ssDNA) parvovirus, but not hepatitis B virus. Although dsRNA staining was primarily observed in the cytoplasm, it was also seen in the nucleus of cells infected with influenza A virus, Nyamanini virus, and MVM. Thus, it is likely that most animal virus infections produce dsRNA species that can be detected by immunofluorescence staining. The apoptosis induced in several uninfected cell lines failed to upregulate dsRNA formation.
IMPORTANCE An effective antiviral host immune response depends on recognition of viral invasion and an intact innate immune system as a first line of defense. Double-stranded RNA (dsRNA) is a viral product essential for the induction of innate immunity, leading to the production of type I interferons (IFNs) and the activation of hundreds of IFN-stimulated genes. The present study demonstrates that infections, including those by ssDNA viruses and positive- and negative-strand RNA viruses, produce dsRNAs detectable by standard immunofluorescence staining. While dsRNA staining was primarily observed in the cytoplasm, nuclear staining was also present in some RNA and DNA virus infections. The nucleus is unlikely to have pathogen-associated molecular pattern (PAMP) receptors for dsRNA because of the presence of host dsRNA molecules. Thus, it is likely that most animal virus infections produce dsRNA species detectable by immunofluorescence staining, which may prove useful in viral discovery as well.
INTRODUCTION
An effective antiviral host response depends on recognition of viral invasion and an intact innate immune system as a first line of defense. Although the mammalian innate immune system responds to other pathogens, the emphasis here is on animal viruses. Double-stranded RNA (dsRNA) is a viral product essential in the induction of innate immunity, leading to the production of type I interferons (IFNs) (1, 2) and activation of hundreds of IFN-stimulated genes (ISGs), including two well-recognized ISG cytoplasmic enzyme systems that are activated by dsRNA (and type I IFNs) and that have broad antiviral activities: the protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase systems (3–6). Together these responses confer resistance to virus (reviewed in reference 7).
Viral infections provide a principal source of dsRNA that is recognized by pathogen-associated molecular pattern (PAMP) receptors. For infection with viruses having dsRNA genomes, the origin may be input dsRNA or dsRNA synthesized in progeny genomes inside the capsid, which is imperfectly hidden from cytoplasmic sensors (8). In single-stranded RNA (ssRNA) virus infections, the source of dsRNA is replicative dsRNA intermediates generated by an RNA-dependent RNA polymerase (RdRp), while in DNA virus infections, convergent transcription from bidirectional promoters results in the formation of overlapping RNAs. Innate immune sensors detect not only the dsRNA structure but also the length, sequence, and cellular location (9, 10). Small RNAs, defined by their length (20 to 30 nucleotides), such as small interfering RNAs (siRNAs; ∼21 nucleotides) and microRNAs (∼22 nucleotides), are not associated with a type I IFN response (11). Thus, recognition of dsRNA is presumed to require a length equal to or greater than ∼30 nucleotides.
Early biochemical studies of viral replication suggested that most viruses produce dsRNAs (12–15). However, in 2006, Weber et al. (16) reported that dsRNA could be detected by immunofluorescence antibody staining in double-stranded DNA (dsDNA) and positive-strand RNA virus infections but not in negative-strand RNA virus infections, suggesting that negative-strand RNA viruses produce little, if any, dsRNA or that more efficient viral countermeasures mask dsRNA in negative-strand RNA virus infections. Recently, two reports of dsRNA production in negative-strand RNA virus infections (9, 17) have challenged the findings of Weber et al. (16). Because of our interest in the use of antibodies to dsRNA for virus discovery, particularly in pathological specimens, we wanted to determine how universal immunostaining for dsRNA might be in animal virus infections. In addition to positive-strand RNA viruses, we also observed the in situ formation of dsRNA in cells infected with vesicular stomatitis virus (VSV), measles virus (MeV), influenza A virus (IAV), and Nyamanini virus (NyaV), which represent different negative-strand RNA virus families. RNA with a dsRNA character was also detected in cells infected with lymphocytic choriomeningitis virus (LCMV), an ambisense RNA virus, and minute virus of mice (MVM), a single-stranded DNA (ssDNA) parvovirus, but not hepatitis B virus (HBV). dsRNA staining was primarily observed in the cytoplasm but was also seen in the nucleus of cells infected with NyaV, MVM, and IAV, generally colocalizing with the expression of viral antigens. Thus, it is likely that immunostaining will detect most animal virus infections and that antibodies to dsRNA may also be used to enrich for viral sequences in cDNA library construction and next-generation sequencing (NGS) for viral discovery.
MATERIALS AND METHODS
Cells.
BHK-21 cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 7.5% tryptose phosphate, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in 5% CO2. Cells of the immature myelomonocytic cell line M1, derived from the SL mouse strain, were induced to differentiate into macrophages with supernatants from mouse L929 fibroblasts and mouse P388D1 macrophages as described previously and designated M1-D cells. (18). Vero B6 cells were grown in DMEM, and Madin-Darby canine kidney cells were grown in minimal essential medium (MEM) supplemented with 10% FBS, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in 5% CO2. DBT (astrocytoma-derived) cells were grown in MEM with 10% FBS and 10% tryptose phosphate, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in 5% CO2.
Viruses.
The origin and passage history of Theiler's murine encephalitis virus (TMEV; BeAn) stock have been reported previously (19). Stocks of VSV were purchased from ATCC (Manassas, VA), and those of enterovirus 71 (EV71), mouse hepatitis virus (MHV; A59), IAV (H3N2, Udorn), MeV (Chicago), LCMV (Armstrong), and NyaV were gifts from Alan Nix (CDC, Atlanta, GA), Tom Gallagher (Loyola School of Medicine, Chicago, IL), Bob Lamb (Northwestern University, Evanston, IL), Mark Burgoon (University of Colorado Denver School of Medicine, Aurora, CO), Robert Fujinami (University of Utah School of Medicine, Salt Lake City, UT), and Robert Tesh (University of Texas Medical Branch, Galveston, TX), respectively. Infection with MVM in A9 mouse fibroblasts was performed in Peter Tattersall's laboratory (Yale University, New Haven, CT). The cells were fixed in neutral formalin and shipped to the University of Illinois at Chicago (UIC; Chicago, IL). Human hepatoblastoma cell line (HepG2) clone 2.2.15, which is persistently infected with hepatitis B virus (HBV; core antigen positive) (20), was grown on coverslips and fixed in 10% neutral formalin in the laboratory of Alan McLachlan (UIC, Chicago, IL).
Virus infections.
After virus adsorption at the multiplicities of infection (MOIs) indicated below for 30 to 60 min at 24°C or 37°C, cell monolayers on coverslips in 6-well plates were washed twice with MEM containing 1 mM CaCl2 and 0.5 mM MgCl2 or remained unwashed and were then incubated for the times designated below in the specific medium containing 1% FBS at 37°C. The virus titers of clarified lysates of infected cells were determined by measurement of the 50% tissue culture infective dose or standard plaque assay. The times postinfection (p.i.) for immunofluorescence studies with each virus were selected after preliminary experiments, and a number of time points were chosen.
Reagents.
The antibodies to dsRNA used were mouse monoclonal antibody (MAb) J2 (Scion, Budapest, Hungary), mouse monoclonal antibody 9D5 (ascitic fluid; David Schnurr, California State Department of Health, Richmond, CA), and rabbit polyclonal antibody 170A (David Stollar, Rutgers, New Brunswick, NJ). Rabbit polyclonal anti-TMEV antibody was raised by intravenous injections of CsCl2 gradient-purified BeAn virus, and mouse immune serum to NyaV was raised by four intraperitoneal injections of virus stock as approved by the Office of Animal Care and Institutional Biosafety Committee. The following antiviral antibodies were purchased or provided: mouse MAb to EV71 (Mybiosource, Inc., San Diego, CA), mouse polyclonal antibody J1.3 to MHV M protein (Tom Gallagher, Loyola University School of Medicine, Chicago, IL), human recombinant 2B4 to MeV NC protein (Mark Burgoon, University of Colorado Denver, Aurora, CO), rabbit polyclonal anti-vesicular stomatitis virus (anti-VSV) glycoprotein antibody (Bethyl Inc., Montgomery, TX), rabbit polyclonal antibody to IAV NC protein (Abcam, Cambridge, MA), rabbit polyclonal antibody to MVM NS1/NS2 (Peter Tattersall, Yale University, New Haven, CT), and mouse MAb to LCMV NP (Michael Buchmeier, University of California, Irvine, CA). Rabbit anti-caspase-3 and rabbit anti-poly(ADP-ribose) polymerase (anti-PARP) were purchased from Cell Signaling Technology (Beverly, MA), goat anti-rabbit IgG-fluorescein isothiocyanate (FITC; 1:500) and anti-mouse IgG-FITC (1:500) were from BD Pharmingen (San Diego, CA), Alexa Fluor 488 (1:2,000) and goat anti-mouse Alexa Fluor 568 (1:2,000) were from Invitrogen (Carlsbad, CA), and goat anti-human IgG-rhodamine was from Santa Cruz (Dallas, TX). RNase One was purchased from Promega (Madison, WI), and RNase III was purchased from Life Technologies (Grand Island, NY).
Immunofluorescence staining.
Cells grown and infected on glass coverslips (Fisher Scientific Co., Pittsburgh, PA) were fixed in 3.7% paraformaldehyde or 10% neutral formalin, permeabilized with phosphate-buffered saline (PBS) containing 0.2% Triton X-100 for 5 min, and washed three times for 5 min each time in PBS. For some IAV infections, the cells were first incubated with proteinase K (20 μg/ml; Sigma, St. Louis, MO) in 50 mM Tris-HCl, pH 8.0, and 5 mM CaCl2 for 1 h at 37°C. Primary antibodies for detection of dsRNA and the different virus antigens in cells were incubated overnight at 4°C (all other antibody incubations were at 24°C), washed three times for 5 min each time in PBS before incubation with secondary antibodies for 30 min, counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) solution in PBS for 3 min, and finally, washed three times with PBS containing 0.05% Triton X-100 for 5 min each time, twice with PBS for 5 min each time, and once with distilled H2O for 1 min. The coverslips were inverted onto gel mount (Biomedia, Foster City, CA) on microscope slides, viewed with a Zeiss digital confocal microscope, and photographed.
RNA duplexes of different sizes.
Small-molecular-size (100- to 1,000-bp) and large-molecular-size (1- to 8-kbp) poly(I·C) (InvivoGen, San Diego, CA) and a 47-mer RNA duplex of BeAn virus nucleotides (5′-GUCUAAGGCCGCUCGGAAUAUGACAGGGUUAUUUUCACCUCUUCUUUU-3′; Integrated DNA Technologies, Skokie, IL) were used in Northwestern blotting.
Northwestern blotting.
Total RNA samples were electrophoresed on a 1% agarose gel in TBE buffer (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA, pH 8.4) and transferred onto a Hybond nylon membrane (GE Healthcare, Piscataway, NJ). The membrane(s) was incubated in 5% nonfat milk in Tris-buffered saline (TBS) at 24°C for 1 h to block nonspecific binding sites, followed by incubation of a 1:6,000 dilution of the 9D5 primary antibody to dsRNA at 24°C for 1 h or 4°C overnight, washed three times in TBS and 0.05% Tween 20 (TBST) for 10 min each time, and then incubated with a 1:6,000 dilution of secondary anti-mouse IgG-horseradish peroxidase (HRP) antibody in TBS at 24°C for 1 h. After washing in TBST three times for 10 min each time, the membranes were blotted with SuperSignal enzyme-linked immunosorbent assay Femto maximum-sensitivity substrate (Pierce, Rockford, IL) at 24°C for 1 to 2 min, exposed, and developed with HyBlot CL autoradiography film (Denville Scientific, Metuchen, NJ).
Immunoblot analysis.
M1-D cells (cells in the monolayer and those shed into the supernatant combined) were washed with PBS and lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) at the times indicated below. Protein samples were electrophoresed on 12% NuPAGE bis-Tris gels (Novex, Life Technologies) with MOPS (morpholinepropanesulfonic acid)-SDS running buffer (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes (GE Healthcare, Piscataway, NJ). The membranes were blocked with Tris-buffered saline containing 3% nonfat dry milk and 0.02% Tween 20 and incubated with primary antibody for 1 h and goat anti-rabbit immunoglobulin-HRP or anti-mouse immunoglobulin-HRP (BD Pharmingen, San Diego, CA) as the secondary antibodies at a 1:2,000 dilution for 1 h. Immunoreactive bands were detected by enhanced chemiluminescence (ECL; PerkinElmer, Inc. Waltham, MA). Mouse antibody to β-actin (Sigma, St. Louis, MO) was used as a loading control.
RESULTS
Specificity of dsRNA antibodies.
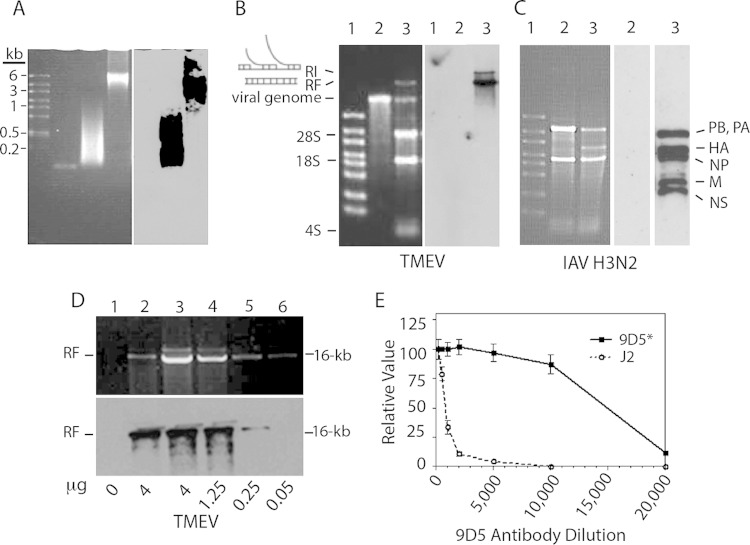
Details of the specificity of rabbit polyclonal antibody 170A and the J2 mouse MAb to dsRNA were reported previously (13, 21). Bonin et al. (22), using scanning force microscopy, found that the J2 MAb recognized only perfect RNA duplexes of ≥40 nucleotides in size. The MAb 9D5, which was initially believed to be a panenterovirus antibody (23), turned out instead to have reactivity to dsRNA. Thus, MAb 9D5 was primarily used in these studies and in Northwestern immunoblotting and reacted with small-molecular-size (100- to 1-kbp) and large-molecular-size (1- to 8-kbp) poly(I·C) but not with a 47-bp dsRNA (Fig. 1A) or with siRNAs (22 bp) (not shown). The replicative form (RF) and a presumed replicative intermediate (RI) of BeAn virus (TMEV) were identified by Northwestern immunoblotting, but the single-stranded viral genomic RNA was not (Fig. 1B). dsRNA RFs of influenza virus genomic segments were also bound by 9D5 (Fig. 1C). However, there was no binding of BHK-21 or MDCK cell 28S and 18S rRNAs, 4S tRNA, or other host cell RNAs. The 9D5 antibody detected as little as 50 ng of TMEV dsRNA (Fig. 1D) and showed positive staining of TMEV-infected M1-D cells at a dilution to 1:10,000, whereas the dilution of J2 required for detection of TMEV dsRNA and positive staining was 1:500 (Fig. 1E). Thus, the 9D5 antibody appears to specifically recognize perfect RNA duplexes ranging in size from ∼100 bp to 16 kbp but not ssRNA, a result also found with the J2 MAb (not shown). The J2 MAb provided identical, albeit weaker staining than MAb 9D5 in infected cells (see below).
FIG 1.
Reactivity of the 9D5 MAb with dsRNA duplexes of various sizes and TMEV and influenza A virus RF RNAs. (A) (Left) Electrophoresis on a 1.7% agarose gel stained with ethidium bromide. The gel contained large molecular size ssRNA markers, a 47-mer RNA duplex, small-molecular-size (100- to 1,000-bp) poly(I·C), and large-molecular-size (1 to 8 kbp) poly(I·C) in the four lanes from left to right, respectively. (Right) Northwestern blot showing the reactivity of MAb 9D5 with 100-bp to 8-kbp poly(I·C). (B) (Left) Electrophoresis on a 1% agarose gel of in vitro-transcribed TMEV RNA and total RNA from TMEV-infected BHK-21 cells (6 h p.i.) stained with ethidium bromide. Lanes: 1, high-range ssRNA markers; 2, in vitro-transcribed TMEV genomic RNA; 3, total RNA from infected cells showing the TMEV positive-strand genome and dsRNA RF along with cellular RNAs. (Right) Northwestern blot of the membrane on the left showing the reactivity of RF and RI RNAs with 9D5 antibodies to dsRNA. (C) (Left) Electrophoresis on a 1% agarose gel of total RNA from uninfected and IAV-infected MDCK cells stained with ethidium bromide. Lanes: 1, ssRNA markers; 2, uninfected MDCK cells; 3, infected MDCK cells. (Middle) Northwestern blot of lane 2 from the panel on the left with no reactivity. (Left) Blot of the membrane of lane 3 from the panel on the left showing the reactivity of IAV genome RF segments. (D) Sensitivity of the 9D5 MAb for the detection of TMEV RF RNA from infected BHK-21 cells. (Top) A 1% agarose gel stained with ethidium bromide showing barely detectable TMEV RF RNA; (bottom) Northwestern blot showing the reactivity of TMEV RF RNA with as little as 50 ng of antibody. (E) Titration of MAb 9D5 (3.27 μg/μl) and MAb J2 (1.2 μg/μl) showing their relative reactivity with TMEV-infected M1-D cells by immunofluorescence intensity.
Intracellular dsRNAs in positive-strand RNA virus infections detected by immunofluorescence staining.
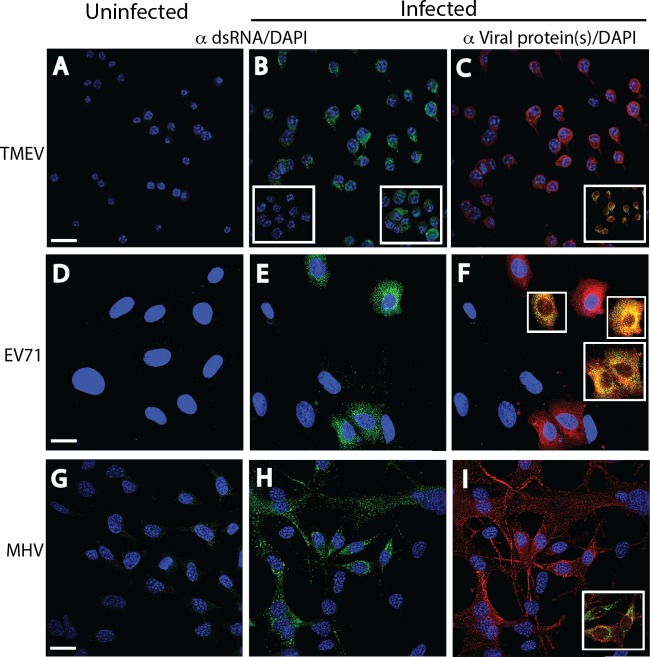
The intracellular localization and confirmation of dsRNA staining with MAb 9D5 by digestion with RNase specific for single-strand RNAs (RNase A) and dsRNAs (RNase III) were initially assessed in several positive-strand RNA virus infections. The dsRNA staining observed in the cytoplasm of TMEV-infected M1-D murine macrophages (MOI = 10) at 6 h p.i. was abrogated when infected monolayers were incubated with RNase III but not RNase A (Fig. 2B, right and left insets, respectively). RNase III and RNase A digestions were performed in cells infected with the other viruses, and the same result described above was obtained (not shown). Similar dsRNA staining results were seen with TMEV infection in BHK-21 cells (not shown). TMEV antigens stained with polyclonal rabbit antisera colocalized with dsRNA, although on different molecules (Fig. 1C, inset), suggesting that dsRNA and a viral nonstructural protein were in close proximity. Similar punctate dsRNA and virus antigen staining was also observed in the cytoplasm of EV71-infected Vero B6 cells (MOI = 5) at 6 h p.i. and MHV-infected astrocytoma-derived DBT cells (MOI = 5) at 6 h p.i. (Fig. 2D to I). EV71 and MHV are two other positive-strand RNA viruses. Colocalization of the two antigens was also observed. While dsRNA remained perinuclear in location in infected DBT cells, the M protein extended from the cell soma into astrocytic processes (linear staining). These results indicate that MAb 9D5 recognized dsRNA in cells infected with these positive-strand RNA viruses, and such staining was abrogated by incubation of the infected cells with RNase III.
FIG 2.
Immunofluorescence analysis of cell monolayers infected with positive-strand RNA viruses. Uninfected (A, D, G) and infected (B, E, H) cell monolayers were stained with a 1:2,000 dilution of either MAb 9D5 or polyclonal antibody 170A to dsRNA, and infected cells were stained with specific antiviral antibodies (C, F, I). The fields in the center (B, E, H) and right (C, F, I) panels are identical. (A to C) Uninfected and TMEV-infected M1-D macrophages. (A) Uninfected cells. (B) TMEV-infected cells showing strong punctate cytoplasmic staining (green). (Insets) A loss of staining after preincubation with RNase III (left) but retention of staining after preincubation with RNase A (right). (C) TMEV-infected cells stained with a 1:3,000 dilution of rabbit polyclonal antibodies to TMEV virions showing cytoplasmic staining (red). (Inset) Colocalization of dsRNA and virus antigen(s). (D to F) Uninfected and EV71-infected Vero B6 cells. Strong punctate cytoplasmic dsRNA staining (green) was shown in uninfected cells (D) but not in infected cells (E). (F) EV71-infected cells exhibited cytoplasmic staining with a 1:2,000 dilution of mouse MAb to EV71 (red; virus protein specificity is not known). (Insets) Colocalization of both antigens. (G to I) Uninfected and MHV-infected DBT astrocytoma cells. Strong punctate cytoplasmic staining (green) with the 170A polyclonal antibody was shown in infected cells (H) but not in uninfected cells (G), while MHV-infected cells exhibited cytoplasmic staining with a 1:500 dilution of mouse MAb to MHV M protein (red) (I). The inset in panel I shows the colocalization of both antigens. The results of incubations of RNases with EV71 and MHV are the same as those for incubations of RNases with TMEV (not shown). Bars, 10 μm. All insets have the same magnification as the full panels.
Weaker dsRNA immunofluorescence was observed in negative-strand RNA virus infections.
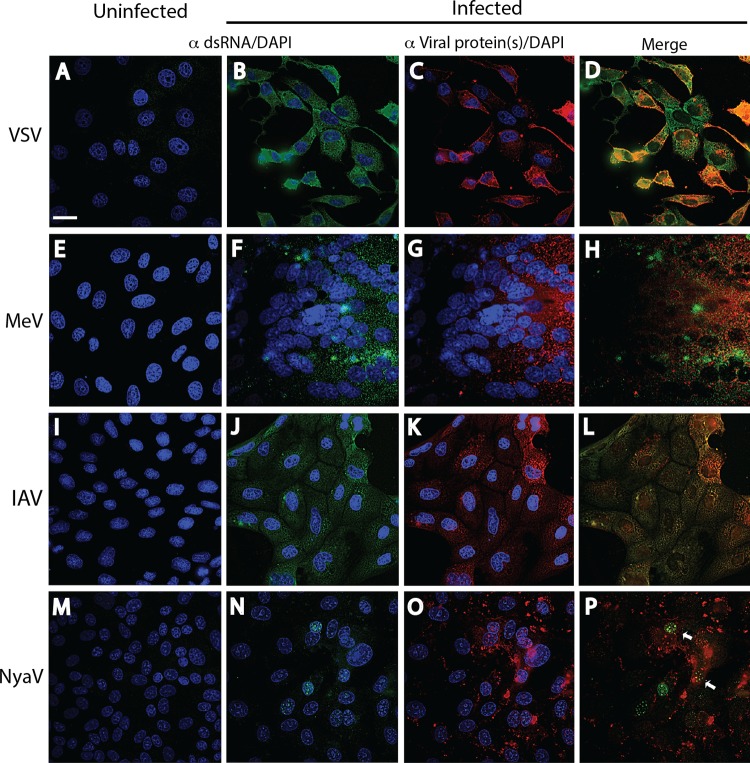
In a previous study, dsRNA was not detected by immunofluorescence staining of IAV and La Crosse virus infections, even though Tyramide signal amplification was used (16). Kato et al. (9) also concluded that dsRNA was not present in IAV infection. Other investigators reported that the expression of dsRNA in wild-type Sendai virus and MeV infections was absent or dsRNA was expressed at much lower concentrations in wild-type Sendai virus and MeV infections than in infections with the same viruses deficient in the C protein (17, 24). In contrast, we have demonstrated dsRNA staining in infections with wild-type viruses of four different negative-strand RNA virus families, including VSV infection in Vero B6 cells (MOI = 2) at 6 h p.i., MeV infection in Vero B6 cells (MOI = 0.1) at 72 h p.i., IAV infection in MDCK cells (MOI = 0.025) at 16 h p.i., and NyaV in Vero B6 cells (MOI = 0.01) at 72 h p.i. Punctate cytoplasmic dsRNA staining was clearly seen in VSV, MeV, and IAV infections (Fig. 3B, F, and J). While the intensity of dsRNA staining in cells with negative-strand RNA virus infections in the selected images in Fig. 3 was qualitatively similar to that in cells with positive-strand RNA virus infections, in most other images (not shown), the dsRNA staining in cells with negative-strand RNA virus infections was less intense. The slightly more prominent staining in MeV-infected cells (Fig. 3F) may be due to an increased amount of cytoplasm of the large giant cell shown; however, single MeV-infected cells had a similar intensity of staining (not shown).
FIG 3.
Immunofluorescence analysis of dsRNA and virus proteins in negative-strand RNA virus infections. Uninfected (A, E, I, M) and infected (B, F, J, N) cell monolayers were stained with a 1:2,000 dilution of MAb 9D5 or polyclonal antibody 170A, and the other infected monolayers were stained with the specific antiviral antibodies (C, G, K, O). (A to D) Vero B6 cells infected or not infected with VSV. (A) Uninfected cells. (B) VSV-infected Vero B6 cells with punctate cytoplasmic staining (green) with MAb 9D5 (B). (C) VSV-infected cells stained with a 1:500 dilution of rabbit polyclonal antibodies to VSV G protein showing copious cytoplasmic staining (red). (D) Merged image of panels B and C. (E to H) Vero B6 cells infected or not infected with MeV. (E) Uninfected cells. (F) MeV-infected Vero B6 cells with punctate cytoplasmic staining with MAb 9D5. Note the large multinucleated giant cell. (G) MeV-infected cells were stained with human recombinant antibody to MeV NC protein and show diffuse cytoplasmic reactivity. (H) Merged image of panels F and G. (I to L) MDCK cells infected or not infected with IAV. (I) Uninfected cells. (J) IAV-infected MDCK cells revealing cytoplasmic staining (green) with MAb 9D5. (K) Cytoplasmic reactivity is seen with a 1:500 dilution of rabbit polyclonal antibody to IAV NC protein. (L) Merged image of panels J and K. (M to P) Vero B6 cells infected or not infected with NyaV. (M) Uninfected cells. (N) NyaV-infected Vero B6 cells revealing punctate areas of staining in the nucleus of infected cells. (O) Cytoplasmic reactivity with a 1:2,000 dilution of mouse antiserum to NyaV proteins is seen. (P) Merged image of panels N and O, with only a few of the stained nuclear dots in two cells showing colocalization (arrows). Bar, 10 μm (the magnification in panel A applies to all panels).
Although IAV replicates in the nucleus, dsRNA was not detected at 3, 4, 8, and 16 h p.i., in spite of a progressive cytopathic effect after 8 h p.i. However, incubation of IAV-infected cells with proteinase K prior to staining revealed spheroidal dots of immunoreactive dsRNA in the nucleus (see Fig. 6B, arrows), indicating that the IAV nucleocapsid complex had prevented staining of the IAV RF. Anti-dsRNA reactivity was present exclusively in the nucleus of NyaV-infected cells (Fig. 3N; see also Fig. 6C). NyaV belongs to a new family in the order Mononegavirales that is closely related to bornaviruses and replicates in the nucleus (25). These results are consistent with the nuclear replication of NyaV, evident here as dsRNA-positive foci of various sizes in the nucleus, suggesting that these are sites of viral replication. Punctate cytoplasmic staining patterns obtained using polyclonal antibodies to VSV G, MeV NP, and IAV NC proteins are shown in Fig. 3C, G, and K. Mouse antiserum to NyaV revealed both diffuse cytoplasmic reactivity and more discrete spots of nuclear reactivity that in some instances colocalized with dsRNA (Fig. 3P, arrows). These results indicate that in negative-strand RNA virus infections, dsRNA is formed in amounts sufficient for detection by immunofluorescence analysis and that staining may be observed in the nucleus. Finally, the J2 MAb also stained negative-strand RNA virus-infected cells, but the staining tended to be weaker than that obtained with MAb 9D5 and in some cells was accompanied by nonspecific background staining, as shown for IAV infection (Fig. 4); photomicrographs of the other negative-strand RNA viruses are not shown.
FIG 6.
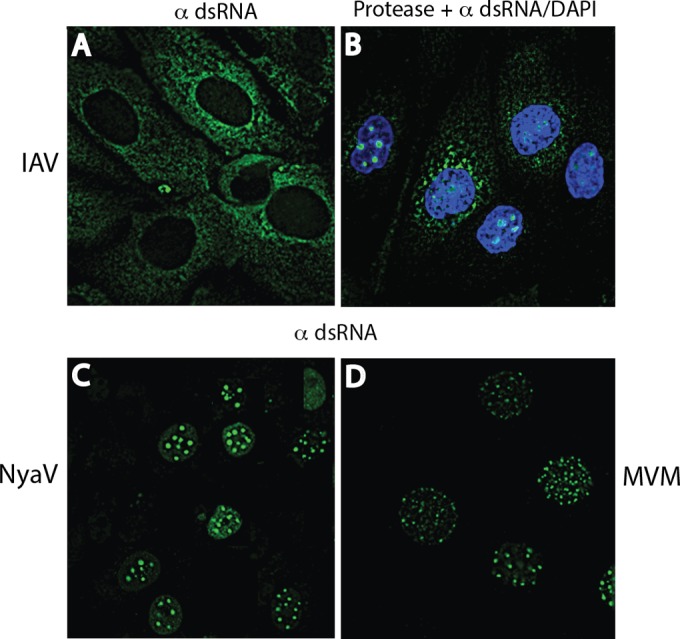
Comparison of nuclear images of cells infected with IAV, NyaV, and MVM stained for dsRNA. (A, B) IAV-infected MDCK cells at 16 and 8 h p.i., respectively, showing cytoplasmic staining (A) and nuclear staining (B); nuclear staining was seen only after the cell monolayers were first incubated with proteinase K. Infected cells with nuclear staining at 8 h did not reveal cytoplasmic staining; however, at 16 h, staining in the nucleus and cytoplasm was seen (not shown). (C) NyaV-infected Vero B6 cells revealed dot-like sites of staining in the nucleus. (D) MVM-infected A9 mouse fibroblasts showing increased numbers of smaller dot-like areas of staining in the nucleus compared with the numbers seen in IAV- and NyaV-infected cells.
FIG 4.
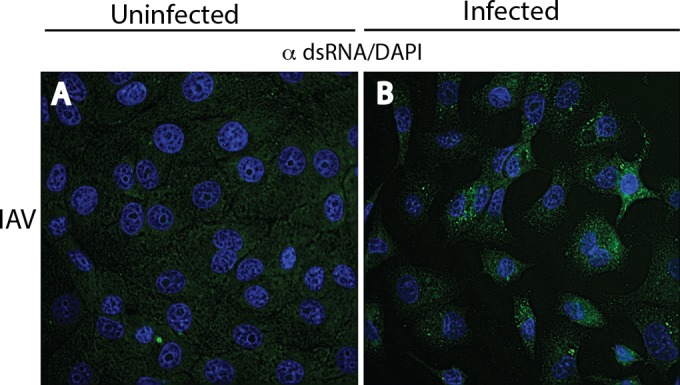
Immunofluorescence analysis of uninfected (A) and IAV-infected (B) Vero B6 cells stained with a 1:500 dilution of the J2 MAb showing that J2 also detects dsRNA in negative-strand RNA virus infections; images of the other negative-strand RNA viruses not shown.
Detection of dsRNA in LCMV and mouse parvovirus infections but not HBV infections.
Cytoplasmic dsRNA staining was also observed in LCMV-infected Vero B6 cells (MOI = 0.01) at 7 days p.i., an expected result, as was staining for the LCMV nucleoprotein, but the nucleoprotein signal colocalized with dsRNA in only some cells (Fig. 5B to D). However, an ssDNA virus like MVM, a mouse parvovirus, was not anticipated to generate dsRNA, yet late in infection (48 h p.i.) we observed a pattern of nuclear staining resembling that seen in NyaV infection (Fig. 5F). However, there were many more small dots of staining in the nucleus, and they varied more in size; a comparison of the nuclear dsRNA staining patterns for MVM, NyaV, and IAV that varied substantially is shown in Fig. 6. Most cells in MVM-infected monolayers were stained with antibody to NS1, and the staining was primarily cytoplasmic, but the intensity of staining varied from cell to cell, probably because the infection could not be synchronized on coverslips (Fig. 5E). Rare nuclei showed discrete areas of NS1 staining in the nucleus, dissimilar to the pattern of dsRNA staining (not shown). Finally, we were unable to detect dsRNA in a HepG2 cell clone persistently infected with HBV and expressing HBV core protein. Collectively, these results indicate that immunofluorescence staining with antibodies to dsRNA probably detects most, but not all, animal virus infections.
FIG 5.
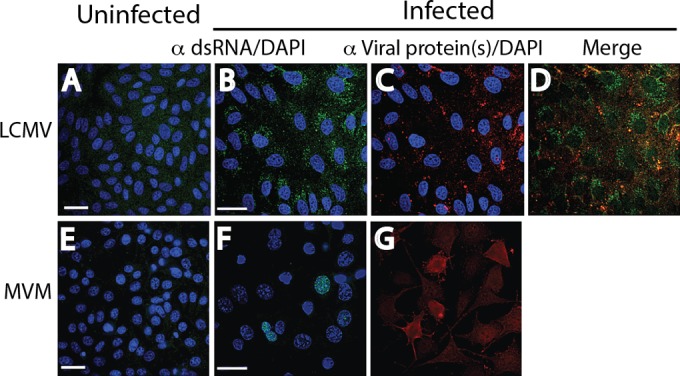
Reactivity of dsRNA and viral proteins in cells infected with an ambisense RNA virus (LCMV) and an ssDNA virus (MVM) by immunofluorescence analysis. Uninfected (A, E) and infected (B, F) cell monolayers were stained with a 1:2,000 dilution of MAb 9D5 to dsRNA, and infected monolayers were stained with the specific antiviral antibodies (C, G). (A to D) LCMV-infected and uninfected Vero B6 cells. (A) Uninfected cells. (B) LCMV-infected Vero B6 cells revealing punctate cytoplasmic staining (green) with MAb 9D5. (C) LCMV-infected cells stained with a 1:100 dilution of MAb to the LCMV NP protein showing cytoplasmic staining (red). (D) Merged image of panels B and C. (E to G) MVM-infected and uninfected mouse A9 cells. (E) Uninfected cells. (F) MVM-infected mouse A9 cells showing punctate areas of various diameters with staining with MAb 9D5 in the nucleus (green). (G) MVM-infected monolayers showed cytoplasmic staining with a 1:200 dilution of polyclonal antibody to MVM NS1/2 (no DAPI counterstaining) (red). The images in panels F and G were not from the same field. Bars, 10 μm (the magnification in the panels with bars applies to all photomicrographs).
Cellular stress resulting in apoptosis did not induce detectable levels of dsRNA staining in uninfected cells.
Since dsRNA staining was not observed in the uninfected cell lines, we were interested in whether cellular stress might result in the upregulation of dsRNA. Initially, uninfected M1-D macrophages were incubated with increasing concentrations of cycloheximide (CHX) and tumor necrosis factor alpha (TNF-α), both of which are known to induce apoptosis (26). As shown in Fig. 7A and C, there was no dsRNA staining in uninfected cell monolayers incubated with buffer or 500 μM CHX, while BeAn virus-infected M1-D cells (MOI = 10) were strongly positive for dsRNA (Fig. 7B). Nuclear fragmentation typical of apoptosis was observed in DAPI-stained M1-D cells (Fig. 7C), and immunoblot analysis showed the appropriate cleavage of PARP and caspase-3 at concentrations of CHX of ≥50 μM, indicative of apoptosis (Fig. 7D). Apoptosis was also induced at lower CHX concentrations when CHX was combined with 10 μg of TNF-α (Fig. 7D) and did not result in dsRNA staining (not shown). Induction of apoptosis in uninfected M1-D, BHK-21, Vero B6, and MDCK cell monolayers with these reagents did not reveal dsRNA staining (not shown). Thus, cellular stress resulting in apoptosis did not induce dsRNA species to levels detectable by immunofluorescence antibody staining.
FIG 7.
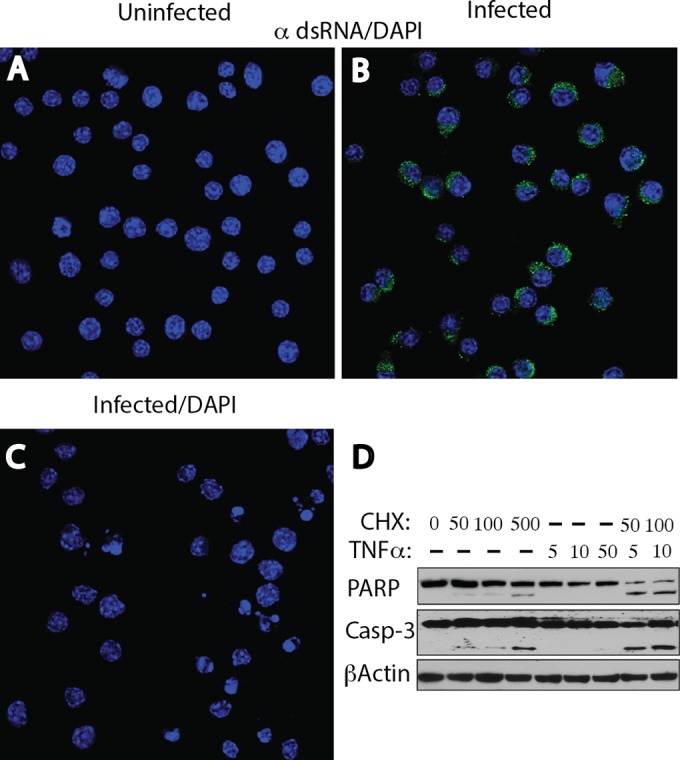
Lack of induction of dsRNA reactivity with CXH and CXH plus TNF-α, which results in apoptosis, in uninfected M1-D macrophages. (A) Uninfected cells were incubated with 500 μM CXH for 6 h before staining with MAb 9D5 and showed no dsRNA reactivity. (B) TMEV-infected cells showing cytoplasmic dsRNA staining as a positive control. (C) DAPI-stained fragmented and blebbing nuclei, indicative of apoptosis, in uninfected cells. (D) Immunoblot analysis showing PARP and caspase-3 (Casp-3) cleavages, indicative of apoptosis, when cells were incubated with CHX at concentrations of ≥50 μM and slightly more robust cleavages when cells were incubated with both CHX at a concentration of 50 or 100 μM and TNF-α at a concentration of 5 or 10 μg.
DISCUSSION
Based on reports in the literature (Table 1) and our present findings, we believe that dsRNA species are produced in sufficient amounts in most animal virus infections to be readily detected by immunofluorescence antibody staining. This is not unexpected, since dsRNA is a major viral trigger of the innate immune response but is nonetheless subject to viral countermeasures, such as the expression of dsRNA binding proteins by viruses that block dsRNA exposure (27–30), yet in a study by Weber et al. (16), little, if any, dsRNA was generated in negative-strand RNA virus infections for detection by immunofluorescence microscopy. This conclusion was reinforced because of the use of Tyramide signal amplification in their studies. However, their lack of success might relate to the production of smaller amounts of dsRNA in negative-strand RNA virus infections, the use of the less sensitive J2 MAb, the choice of less than optimum times for p.i. sampling, or, possibly, technical issues. In the present study, we have shown that negative-strand RNA viruses from four different virus families, VSV (Rhabdoviridae), IAV (Myxoviridae), MeV (Paramyxoviridae), and NyaV, generated RNAs of double-stranded character detectable by routine immunofluorescence antibody staining. NyaV belongs to a novel genus closely related to bornaviruses in the order Mononegavirales (31, 32). Interestingly, the intensity of staining tended to be weaker in VSV-, IAV-, and MeV-infected cells than in positive-strand RNA virus-infected cells; however, the optimum dose and the timing p.i. of dsRNA formation by the different viruses were not examined in detail, nor were viral countermeasures measured. Pfaller et al. (17) also found that dsRNA expression was much lower in cells infected with wild-type MeV than in cells infected with an MeV mutant with a knockout of the C protein (CKO), indicating that the C protein, which is known to control genome replication and transcription, also limits or masks dsRNA production. Similarly, Takeuchi et al. (24) observed dsRNA formation in cells infected with Sendai virus with CKO but failed to detect dsRNA in wild-type Sendai virus-infected cells by immunofluorescence antibody staining.
TABLE 1.
Detection of dsRNA produced in virus infections by immunofluorescence antibody staining
| Virus family and virusa | Nature of genome | Reference(s) |
|---|---|---|
| Adenoviridae (adenovirus) | dsDNA | 16, 39 |
| Herpesviridae | ||
| HSV-1 | dsDNA | 16 |
| CMV | dsDNA | 39 |
| Poxviridae, vaccinia virus | dsDNA | 16 |
| Reoviridae | ||
| Reovirus | dsRNA | 16 |
| Rotavirus | dsRNA | 39 |
| Togaviridae | ||
| Rubella virus | ssRNA, positive strand | 40, 41 |
| Sindbis virus | ssRNA, positive strand | 13 |
| Flaviviridae | ||
| Kunjin virus | ssRNA, positive strand | 42 |
| West Nile virus | ssRNA, positive strand | 43 |
| Dengue virus | ssRNA, positive strand | 44 |
| Japanese B encephalitis virus | ssRNA, positive strand | 44 |
| Hepatitis C virus | ssRNA, positive strand | 45 |
| Coronaviridae, SARS-CoV | ssRNA, positive strand | 16 |
| Picornaviridae | ||
| EMCV | ssRNA, positive strand | 9, 16 |
| CVB types 1 to 5, echovirus types 3, 6, and 9; poliovirus type 3, HPev1b | ssRNA, positive strand | 39 |
| Paramyxoviridae | ||
| MeV | ssRNA, negative strand | 17 |
| Newcastle disease virus | ssRNA, negative strand | 46 |
| Rhabdoviridae, VSV | ssRNA, negative strand | 9 |
HSV-1, herpes simplex virus 1; CMV, cytomegalovirus; SARS-CoV, severe acute respiratory syndrome coronavirus; EMCV, encephalomyocarditis virus; CVB, coxsackie B virus; HPev1, human parechovirus 1; MeV, measles virus.
Detected by the immunohistochemical method rather than immunofluorescence.
In most of the virus infections studied here and those described in the literature (Table 1), dsRNA was found in the cytoplasm, was frequently perinuclear in location, and was found in areas where the viroplasm is found. However, in NyaV- and MVM-infected cells that replicate in the nucleus, dsRNA was exclusively nuclear. We are unaware of prior reports of dsRNA staining in the nucleus for any virus infection. Currently, there is insufficient knowledge of NyaV RNA replication, and we can only speculate that the dsRNA is likely to be that of NyaV RF RNA. Recently, Matsumoto et al. (33) reported images obtained by confocal microscopy showing a pattern of nuclear staining (nuclear dots) of Borna disease virus N and P proteins that were closely associated with host chromosomes in infected cells, indicating that these were sites of viral factories. In rare instances, NyaV proteins stained with mouse antisera colocalized with dsRNA in the nucleus (Fig. 3P), but in MVM infections, antisera to the NS1 and NS2 proteins (that share an N-terminal peptide) did not appear to colocalize with dsRNA. Since MVM has an ssDNA genome, we could speculate that the failure of transcription termination close to the poly(A) site might produce transcripts capable of base pairing with the initial VP1/VP2 transcript. However, an understanding of the mechanism(s) of generation of overlapping transcripts in parvovirus infection awaits further study.
IAV transcription and replication occur in the nucleus inside a helical nucleocapsid. While diffuse dsRNA immunoreactivity was observed in the cytoplasm, infected cells grown on coverslips required protease digestion to elicit nuclear staining. After protease digestion, dsRNA antibodies most likely bound the RF RNAs of the IAV genome within nucleocapsids. The dsRNA target in the cytoplasm remains a mystery, and we can only speculate that it is also RF RNA exported from the nucleus to the cytoplasm in nucleocapsid particles that are inefficiently assembled or subjected to proteolysis in the cytoplasm. On the other hand, nuclear trafficking of the IAV nucleocapsid complex is a tightly regulated process (34), so the possibility that the upregulation of host dsRNA accounts for cytoplasmic staining cannot be excluded (see below).
The issue of dsRNA as a cellular product upregulated during viral infections was raised in a detailed study of the activation of PKR in MeV infection (17). In those studies, dsRNA was inhibited by CHX but not actinomycin D, indicating that RNA synthesis was due to the MeV RNA-dependent RNA polymerase and not the host cell dsDNA-dependent RNA polymerase. Recently, toxicity from accumulation of dsRNA in the cell cytoplasm was highlighted in publications describing studies of the pathogenesis of geographic atrophy, a form of macular degeneration (35, 36). Those investigators found that the accumulation of Alu dsRNA in the cytoplasm of retinal pigment epithelial cells was due to the reduction of DICER1, a type III RNase, leading to apoptosis of retinal ganglion cells (36). It has also been shown that cellular levels of RNAs for Alu, the most abundant repetitive element in the human genome capable of forming intramolecular dsRNAs, may increase by as much as 20-fold under various stress conditions (37, 38). Although we did not observe dsRNA staining of uninfected cell lines in this study, we wondered whether cellular stress might upregulate dsRNA species to a detectable level. However, the cellular stress from apoptosis induced by incubation of uninfected BHK-21, M1-D, MDCK, and Vero B6 cell monolayers with CHX alone or both CHX and TNF-α did not result in observable intracellular dsRNA reactivity. The results of this experiment, however, do not preclude the possibility that other stresses might induce the accumulation of dsRNA that reaches the cytoplasm.
Finally, since antibodies to dsRNA should detect most virus infections, these antibodies may have a place in viral discovery, particularly for immunofluorescence and immunohistochemistry methods in cases suspected of having a viral cause. In fact, antibodies to dsRNA were reported to identify viruses in acute central nervous system infections (39). Application of immunohistochemistry to archival formalin-fixed paraffin-embedded tissue suffers post hoc from the marked variability, timing, and duration of tissue fixation. dsRNA antibodies might also have a place in viral discovery using next-generation sequencing (NGS). A major limitation of cDNA libraries made from ssRNA molecules for NGS is that viral sequences comprise only a tiny fraction of a massive, complex, and largely irrelevant host RNA data set. This is especially problematic for chronic viral infections where the nucleic acid of a virus may be present in ultralow abundance compared to the levels in acute infections. In contrast, cDNA libraries constructed from dsRNA are enriched for viral sequences because of the lower host dsRNA background (unpublished data). Thus, dsRNA antibodies should have a role in viral discovery using NGS because of the exclusion of ssRNA in host tissues.
ACKNOWLEDGMENTS
We thank Patricia Kallio for expert technical help; Steve Ro, a UIC medical student, for starting this project; and David Schnurr (deceased), Viral and Rickettsial Disease Laboratory, California State Department of Public Health, for insightful discussions about dsRNA monoclonal antibodies and their use in viral diagnostics.
This work was supported by NMSS RG 4928A, NIH grant NS 077755, and the Modestus Bauer Foundation
REFERENCES
- 1.Vilcek J, Ng MH, Friedman-Kien AE, Krawciw T. 1968. Induction of interferon synthesis by synthetic double-stranded polynucleotides. J Virol 2:648–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long WF, Burke DC. 1971. Interferon production by double-stranded RNA: a comparison of induction by reovirus to that by a synthetic double-stranded polynucleotide. J Gen Virol 12:1–11. doi: 10.1099/0022-1317-12-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Williams BR. 2001. Signal integration via PKR. Sci STKE 2001:re2. [DOI] [PubMed] [Google Scholar]
- 4.Munir M, Berg M. 2014. The multiple faces of proteinkinase R in antiviral defense. Virulence 13:85–89. doi: 10.4161/viru.23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman RH. 2007. Viral encounters with 2′5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. 2011. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res 31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 7.Schneider WM, Dittmann Chevillote M, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dermody TS, Parker JSL, Sherry B. 2013. Orthoreoviruses, p 1304–1346. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peisley A, Hur S. 2013. Multi-level regulation of cellular recognition of viral dsRNA. Cell Mol Life Sci 70:1949–1963. doi: 10.1007/s00018-012-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha M, Kim N. 2014. Regulation of microRNA biogenesis. Nat Mol Cell Biol 15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 12.Ralph RK. 1969. Double-stranded viral RNA. Adv Virus Res 15:61–158. doi: 10.1016/S0065-3527(08)60874-X. [DOI] [PubMed] [Google Scholar]
- 13.Stollar V, Stollar BD. 1970. Immunochemical measurement of double-stranded RNA of uninfected and arbovirus-infected mammalian cells. Proc Natl Acad Sci U S A 65:993–1000. doi: 10.1073/pnas.65.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colby C, Jurale C, Kates JR. 1971. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J Virol 7:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kisseljov FL, Semjonova IS, Irlin IS, Shatalova GG. 1972. Partially double-stranded RNA in mouse spleen cells: the effect of Rauscher virus infection. Arch Virol 36:265–274. [DOI] [PubMed] [Google Scholar]
- 16.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand viruses. J Virol 80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller CK, Radeke MJ, Cattaneo R, Samuel CE. 2014. Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J Virol 88:456–468. doi: 10.1128/JVI.02572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jelachich ML, Lipton HL. 1999. Restricted Theiler's murine encephalomyelitis virus infection in murine macrophages induces apoptosis. J Gen Virol 80:1701–1705. [DOI] [PubMed] [Google Scholar]
- 19.Rozhon EJ, Kratochvil JD, Lipton HL. 1983. Analysis of genetic variation in Theiler's virus during persistent infection in the mouse central nervous system. Virology 128:16–32. doi: 10.1016/0042-6822(83)90315-X. [DOI] [PubMed] [Google Scholar]
- 20.Sells MA, Chen M-L, Acs G. 1987. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stollar BD, Stollar V. 1970. Immunofluorescent demonstration of double-stranded RNA in the cytoplasm of Sindbis virus-infected cells. Virology 42:276–280. doi: 10.1016/0042-6822(70)90270-9. [DOI] [PubMed] [Google Scholar]
- 22.Bonin M, Oberstrab J, Lukacs N, Ewert K, Oesterschulze E, Kassing R, Nellen W. 2000. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA 6:563–570. doi: 10.1017/S1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi S, Schnurr D, Lin J. 1992. Spectrum of monoclonal antibodies to coxsackievirus B-3 includes type- and group-specific antibodies. J Clin Virol 30:2498–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J Virol 82:10102–10110. doi: 10.1128/JVI.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrel M, Hoefs N, Staeheli P, Schneider U. 2012. Tick-borne Nyamanini virus replicates in the nucleus and exhibits unusual genome and matrix protein properties. J Virol 86:10739–10747. doi: 10.1128/JVI.00571-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujikura D, Uto M, Chiba S, Harada T, Perez F, Reed JC, Uede T, Miyazaki T. 2012. CLIPR-59 regulates TNF-α-induced apoptosis by controlling ubiquitination of RIP1. Cell Death Dis 3:e264. doi: 10.1038/cddis.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min J-Y, Krug RM. 2006. The primary function of RNA binding by the influenza A NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A 103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olland AM, Jane-Valbuena J, Schiff LA, Nibert MI, Harrison SC. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein sigma 3 at 1.8 Å resolution. EMBO J 20:979–989. doi: 10.1093/emboj/20.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry B. 2009. Rotavirus and reovirus modulation of the interferon response. J Interferon Cytokine Res 29:559–567. doi: 10.1089/jir.2009.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye C, Jia L, Sun Y, Hu B, Wang L, Lu X, Zhou J. 2014. Inhibition of antiviral innate immunity by birnavirus VP3 protein via blockage of viral double-stranded RNA to the host cytoplasmic RNA detector MDA5. J Virol 88:11154–11165. doi: 10.1128/JVI.01115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mihindukulasuriya KA, Nguyen NL, Wu G, Huang HV, Travassos da Rosa APA, Popov VL, Tesh RB, Wang D. 2009. Nyamanini and Midway viruses define a novel taxon of RNA viruses in the order Mononegavirales. J Virol 83:5109–5116. doi: 10.1128/JVI.02667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn JH, Bekal S, Cai Y, Clawson AN, Domier LL, Herrel M, Jahrling PB, Kondo H, Lambert KN, Mihindukulasuriya KA, Nowotny N, Radoshitzky SR, Schneider U, Staeheli P, Suzuki N, Tesh RB, Wand D, Wang L-F, Dietzgen RG. 2013. Nyamiviridae: proposal for a new family in the order Mononegavirales. Arch Virol 158:2209–2226. doi: 10.1007/s00705-013-1674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto Y, Hayashi Y, Omori H, Honda T, Saito T, Horie M, Ikuta K, Fujino K, Nakamura S, Schneider U, Chase G, Yoshimori T, Schwemmie M, Tomonaga K. 2012. Bornavirus closely associates and segregates with host chromosome to ensure persistent intranuclear infection. Cell Host Microbe 11:492–503. doi: 10.1016/j.chom.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Shaw ML, Palese P. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 35.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Karikó K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471:325–332. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimj Y, Tarallo V, Kerur N, Yasuma T, Gelfand BD, Bastos-Carvalho A, Hirano Y, Yasuma R, Mizutani T, Fowler BJL, Kaneko SH, Boddanovich S, Ambati BK, Hinton DR, Hauswirth WW, Hakem R, Wright C, Ambati J. 2014. DICER1/Alu dysmetabolism induces caspase-8-mediated cell death in age-related macular degeneration. Proc Natl Acad Sci U S A 111:16082–16097. doi: 10.1073/pnas.1403814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu WM, Chu MW, Choudary PV, Schmid CW. 1995. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res 23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li TH, Schmid CW. 2001. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene 276:135–141. doi: 10.1016/S0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 39.Richardson SJ, Willcox A, Hilton DA, Tauriainen S, Hyoty H, Bone AJ, Foulis AK, Morgan NG. 2010. Use of antisera directed against dsRNA to detect infections in formalin-fixed paraffin-embedded tissue. J Clin Virol 49:180–185. doi: 10.1016/j.jcv.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 40.Bowden DS, Pedersent JS, Toh BH, Westaway EG. 1987. Distribution of immunofluorescence of viral products and actin-containing cytoskeletal filaments in rubella virus-infected cells. Arch Virol 92:211–219. doi: 10.1007/BF01317478. [DOI] [PubMed] [Google Scholar]
- 41.Lee JY, Marshall JA, Bowden DS. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- 42.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B and NS3, in virus-induced membrane structures. J Virol 71:6650–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeWitte-Orr SJ, Mehta DR, Collins SE, Suthar MS, Gale M Jr, Mossman KL. 2009. Long double-stranded RNA induces an antiviral response independent of FIN regulatory factor 3, IFN-beta promoter stimulator 1, and IFN. J Immunol 183:6545–6553. doi: 10.4049/jimmunol.0900867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchida L, Espada-Murao LA, Takamatsu Y, Okamoto K, Hayasaka D, Yu F, Nabeshima T, Buerano CC, Marita K. 2014. The dengue virus conceals double-stranded RNA in the intracellular membrane to escape from an interferon response. Sci Rep 4:7395. doi: 10.1038/srep07395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Targett-Adams P, Boulant S, McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol 82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Sun Y, Chen H, Dai Y, Zhan Y, Yu S, Qiu X, Tan L, Song C, Ding C. 2014. Activation of the PKR/eIF2a signaling cascade inhibits replication of Newcastle disease virus. Virol J 11:62. doi: 10.1186/1743-422X-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]