ABSTRACT
Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) is a host protein with reported cell-intrinsic antiviral activity against several RNA viruses. The proposed basis for the activity against negative-sense RNA viruses is the binding to exposed 5′-triphosphates (5′-ppp) on the genome of viral RNA. However, recent studies reported relatively low binding affinities of IFIT1 for 5′-ppp RNA, suggesting that IFIT1 may not interact efficiently with this moiety under physiological conditions. To evaluate the ability of IFIT1 to have an impact on negative-sense RNA viruses, we infected Ifit1−/− and wild-type control mice and primary cells with four negative-sense RNA viruses (influenza A virus [IAV], La Crosse virus [LACV], Oropouche virus [OROV], and Ebola virus) corresponding to three distinct families. Unexpectedly, a lack of Ifit1 gene expression did not result in increased infection by any of these viruses in cell culture. Analogously, morbidity, mortality, and viral burdens in tissues were identical between Ifit1−/− and control mice after infection with IAV, LACV, or OROV. Finally, deletion of the human IFIT1 protein in A549 cells did not affect IAV replication or infection, and reciprocally, ectopic expression of IFIT1 in HEK293T cells did not inhibit IAV infection. To explain the lack of antiviral activity against IAV, we measured the binding affinity of IFIT1 for RNA oligonucleotides resembling the 5′ ends of IAV gene segments. The affinity for 5′-ppp RNA was approximately 10-fold lower than that for non-2′-O-methylated (cap 0) RNA oligonucleotides. Based on this analysis, we conclude that IFIT1 is not a dominant restriction factor against negative-sense RNA viruses.
IMPORTANCE Negative-sense RNA viruses, including influenza virus and Ebola virus, have been responsible for some of the most deadly outbreaks in recent history. The host interferon response and induction of antiviral genes contribute to the control of infections by these viruses. IFIT1 is highly induced after virus infection and reportedly has antiviral activity against several RNA and DNA viruses. However, its role in restricting infection by negative-sense RNA viruses remains unclear. In this study, we evaluated the ability of IFIT1 to inhibit negative-sense RNA virus replication and pathogenesis both in vitro and in vivo. Detailed cell culture and animal studies demonstrated that IFIT1 is not a dominant restriction factor against three different families of negative-sense RNA viruses.
INTRODUCTION
To protect against pathogenic microbial agents, host cells have evolved cell-intrinsic and cell-extrinsic innate immune defense mechanisms. Type I interferon (IFN) is an essential component of the host innate immune response against viruses. The type I IFN response pathway is activated after the detection of nonself pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), such as retinoic-acid inducible gene I (RIG-I)-like receptors, DNA sensors, and Toll-like receptors. The binding of PAMPs triggers a signaling cascade that results in the transcriptional activation of type I IFN and its secretion from infected cells. Type I IFN can bind to its cognate receptor on both infected and uninfected cells and can induce hundreds of interferon-stimulated genes (ISGs), many of which may have antiviral effects.
Members of the IFN-induced protein with tetratricopeptide repeats (IFIT) gene family are induced to high levels in cells in response to IFN signaling or viral infection (1). IFIT proteins reside in the cytoplasm and contain multiple tetratricopeptide repeat (TPR) motifs enabling interactions with other proteins and molecules. IFIT genes are present in many vertebrate species, but the number of genes and their sequence identity vary between species, suggesting the presence of a positive evolutionary selection pressure (2). The human IFIT family contains four characterized (IFIT1, IFIT2, IFIT3, and IFIT5) and two uncharacterized (IFIT1B and IFIT1P1) genes. Mice also have four characterized (Ifit1, Ifit1c, Ifit2, and Ifit3) and two uncharacterized (Ifit1b and Ifit3b) genes (1–3). The first reported activities of human IFIT1 were related to interactions with host and viral proteins. The binding of IFIT1 to eukaryotic initiation factor 3 (eIF3) and the 40S ribosome subunit (4–6) inhibited protein translation and hepatitis C virus replication (7). IFIT1 also binds to the E1 protein of human papillomavirus, inhibiting its replication (8, 9). More recently, human and mouse IFIT1 proteins were identified as proteins that distinguish between self and nonself RNA species. Specifically, IFIT1 restricted infections by flaviviruses, poxviruses, and coronaviruses that were deficient in 2′-O methyltransferase activity (10–15). Loss-of-function mutations in viral 2′-O methyltransferases resulted in an inability to generate cap 1 (n7mGpppNm) mRNA structures, which rendered the cap 0 (n7mGpppN) viral RNA susceptible to IFIT1-mediated inhibition of translation (11, 15, 16). IFIT1 was also shown to restrict attenuated alphaviruses containing a single nucleotide change, at position 3 of the 5′ end of the untranslated region (UTR) (17). This residue modulates the thermostability of a secondary structure element that allows alphavirus mRNA, which naturally lacks cap 1 structures, to evade IFIT1 restriction. The mechanism for how IFIT1 distinguishes between host (self) and viral RNAs is not yet fully understood, but the atomic structure suggests that the TPR motifs create a positively charged pocket that is responsible for direct RNA binding (18).
Human and mouse IFIT1 proteins can also interact with the 5′-triphosphate (5′-ppp) moiety present in the genomes of negative-sense RNA viruses. Previous studies suggested that this interaction inhibits infections by vesicular stomatitis virus (VSV) and influenza A virus (IAV) (18, 19), possibly by sequestering viral RNA from the replicating pool (19). However, the antiviral effect of mouse Ifit1 on VSV replication and pathogenesis was not confirmed in a subsequent study (20). Furthermore, binding studies with the various RNA ligands of human and rabbit IFIT1 proteins (reviewed in reference 21) demonstrated that IFIT1 has a higher affinity for cap 0 RNA than for 5′-ppp RNA or cap 1 RNA.
To evaluate the importance of 5′-ppp RNA recognition by human and mouse IFIT1 proteins in the replication and pathogenesis caused by negative-strand RNA viruses, we infected human and mouse cells deficient in IFIT1 protein expression with four different negative-sense RNA viruses corresponding to three distinct families. We also performed an in vivo analysis of wild-type (WT) and Ifit1−/− mice after inoculation with IAV (Orthomyxoviridae), La Crosse virus (LACV) (Bunyaviridae), and Oropouche virus (OROV) (Bunyaviridae). Our studies indicate that human and murine IFIT1 proteins do not efficiently restrict infection by IAV, LACV, OROV, or Ebola virus (EBOV).
MATERIALS AND METHODS
Mice.
WT and congenic Ifit1−/− (gene ID 15957) (13) C57BL/6 mice were bred under specific-pathogen-free conditions at the Washington University School of Medicine. All animal studies were approved and performed in accordance with protocols approved by the Washington University School of Medicine Institutional Animal Care and Use Committees.
Cells.
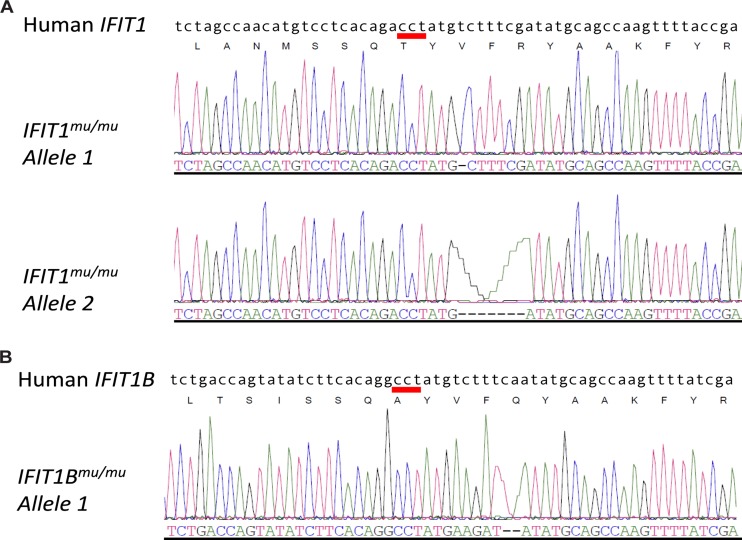
Deletion of IFIT1 (gene ID 3434) and IFIT1B (gene ID 439996) protein expression in the A549 human lung epithelial cell line was achieved by using CRISPR/Cas9 gene-editing technology (22). A guide RNA specific for human IFIT1 (GCTGCATATCGAAAGACAT) was cloned into the gRNA expression plasmid and cotransfected into A549 cells by using Lipofectamine LTX, together with a human-codon-optimized Cas9 expression plasmid, and an empty vector containing a puromycin resistance gene. After 24 h, transfected cells were treated with 2 μg/ml of puromycin for 2 days and cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), penicillin, streptomycin, l-glutamine, 25 mM HEPES, and nonessential amino acids. IFIT1 gene modification was evaluated by DNA sequencing of a PCR product containing the gRNA target site. Next, we performed a limiting dilution assay to generate clonal cell lines. DNAs were extracted from these clonal A549 cells, and the target area was amplified by PCR (primer sequences are available upon request). The PCR product was cloned into the TOPO-blunt vector (Life Technologies) and sequenced using an M13 primer (IDT Technologies). One cell line harboring two different modifications (at nucleotide position 757 from the start codon) in the IFIT1 gene was selected for further evaluation (IFIT1mu/mu A549) (Fig. 1). The modification resulted in a frameshift and the generation of a stop codon approximately 60 nucleotides downstream from the target site. Due to the similarity in target sequence between the IFIT1 and IFIT1B genes, we sequenced the IFIT1B gene and identified one modified IFIT1B gene. The modification in IFIT1B resulted in a frameshift and the generation of a stop codon 39 nucleotides downstream from the target site. As a control, we selected a clonal A549 cell line from the procedure described above that had no modifications in both IFIT1 and IFIT1B (referred to as A549-CRISPR ctrl).
FIG 1.
CRISPR-Cas9 editing of IFIT1 and IFIT1B genes in A549 cells. (A and B) Representative electropherograms of individual TOPO plasmids containing PCR fragments of the IFIT1 (A) and IFIT1B (B) genes from IFIT1mu/mu A549 cells. Two distinct alleles were identified for IFIT1, and only a single allele was identified for IFIT1B. The red bar indicates the NGG motif required for SpCas9 activity.
Bone marrow-derived macrophages (Mϕ) were generated as described previously (23). Briefly, bone marrow was isolated from WT or Ifit1−/− mice and cultured for 7 days in the presence of 40 ng/ml macrophage colony-stimulating factor (M-CSF; PeproTech). Murine embryonic fibroblasts (MEFs) were generated from day 15 WT or Ifit1−/− embryos and maintained in DMEM supplemented with 10% FBS, l-glutamine, and nonessential amino acids as described previously (24). Primary mouse tracheal epithelial cells (mTECs) were harvested from 6- to 8-week-old WT and Ifit1−/− mice and cultured in growth factor-enriched medium on semipermeable membranes (Transwell; Corning-Costar, Corning, NY) (25). Medium was maintained in the upper and lower chambers until the transmembrane resistance increased (>1,000 Ω/cm2), indicating tight junction formation. Medium was then removed from the upper chamber to establish an air-liquid interface (ALI), and cells were allowed to differentiate for 14 days before they were used.
Viruses.
The 2009 pandemic influenza A virus A/California/04/2009 H1N1 (IAV-Cal) and A/Puerto Rico/8/1934 (IAV-PR8) were obtained from St. Jude Children's Research Hospital and propagated in the allantoic cavities of 10-day-old embryonic chicken eggs (26). WT West Nile virus (WNV-WT) and WNV-E218A (27) were generated from an infectious cDNA clone of the New York 1999 strain and propagated in BHK clone 13 cells (ATCC). Mouse-adapted Ebola virus (maEBOV; adapted from Ebola virus [Zaire ebolavirus] isolate Mayinga as described previously [28]) was propagated in Vero E6 cells. All work with maEBOV was performed under biosafety level 4 conditions at the Integrated Research Facility at the Rocky Mountain Laboratories, NIAID, NIH, Hamilton, MT. Sample inactivation/removal was performed according to standard operating protocols approved by the local institutional biosafety committee. LACV (original strain) and OROV (strain Bean 19991) were provided by Andrew Pekosz (Johns Hopkins University, Baltimore, MD) and Eurico Arruda (São Paulo University, Ribeirão Preto, Brazil), respectively, and were passaged in Vero cells (29). All experiments with OROV were conducted in a biosafety level 3 facility with appropriate personal protective equipment and approval from the U.S. Department of Agriculture.
Morbidity and mortality analyses.
Seven- to 8-week-old WT and Ifit1−/− female mice were inoculated with 104 50% egg infective doses (EID50) of IAV-Cal intranasally in 30 μl of sterile phosphate-buffered saline (PBS) after sedation with 2,2,2-tribromoethanol (Avertin; Sigma-Aldrich). Six- or 8-week-old WT and Ifit1−/− mice were inoculated subcutaneously in the footpad with 106 or 105 focus-forming units (FFU) of OROV or LACV, respectively, in a volume of 50 μl after sedation with ketamine hydrochloride (80 mg/kg of body weight) and xylazine (15 mg/kg). Morbidity and mortality were monitored for 21 days.
Tissue viral titers.
Lungs were collected on day 3, 6, or 9 after inoculation with 104 EID50 of IAV-Cal. Tissues were homogenized in 1 ml of minimal essential medium, centrifuged for 5 min at 1,000 × g to remove cellular debris, and stored at −80°C. The supernatant was used to quantify the amount of infectious virus present in the lungs. Virus titers were determined on Madin-Darby canine kidney (MDCK) cells as described previously (26). Livers, spleens, brains, and sera of WT and Ifit1−/− mice inoculated with either 106 FFU of OROV or 105 FFU of LACV were collected on day 4 or 8, respectively, after perfusion (20 ml) with PBS. Organs were weighed and homogenized. Sera from all infected animals were collected prior to perfusion and stored at −80°C. Total RNAs from organs were extracted by using a Qiagen RNeasy kit and were treated with DNase to remove genomic DNA. Quantitative reverse transcription-PCR (qRT-PCR) was performed using One-Step RT-PCR master mix and a model 7500 Fast real-time PCR system (Applied Biosystems). Viral burdens in sera and organs were measured using OROV- and LACV-specific primers and probes, and the level of viral RNA was normalized to Gapdh gene expression in the tissue samples (29). The viral burden was recorded as the number of viral genome equivalents per gram of tissue for the organs and as the number of viral genome equivalents per milliliter for serum.
Viral growth kinetics in cell culture.
The growth of IAV-PR8 was assessed in wild-type A549, IFIT1mu/mu A549, and A549-CRISPR ctrl cells. Cells were infected at a multiplicity of infection (MOI) of 0.01, and the supernatant was collected at 24, 48, and 72 h postinoculation. The virus titer in the supernatant was quantified on MDCK cells, and the 50% tissue culture infective dose (TCID50) was calculated using the Reed and Muench method. OROV and LACV growth curve studies were performed by using WT and Ifit1−/− MEFs as described previously (29). Briefly, multistep virus growth curve studies were performed after infection of cells at an MOI of 0.01, and the viral titer in the cell-free supernatant was determined by FFU assay of Vero E6 cells at the following time points after infection: 0, 1, 4, 12, 24, 36, 48, and 60 h. The infected-cell foci in the FFU assay were detected at 22 to 24 h of infection by using a polyclonal mouse anti-OROV ascites fluid (VR1228AF; ATCC) or a 1:100 dilution of hybridoma cell supernatants containing monoclonal antibodies (MAbs) against LACV (807-31 and 807-33; provided by Andrew Pekosz). For maEBOV, 1.5 × 105 Mϕ were seeded in a 12-well plate 7 to 8 days prior to infection. Eight hours prior to infection, Mϕ were pretreated with 100 U/ml of mouse IFN-β. Mϕ were infected with maEBOV at an MOI of 0.1 for 1 h at 37°C before the inoculum was removed and the cells were washed with PBS. Supernatant containing maEBOV was harvested at 24, 48, and 72 h postinfection, and viral titers were determined by a TCID50 assay on Vero E6 cells, using the Spearman-Karber method.
Analysis of bronchoalveolar cellular infiltrate after IAV infection.
To assess the levels of cellular infiltrate 3, 5, 7, and 9 days after IAV-Cal infection, bronchoalveolar lavage (BAL) was performed by lavaging the trachea and lungs three times by injecting a total of 2.5 ml of Hanks balanced salt solution (HBSS). Cells were collected by centrifugation and stained with MAbs specific for CD3, CD8, CD4, CD11b, CD11c, Ly6G, major histocompatibility complex (MHC) class II, and B220 to define cell types. All samples were processed on an LSRII machine (BD Biosciences). The resulting data were analyzed using FlowJo software (Treestar).
Western blotting.
WT and IFIT1mu/mu A549 cells that were left untreated or treated with 100 U/ml human IFN-β for 24 h were lysed in RIPA buffer (10 mM Tris, 150 mM NaCl, 0.02% sodium azide, 1% sodium deoxycholate, 1% Triton X-100, and 0.1% SDS, pH 7.4) containing protease inhibitors. Samples were resolved by electrophoresis on 10% or 12% SDS-polyacrylamide gels. Following transfer of proteins, membranes were blocked and probed with the following panel of primary antibodies: rabbit anti-β-actin (Abcam), mouse anti-human IFIT1 (Pierce), and mouse anti-mouse Ifit3, which cross-reacts with human IFIT3 (M. S. Diamond, unpublished result). The secondary antibodies were anti-rabbit–Alexa 690 and anti-mouse–Alexa 800 (Li-Cor). The Western blots were imaged on a Li-Cor Odyssey infrared imager and then analyzed with Image Studio Lite.
Measurement of ISG expression by qRT-PCR.
RNAs were isolated from control and IFN-β-treated A549 cells or mTECs by using an RNA isolation kit (Qiagen), and expression was measured by fluorogenic qRT-PCR using primers and probes (sequences available upon request) specific to human and murine IFIT1, IFIT2, IFIT3, and RSAD2, using One-Step RT-PCR master mix and a model 7500 Fast real-time PCR system (Applied Biosystems).
IAV and WNV antigen flow cytometry.
HEK293T cells (2 × 105) were transfected with 500 ng of the indicated plasmids by using Trans-IT LTI (Mirus Bio LLC) according to the manufacturer's instructions. Sixteen hours later, cells were infected with IAV-PR8 at an MOI of 5 for 6 h. For WNV infections, HEK293T cells were transiently transfected with the IFIT1 or control vector. After 24 h, cells were infected with WNV-WT or WNV-E218A (MOI of 10), and 24 h later the cells were processed for relative infection detection by flow cytometry. For both IAV and WNV, cells were collected, fixed, and permeabilized prior to intracellular antibody staining with an anti-Flag-tag MAb and a biotinylated anti-IAV NP (MAb 8258B; EMD Millipore) or anti-WNV E antibody (human E16) (30). NP- or E-positive cells were visualized by use of Alexa 647-labeled streptavidin. All flow cytometry studies were performed using a FACSCalibur flow cytometer (BD Biosciences), and data were analyzed with FlowJo software (TreeStar). The relative infection level was calculated as the percent infection of transfected cells divided by the percent infection of untransfected cells. The percent infection of transfected cells was determined by dividing the percentage of the viral antigen+ Flag+ population by that of the Flag+ population. The percent infection of untransfected cells was determined by dividing the percentage of the viral antigen+ Flag− population by that of the Flag− population. The calculation was done for each sample and then normalized to the value for luciferase control-transfected cells.
RNA binding and competition assays.
Oligonucleotides representing the 5′ ends of the PB2, PB1, and NS gene segments of IAV (A/California/04/09) were left untreated (5′-ppp RNA), treated with a ScriptCap m7G capping system (Epicentre) alone (cap 0 [7mGpppNp]) or in conjunction with a ScriptCap 2′-O methyltransferase kit (Epicentre) (cap 1 [7mGpppNm]), or treated with calf intestinal phosphatase (NEB) (5′-OH RNA). Following radiolabeling with [α-32P]ATP by using T4 polynucleotide kinase (NEB), the RNAs were purified using 12% urea-PAGE. Labeled RNAs (5 nM) were incubated with increasing concentrations of purified murine Ifit1 protein (GenScript). In the competition assay, unlabeled RNA with cap 0 or 5′-OH (125 nM) was added to the murine Ifit1 protein (0.3 μM) and radiolabeled 5′-ppp RNA (5 nM). After 15 min at room temperature, samples were applied to a dot blot apparatus (Whatman) with one nitrocellulose (NC) membrane on top of one nylon (NY) membrane. Radiolabeled RNA bound to the NC and NY membranes was quantified using a Typhoon 9410 variable-mode imager, and the fraction of RNA bound to Ifit1 was calculated using the following equation: fraction bound = RNA signal on NC/(RNA signal on NC + RNA signal on NY). Data were fitted to the Hill equation by using Origin, and the dissociation constants (KD) were calculated.
Cytokine Bio-Plex assay.
WT or Ifit1−/− mice were infected with IAV, and at specified times, lungs were collected and homogenized as described above. The Bio-Plex Pro assay was performed according to the manufacturer's protocol (Bio-Rad). The cytokine screen included interleukin-1α (IL-1α), IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-17, eotaxin, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, KC, CCL2, CCL3, CCL4, CCL5, and tumor necrosis factor alpha (TNF-α).
Statistical analysis.
All data were analyzed using Prism software (GraphPad6). Kaplan-Meier survival curves were analyzed by the log rank test. Differences in viral titers, cytokine levels, gene expression data, and cell numbers were analyzed by the Mann-Whitney test. Differences in the percentages of NP+ A549-WT, A549-IFIT1mu/mu, and A549-CRISPR ctrl cells following pretreatment with increasing doses of human IFN-β were determined by linear regression analysis. Other specific tests are indicated in the text.
RESULTS
Effect of IFIT1 on influenza virus infection in cell culture.
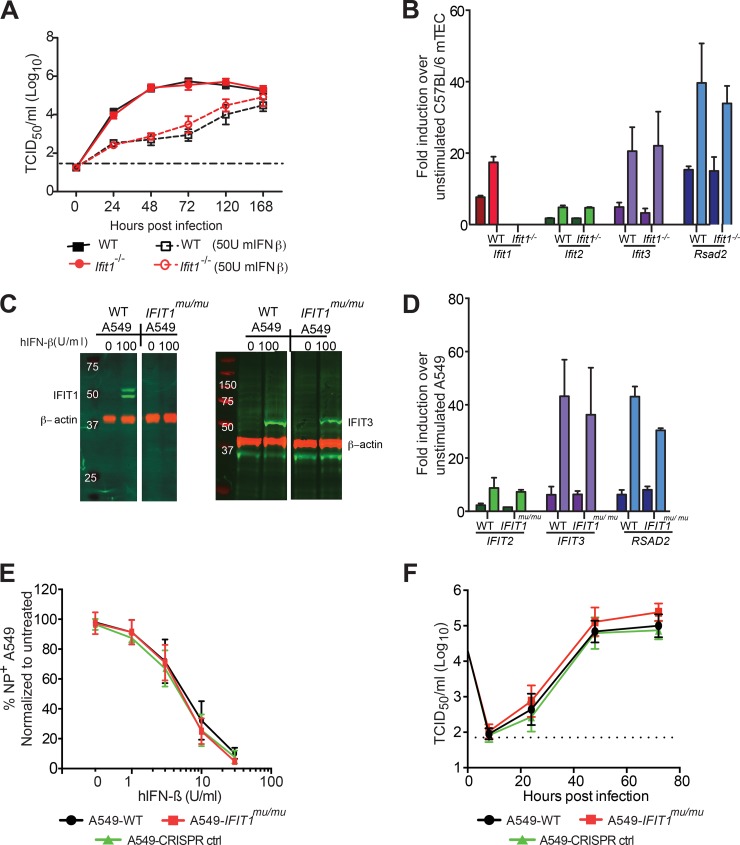
We first established an in vitro growth system with Ifit1-sufficient and -deficient mouse and human respiratory epithelial cells that are permissive for IAV infection. We used differentiated primary mouse tracheal epithelial cells (mTECs) generated from WT or Ifit1−/− C57BL/6 mice (31). These cells were left unmanipulated or pretreated with 50 U/ml of recombinant mouse IFN-β prior to infection with IAV-Cal (A/California/04/2009; H1N1) at an MOI of 0.01. The virus was harvested at different times after IAV infection, and the viral yield was quantified. Unexpectedly, we observed no difference in the growth of IAV in untreated or IFN-β-pretreated WT and Ifit1−/− mTECs at any of the time points (Fig. 2A) (P > 0.6). To confirm that the mTECs responded to recombinant IFN-β, we analyzed gene expression data from both WT and Ifit1−/− mTECs stimulated with 50 U/ml or 1,000 U/ml IFN-β (Fig. 2B). Following IFN-β stimulation, WT mTECs expressed high levels of Ifit1 and Rsad2 mRNAs, whereas Ifit1−/− cells expressed high levels of Rsad2 mRNA but no Ifit1 mRNA, as expected. Since Ifit1 can oligomerize with other IFIT proteins (16, 19), we assessed the expression of Ifit2 and Ifit3. Notably, Ifit2 and Ifit3 were upregulated equivalently in both WT and Ifit1−/− mTECs following IFN-β stimulation (Fig. 2B), suggesting that the loss of Ifit1 did not affect the expression of other IFIT gene family members. Thus, the absence of an antiviral effect of Ifit1 in mTECs against IAV was not due to an absence of expression of other Ifit genes.
FIG 2.
Effects of human and mouse IFIT1 proteins on influenza A virus infection and replication. (A) Kinetics of IAV-Cal replication in WT and Ifit1−/− primary mTECs after infection at an MOI of 0.01, with or without IFN-β stimulation. The virus was harvested at 0, 24, 48, 72, 120, and 168 h postinfection, and viral yields were quantified by a TCID50 assay on MDCK cells. Data were pooled from two independent experiments performed in triplicate. (B) Expression of ISGs in WT and Ifit1−/− mTECs after IFN-β stimulation. mTECs from WT and Ifit1−/− mice were treated with 50 U/ml (dark colors) or 1,000 U/ml (light colors) of recombinant IFN-β or mock treated for 18 h before cellular RNA was extracted and used to quantify Ifit1, Ifit2, Ifit3, and Rsad2 gene expression by quantitative RT-PCR. Data were pooled from three independent experiments performed in duplicate. (C and D) Expression of ISGs in WT and IFIT1mu/mu A549 cells after IFN-β stimulation. (C) Western blots of human IFIT1, IFIT3, and β-actin on cell lysates from WT and IFIT1mu/mu A549 cells stimulated with 0 or 100 U/ml of human IFN-β for 18 h. (D) A549-WT and A549-IFIT1mu/mu cells were treated with 100 U/ml (dark colors) or 1,000 U/ml (light colors) of recombinant human IFN-β or mock treated for 18 h before cellular RNA was extracted and used to quantify IFIT2, IFIT3, and RSAD2 gene expression by quantitative RT-PCR. Data were pooled from two independent experiments performed in triplicate. (E and F) Kinetics of IAV-PR8 replication in WT and IFIT1mu/mu A549 cells. (E) Viral replication was measured in WT and IFIT1mu/mu A549 cells that were pretreated for 18 h with human IFN-β (hIFN-β). (F) WT and IFIT1mu/mu A549 cells were infected, and the virus titers in supernatants were determined at 1, 8, 24, 48, and 72 h postinfection. The data represent the means ± standard deviations (SD) for two independent experiments performed in triplicate.
As the antiviral activities of murine and human IFIT1 proteins may differ (reviewed in reference 32), we questioned whether human IFIT1 could inhibit IAV replication, as suggested previously (19). To evaluate this, we generated A549 cells deficient in IFIT1 and IFIT1B protein expression (IFIT1mu/mu cells) by using CRISPR/Cas9 gene-editing technology. IFIT1 and IFIT1B gene modification was confirmed by sequencing of individual PCR products (Fig. 1). The two alleles of IFIT1 in IFIT1mu/mu A549 cells contained 1- and 7-nucleotide deletions at position 757 from the translation start site, resulting in a premature stop codon approximately 60 nucleotides downstream of the modification. Only a single allele of IFIT1B was identified, and the 2-nucleotide deletion resulted in a premature stop codon 39 nucleotides downstream of the modification. The loss of expression of IFIT1 proteins was corroborated by Western blotting of A549 cells treated with human IFN-β with an antibody that recognizes IFIT1 and IFIT1B (Fig. 2C). However, IFIT1mu/mu A549 cells expressed WT levels of IFIT3 protein and IFIT2, IFIT3, and RSAD2 mRNAs after IFN-β treatment (Fig. 2D).
To assess the role of IFIT1 in restriction of IAV infection, IFIT1mu/mu A549 cells were pretreated with increasing doses of human IFN-β and infected with A/Puerto Rico/8/1934 H1N1 (IAV-PR8) at an MOI of 5 for 8 h (Fig. 2E). Infected cells were analyzed for expression of nucleoprotein (NP) by flow cytometry. As the dose of human IFN-β was decreased, infection became apparent in both WT and IFIT1mu/mu A549 cells, and the levels of NP+ cells were equivalent (P > 0.3). To assess whether IFIT1 would have a greater impact over several rounds of replication, we performed a multistep growth analysis by infecting cells at an MOI of 0.01 and monitoring viral growth over 72 h. Again, we saw no statistically significant differences in replication of IAV in WT and IFIT1mu/mu A549 cells (Fig. 2F) (P > 0.1).
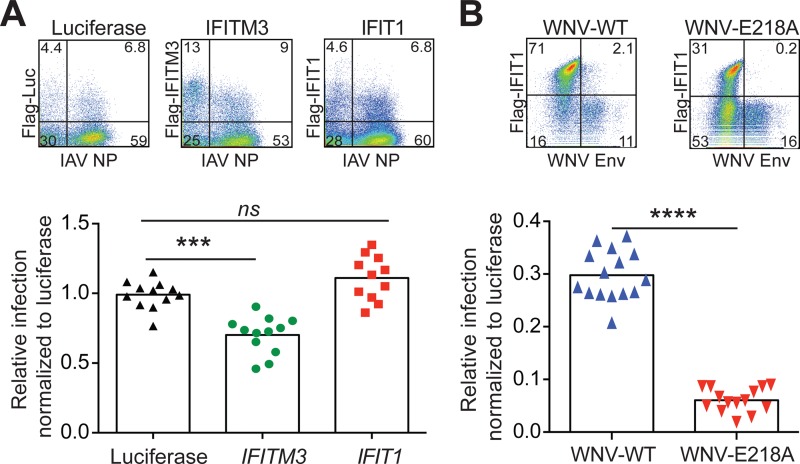
An absence of an effect of murine and human IFIT1 on IAV infection was unexpected, because a previous report suggested that IFIT1 bound to viral RNA displaying 5′-ppp moieties and inhibited replication of negative-strand RNA viruses (19). One possible explanation for the disparity in our results was that IFN-β treatment of mTECs or A549 cells could have induced multiple other antiviral ISGs against IAV, such that the impact of any single ISG, including IFIT1, was masked. To evaluate this, we tested whether ectopic expression of N-terminally Flag-tagged human IFIT1 could inhibit IAV infection directly. Human IFIT1 was transfected into HEK293T cells, and 16 h later the cells were infected with IAV-PR8 at an MOI of 5. After 6 h, the cells were costained with an anti-Flag-tag antibody to identify transfected cells and an anti-NP antibody to define IAV-infected cells (Fig. 3A). As a positive control, we transfected the HEK293T cells with human IFITM3, which inhibits IAV entry (33). In contrast to IFITM3, ectopic expression of human IFIT1 did not affect IAV-PR8 infection and replication in these cells. Human IFIT1 was active functionally in HEK293T cells, as it preferentially inhibited a mutant strain of WNV (WNV-E218A) lacking 2′-O methylation (Fig. 3B).
FIG 3.
Ectopic expression of human IFIT1 does not affect IAV infection. (A) Impact of luciferase, IFITM3, or IFIT1 expression in HEK293T cells infected with IAV-PR8. HEK293T cells were transfected with Flag-tagged luciferase, IFITM3, or IFIT1 and then, 16 h later, infected with IAV-PR8 at an MOI of 5. Cells were harvested at 6 h postinfection and stained with MAbs specific for IAV-NP and Flag. Representative flow cytometry plots are shown for luciferase, IFITM3, and IFIT1. The relative infection level was calculated as the percent infection of transfected cells divided by the percent infection of nontransfected cells for each sample and then normalized to that of control (luciferase) transfected cells. The data represent the mean relative infection levels ± SD for three independent experiments with three or four technical replicates. (B) Expression of IFIT1 preferentially inhibits WNV-E218A. HEK293T cells were transiently transfected with Flag-tagged IFIT1 or a Flag-tagged luciferase control vector. After 48 h, cells were infected with WNV-WT or WNV-E218A (MOI of 10). The cells were harvested 24 h after infection and stained with MAbs specific for WNV E and Flag. Representative flow cytometry plots are shown for IFIT1-transfected cells infected with either WNV-WT or WNV-E218A. The relative infection level was calculated as the percent infection of transfected cells divided by the percent infection of untransfected cells. The percent infection of transfected cells was determined by dividing the percentage of the viral antigen+ Flag+ population by the percentage of the Flag+ population. The percent infection of the untransfected cells was determined by dividing the percentage of the viral antigen+ Flag− population by the percentage of the Flag− population. The calculation was done for each sample and then normalized to that for luciferase control-transfected cells to adjust for interassay variation. The data represent the mean relative infection levels ± SD for two independent experiments performed in triplicate. ***, P < 0.001; ****, P < 0.0001; ns, no significant difference.
Effect of Ifit1 on bunyavirus and filovirus replication in cell culture.
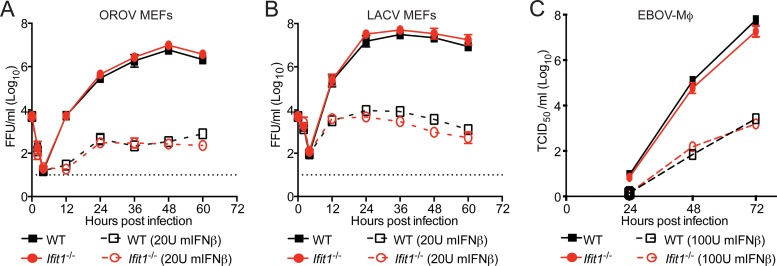
Given the results obtained with IAV, we next assessed whether Ifit1 restricted infection by three other negative-sense RNA viruses, including two bunyaviruses (OROV and LACV) and a filovirus (maEBOV). Bunyaviruses were chosen for study because ectopic expression of IFIT1 reportedly inhibited infection by Rift Valley fever virus (19), a phlebovirus in the Bunyaviridae family. Since bunyavirus mRNA contains a cap 1 structure that is acquired through cap snatching (34, 35), IFIT1 restriction presumably would occur through recognition of the 5′-ppp moiety on the negative-sense genomic strand of viral RNA. We performed a multistep growth analysis with LACV and OROV in WT and Ifit1−/− murine embryonic fibroblasts (MEFs), with and without IFN-β pretreatment (Fig. 4A and B). In the absence of IFN-β treatment, LACV and OROV both replicated to high titers in WT and Ifit1−/− MEFs. As expected, pretreatment of MEFs with IFN-β inhibited replication of LACV and OROV, but we saw no significant increase in replication in Ifit1−/− MEFs, suggesting that in the context of an antiviral type I IFN response, Ifit1 did not have a dominant inhibitory phenotype against these two orthobunyaviruses.
FIG 4.
Murine Ifit1 does not restrict infection by OROV, LACV, and maEBOV. (A and B) Kinetics of OROV (A) and LACV (B) replication in WT and Ifit1−/− MEFs, with or without 18 h of IFN-β (20 U/ml) stimulation prior to infection at an MOI of 0.01. Supernatants were harvested at the indicated times for titration by a focus-forming assay. Two independent experiments were performed, each with seven technical replicates. (C) Kinetics of maEBOV replication in WT and Ifit1−/− Mϕ, with or without 18 h of IFN-β (100 U/ml) stimulation before infection at an MOI of 0.01. Supernatants were harvested at the indicated times for titration by a TCID50 assay. A dotted line represents the limit of detection of the assay. The data represent the means ± SD for two independent experiments performed in triplicate. No statistical differences were observed as judged by the Mann-Whitney test.
As an additional test of the antiviral function of Ifit1 against negative-strand RNA viruses, we assessed its activity against a filovirus, maEBOV. Filoviruses contain a genomic RNA with a 5′-ppp RNA moiety and likely encode viral methyltransferases to generate cap 1 structures on their mRNA (36), although this has not been demonstrated formally (37). We analyzed the multistep growth kinetics of maEBOV in WT and Ifit1−/− bone marrow-derived macrophages (Mϕ) that were left untreated or pretreated with 100 U of murine IFN-β (Fig. 4C). In the absence of IFN-β pretreatment, there was no difference in the growth of maEBOV in WT and Ifit1−/− Mϕ (P > 0.2). Analogously, following IFN-β pretreatment, we observed no difference in the growth of maEBOV in WT and Ifit1−/− Mϕ at any of the time points tested, although IFN-β inhibited infection. Thus, in the context of an antiviral type I IFN response, Ifit1 did not have potent antiviral activity against viruses from three different families of negative-strand RNA viruses, all of which display 5′-ppp on their genomic RNA.
Relative binding of Ifit1 to 5′-OH, capped, and 5′-ppp RNA structures.
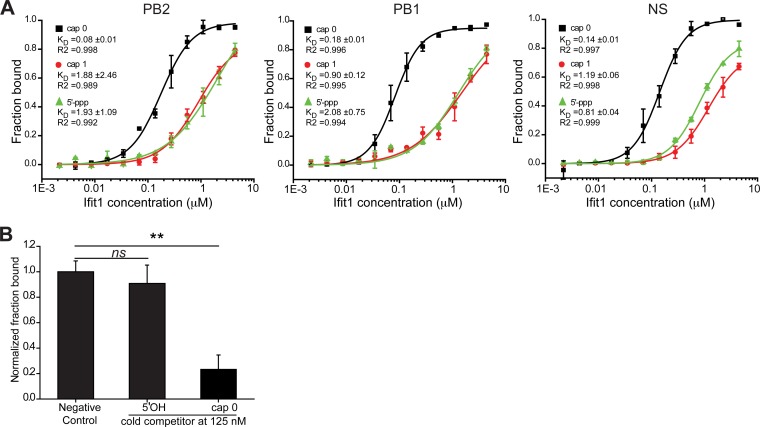
Given the absence of an antiviral effect of Ifit1 against IAV, OROV, LACV, and maEBOV, we considered whether Ifit1 was capable of recognizing the 5′-UTR of IAV. Oligonucleotides representing the first 41 bases of the viral PB2, PB1, and NS gene segments were synthesized and used to generate 5′-OH, cap 0, cap 1, and 5′-ppp RNA structures. Mouse Ifit1 binding to the 5′-UTRs of all three gene segments was detected when they displayed cap 0, cap 1, and 5′-ppp RNA structures (Fig. 5A). However, Ifit1 bound 5′-ppp RNA and cap 1 RNA less avidly than it bound cap 0 RNA (5- to 17-fold lower affinity) (Table 1), as also suggested by another group (16). To validate the specificity of our assay, we performed a competition assay between radiolabeled 5′-ppp RNA and nonradiolabeled cap 0 or 5′-OH RNA of the PB2 gene segment. The cap 0 RNA outcompeted the 5′-ppp RNA (P < 0.02) (Fig. 5B), whereas the 5′-OH RNA did not (P > 0.2). Thus, the lower-affinity binding of Ifit1 to 5′-ppp RNA than to cap 0 RNA may explain the absence of antiviral effects against negative-strand RNA viruses that we observed in cell culture.
FIG 5.
Binding of murine Ifit1 to the 5′-UTR of IAV. (A) A filter-binding assay was used to measure the binding affinity of murine Ifit1 for the viral RNA (vRNA) of influenza A virus (IAV-Cal). Oligonucleotides representing the first 41 nucleotides of the PB2, PB1, and NS vRNAs were modified to contain a cap 0 (black) or cap 1 (red) structure or left untreated (5′-ppp RNA) (green). Dissociation constants (KD) were calculated by using the nonlinear Hill equation, using Origin. The data are means ± SD and are representative of two independent experiments performed in triplicate. (B) Ifit1 has a lower affinity for RNA with 5′-OH than for 5′-ppp and cap 0 RNAs. A nonradiolabeled RNA (125 nM) representing the 5′ end of PB2 of IAV-Cal was modified to contain a cap 0 or 5′-OH structure and added to the Ifit1 protein (0.3 μM) in the presence of radiolabeled 5′-ppp RNA from PB2 (5 nM). The data represent the means ± SD for two independent experiments performed in triplicate. **, P < 0.02; ns, no significant difference.
TABLE 1.
Difference in dissociation constants of murine Ifit1 for RNA oligonucleotides representing influenza virus genomic RNAa
| 5′-End modification | Fold difference in KD of murine Ifit1 for IAV gene segment |
||
|---|---|---|---|
| PB2 | PB1 | NS | |
| cap 0 → cap 1 | 5 | 16 | 7 |
| cap 0 → 5′-ppp | 11 | 17 | 6 |
A filter-binding assay was used to measure the binding affinity of murine Ifit1 for IAV 5′-UTR cap 0, cap 1, and 5′-ppp. The KD (micromolar) was calculated by using the nonlinear Hill equation, using Origin. The data presented are fold differences in KD values between cap 0 and cap 1 or cap 0 and 5′-ppp RNA, calculated from two independently repeated assays.
Effect of Ifit1 on IAV, OROV, and LACV pathogenesis in vivo.
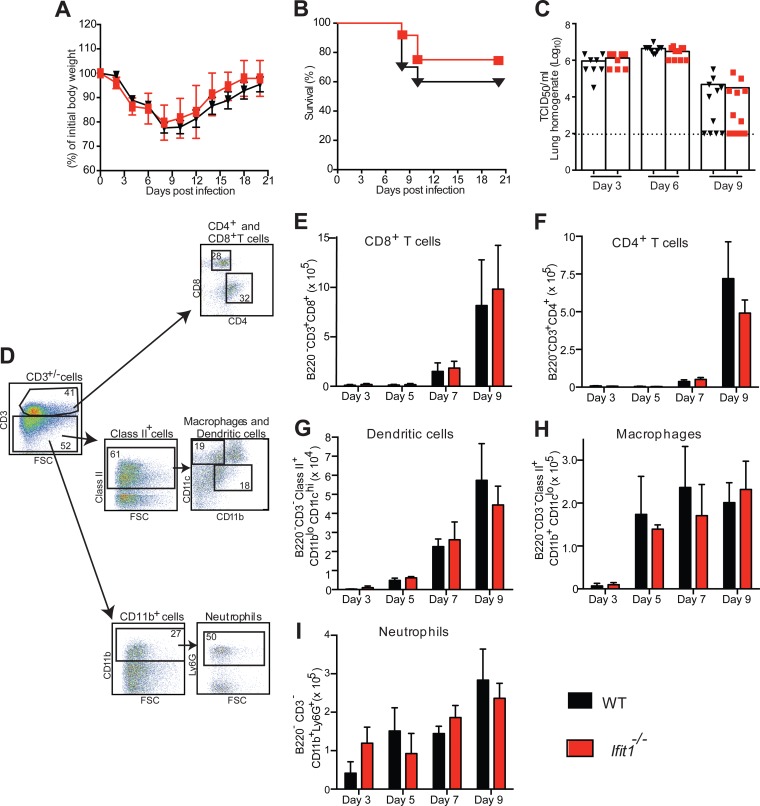
Although an antiviral effect of Ifit1 against several negative-strand RNA viruses was not observed in cell culture, it remained possible that one could exist in vivo. To evaluate this hypothesis, we infected WT and Ifit1−/− mice with IAV, LACV, or OROV and monitored morbidity, mortality, and viral burden. First, we inoculated mice via the intranasal route with 104 EID50 of IAV-Cal (Fig. 6A and B). Weight loss and lethality data between IAV-infected WT and Ifit1−/− mice were not significantly different in magnitude, rate, or duration (P > 0.1). Correspondingly, no difference in viral burden in the lungs was observed on days 3, 6, and 9 after IAV infection of WT and Ifit1−/− mice (P > 0.2) (Fig. 6C). Consistent with the virologic results, cytokine levels in the lungs at these time points revealed no differences between IAV-infected WT and Ifit1−/− mice (Table 2) (P > 0.08). We also observed no differences in the recruitment of dendritic cells, macrophages, neutrophils, or lymphocytes into the lungs on days 3, 5, 7, and 9 after infection (Fig. 6D to I) (P > 0.08).
FIG 6.
Murine Ifit1 does not affect virus infection, pathogenesis, or cellular infiltrates in vivo after infection with IAV. (A and B) Morbidity and survival analyses of 6- to 7-week-old female mice after intranasal inoculation of WT (n = 19) and Ifit1−/− mice (n = 10 for morbidity study and n = 19 for mortality study) with 104 EID50 of IAV-Cal. Data were pooled from four independent experiments. (C) Viral burdens in lung homogenates on days 3, 6, and 9 after IAV infection of WT and Ifit1−/− mice were measured by a TCID50 assay on MDCK cells. A scatterplot of the data is shown, with each individual point representing a single animal (n = 8 to 12). The dashed line represents the limit of detection of the assay. Statistical significance was judged by the Mann-Whitney test. (D) Representative flow plots showing the gating strategy for BAL fluid immune cell analysis. (E to I) BAL fluid cell composition in WT and Ifit1−/− mice after infection with IAV-Cal. Three, 5, 7, and 9 days after inoculation with 104 EID50 of IAV-Cal, BAL was performed, and the cellular composition of the fluid was determined by flow cytometry, using monoclonal antibodies against mouse CD3, CD4, CD8, CD11c, CD11b, Ly6G, MHC class II, and B220. The data shown are the numbers of CD8+ T cells (B220− CD3+ CD8+) (E), CD4+ T cells (B220− CD3+ CD4+) (F), dendritic cells (B220− CD3− MHC class II+ CD11blo CD11chi) (G), macrophages (B220− CD3− MHC class II+ CD11b+ CD11clo) (H), and neutrophils (B220− CD3− CD11b+ Ly6G+) (I). The data were pooled from three experiments, each containing at least three WT and Ifit1−/− mice. No statistical differences were observed as judged by the Mann-Whitney test.
TABLE 2.
Cytokine and chemokine levels in lungs of WT and Ifit1−/− mice after influenza virus infectiona
| Cytokine | Concn (pg/ml) (mean ± SD) in mouse lung |
|||||
|---|---|---|---|---|---|---|
| Day 3 |
Day 6 |
Day 9 |
||||
| WT | Ifit1−/− | WT | Ifit1−/− | WT | Ifit1−/− | |
| IL-1α | 31 ± 3 | 33 ± 5 | 52 ± 15 | 45 ± 3 | 16 ± 3 | 16 ± 3 |
| IL-1β | 319 ± 13 | 326 ± 30 | 787 ± 76 | 852 ± 197 | 214 ± 25 | 227 ± 26 |
| IL-2 | 23 ± 11 | 24 ± 11 | 35 ± 12 | 35 ± 12 | 29 ± 13 | 33 ± 13 |
| IL-3 | 4 ± 10 | 5 ± 10 | 14 ± 12 | 13 ± 11 | 8 ± 12 | 8 ± 10 |
| IL-4 | 4 ± 10 | 5 ± 11 | 36 ± 3 | 37 ± 3 | 23 ± 5 | 22 ± 6 |
| IL-5 | 65 ± 9 | 67 ± 18 | 214 ± 33 | 397 ± 73 | 21 ± 9 | 21 ± 6 |
| IL-6 | 297 ± 32 | 231 ± 37 | 413 ± 40 | 416 ± 178 | 127 ± 7 | 57 ± 4 |
| IL-10 | 21 ± 1 | 23 ± 2 | 43 ± 7 | 67 ± 38 | 85 ± 13 | 82 ± 20 |
| IL-12(p40) | 253 ± 18 | 270 ± 22 | 49 ± 13 | 77 ± 67 | 38 ± 8 | 22 ± 3 |
| IL-12(p70) | 39 ± 4 | 32 ± 2 | 821 ± 76 | 772 ± 51 | 235 ± 49 | 259 ± 45 |
| IL-13 | ND | ND | 79 ± 32 | 92 ± 22 | 90 ± 28 | 74 ± 26 |
| IL-17 | ND | ND | 8 ± 21 | 12 ± 11 | 37 ± 34 | 33 ± 34 |
| Eotaxin | 613 ± 76 | 455 ± 79 | 489 ± 58 | 657 ± 50 | 6 ± 11 | 5 ± 14 |
| G-CSF | 294 ± 41 | 315 ± 67 | 743 ± 70 | 821 ± 89 | 299 ± 63 | 114 ± 23 |
| GM-CSF | 63 ± 31 | 64 ± 4 | 52 ± 41 | 55 ± 21 | 38 ± 53 | 33 ± 21 |
| IFN-γ | 9 ± 21 | 8 ± 11 | 40 ± 11 | 93 ± 11 | ND | ND |
| KC | 3,303 ± 242 | 2,985 ± 588 | 1,635 ± 155 | 1,383 ± 168 | 107 ± 22 | 151 ± 30 |
| CCL2 | 1,272 ± 178 | 1,018 ± 122 | 1,357 ± 464 | 2,216 ± 243 | 698 ± 111 | 474 ± 114 |
| CCL3 | 182 ± 29 | 145 ± 19 | 767 ± 42 | 931 ± 158 | 111 ± 28 | 87 ± 25 |
| CCL4 | 134 ± 18 | 128 ± 24 | 121 ± 12 | 267 ± 39 | 58 ± 11 | 53 ± 8 |
| CCL5 | 395 ± 57 | 284 ± 55 | 513 ± 36 | 515 ± 53 | 120 ± 20 | 152 ± 44 |
| TNF-α | 296 ± 40 | 254 ± 46 | 323 ± 23 | 332 ± 23 | 239 ± 42 | 282 ± 44 |
WT and Ifit1−/− mice (n = 9) were infected with 104 EID50 of IAV-Cal, and 3, 6, or 9 days after infection, lungs were harvested, homogenized, and analyzed by the Bio-Plex Pro 23-plex mouse cytokine and chemokine assay. Data were pooled from three independent experiments. ND, levels were below the threshold of detection. No significant differences (P > 0.08) were observed between WT and Ifit1−/− mice for any of the cytokines or chemokines for any day tested.
We next assessed the effect of Ifit1 on OROV and LACV pathogenesis. We infected WT and Ifit1−/− mice with 105 FFU of LACV (Fig. 7A and B) or 106 FFU of OROV (Fig. 7D and E) and monitored the mice for 21 days. We observed no difference in weight loss or mortality between WT and Ifit1−/− mice for either LACV or OROV (P > 0.4). Analysis of viral RNA levels in the serum, spleen, liver, and brain, on day 8 for LACV (Fig. 7C) and on day 4 for OROV (Fig. 7F), also revealed no significant differences between WT and Ifit1−/− mice (P > 0.07). These experiments indicate that Ifit1 does not have a major role in restricting infection or pathogenesis of several negative-strand RNA viruses.
FIG 7.
Murine Ifit1 does not affect virus infection or pathogenesis in vivo after infection with OROV or LACV. (A and B) Morbidity and survival analyses of 8-week-old WT (n = 15) and Ifit1−/− (n = 13) mice after inoculation with 105 FFU of LACV by subcutaneous injection in the footpad. Data were pooled from three independent experiments. (C) Viral burdens in the liver, spleen, brain, and serum on day 8 after LACV infection of WT and Ifit1−/− mice were measured by quantitative RT-PCR. A scatterplot of the data is shown, with each individual point representing a single animal (n = 10 to 12). The dashed line represents the limit of sensitivity of the assay. Statistical significance was judged by the Mann-Whitney test. (D and E) Morbidity and survival analyses of 6-week-old WT (n = 23) and Ifit1−/− (n = 10) mice after inoculation with 106 FFU of OROV by subcutaneous injection in the footpad. Data were pooled from three independent experiments. (F) Viral burdens in the liver, spleen, brain, and serum on day 4 after OROV infection of WT and Ifit1−/− mice were measured by quantitative RT-PCR. Viral burdens are recorded as per-gram values for organs (left axis) and per-milliliter values for serum (right axis). A scatterplot of the data is shown, with each individual point representing a single animal (n = 10 to 13). The dashed line represents the limit of sensitivity of the assay. No statistical differences were observed as judged by the Mann-Whitney test.
DISCUSSION
To evaluate the possible role of IFIT1 in the recognition and restriction of negative-sense RNA viruses, we performed viral infection studies in cells that were deficient in or ectopically expressed IFIT1, as well as pathogenesis studies in Ifit1−/− mice. Our results revealed no ostensible antiviral effect of Ifit1 on viruses from three different negative-strand RNA virus families, including orthomyxoviruses, bunyaviruses, and filoviruses. These results were unexpected but might be explained by the relatively low binding affinity of Ifit1 for 5′-ppp RNA. Thus, although IFIT1 can bind to 5′-ppp RNA, its relevance for controlling negative-sense RNA viruses may be limited.
The genome of IAV is composed of eight single-stranded, negative-sense RNA gene segments. The 5′ end of each segment displays a triphosphate motif that could be recognized and sequestered by IFIT1 during trafficking from the endosome to the nucleus or from the nucleus to the cell membrane prior to budding of new virus particles. Given that our results suggest that human and murine IFIT1 proteins do not restrict IAV infection, IFIT1 may not bind avidly enough to 5′-ppp RNA to block viral replication within a cellular context. It is possible that the 5′-ppp moiety of IAV segments is not exposed because of masking by the polymerase complex that sits upon each gene segment (38, 39). Alternatively, the 5′ end of each segment may form secondary structures (40, 41) to limit binding of IFIT1 to the genome of IAV. Because IAV has a host mRNA cap-snatching mechanism, it disguises its mRNA as host RNA, which is less susceptible to IFIT1-mediated restriction. Finally, IAV replicates in the nucleus, evading the possible detection of its complementary, positive-sense RNA by IFIT1, which resides in the cytoplasm.
Bunyaviruses, including LACV and OROV, also have a segmented genome, with each segment displaying a 5′-ppp moiety. Unlike IAV, bunyaviruses replicate in the cytoplasm, which could facilitate possible interactions between IFIT1 and 5′-ppp RNA. Nonetheless, LACV and OROV were not inhibited by Ifit1 in vitro or in vivo. Similar to IAV, LACV and OROV also have a cap-snatching mechanism (34, 35), which likely contributes to evasion of IFIT1 restriction. Although the relatively low-affinity binding of 5′-ppp RNA likely limits IFIT1-mediated sequestration of LACV and OROV RNAs, it is possible that bunyaviruses have evolved alternate evasion mechanisms, including subcellular localization, actions of viral proteins, or the formation of secondary structure elements at the 5′ end of the genome. As part of our analysis, we also tested the role of Ifit1 in a mouse-adapted strain of EBOV, a negative-sense RNA virus that replicates in the cytoplasm. Unlike IAV, LACV, and OROV, the maEBOV genome encodes a polymerase protein with a postulated capping activity. Therefore, only the 5′-ppp moiety at the end of the genomic negative-sense RNA and the complementary, positive-sense RNA are likely targets for Ifit1 binding and restriction (42). We did not find any evidence for Ifit1-mediated restriction of maEBOV infection. Thus, despite its ability to bind 5′-ppp viral RNA, our data suggest that IFIT1 does not efficiently inhibit RNA viruses with a negative genome polarity.
A lack of IFIT1-mediated restriction of negative-sense RNA viruses is consistent with published data showing no effect of murine Ifit1 on VSV replication and pathogenesis (17). Our data contrast with those from studies showing antiviral activity of the IFIT1/2/3 protein complex on VSV infection and, to a lesser extent, on IAV replication (18, 19). This study did not include in vivo data for IAV, making a direct comparison challenging. It remains possible that ectopic expression of IFIT1 at very high levels overcomes the lower-affinity binding to 5′-ppp RNA and inhibits infection of some negative-sense RNA viruses. Alternatively, high-level expression of IFIT1 could have antiviral activity more generally, through its reported effects on eIF3 binding and inhibition of protein translation (5–7). Finally, it is possible that IFIT1 expression or activity toward negative-sense RNA viruses is greater in HeLa cells (19) than in the A549 cells or mTECs used in our study.
Although in our experiments IFIT1 did not have a major impact on infection by negative-strand RNA viruses, several groups have reported significant inhibitory effects on positive-strand RNA (flaviviruses, alphaviruses, and coronaviruses) and DNA (poxvirus) viruses lacking 2′-O methylation of their viral RNAs (10–15, 17, 43, 44). Moreover, we and others have measured a higher affinity of IFIT1 for RNA containing cap 0 than for cap 1 or 5′-ppp structures (11, 15–17). Consistent with these observations, flavivirus, alphavirus, or coronavirus mutants lacking 2′-O methylation of their viral RNAs replicate to higher levels and cause greater pathogenesis in Ifit1−/− mice and are attenuated in WT mice.
It is possible that other members of the human and mouse IFIT gene family restrict infection and pathogenesis by IAV, LACV, OROV, and EBOV. For example, murine Ifit2 inhibited the replication of several positive- and negative-sense RNA viruses in specific tissues, including the brain and lungs (20, 45). Future studies are needed to determine if human and mouse IFIT2 proteins restrict IAV, LACV, OROV, and EBOV. Although IFIT3 has been implicated in innate immune signaling (46), determination of its precise role in viral pathogenesis and innate immunity awaits the generation of Ifit3−/− mice and cell lines. Our study shows that human and murine IFIT1 proteins do not efficiently restrict infection by negative-sense RNA viruses from multiple families. This may be related to the relatively low affinity of IFIT1 proteins for the 5′-ppp moiety present in negative-sense RNA viruses.
ACKNOWLEDGMENTS
This work was supported in part by grant U54 AI057160 to A.C.M.B., by grants R01 AI104972 and R01 AI104002 to M.S.D., and by the Division of Intramural Research, NIAID, NIH, to H.E. G.D.W. is supported by training grant GM: 007067. J.P.W. is supported by an F32 AI112274 NIH grant.
We thank S. Brody and J. Xu for assistance with growing murine tracheal epithelial cells. Finally, we are thankful for the technical assistance of Brittany DesRochers, Jennifer Govero, and Michelle Noll.
REFERENCES
- 1.Fensterl V, Sen GC. 2011. The ISG56/IFIT1 gene family. J Interferon Cytokine Res 31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou X, Michal JJ, Zhang L, Ding B, Lunney JK, Liu B, Jiang Z. 2013. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci 9:200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Zhang YB, Liu TK, Gui JF. 2013. Lineage-specific expansion of IFIT gene family: an insight into coevolution with IFN gene family. PLoS One 8:e66859. doi: 10.1371/journal.pone.0066859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J, Hui DJ, Merrick WC, Sen GC. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J 19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui DJ, Terenzi F, Merrick WC, Sen GC. 2005. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J Biol Chem 280:3433–3440. doi: 10.1074/jbc.M406700200. [DOI] [PubMed] [Google Scholar]
- 6.Hui DJ, Bhasker CR, Merrick WC, Sen GC. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J Biol Chem 278:39477–39482. doi: 10.1074/jbc.M305038200. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Pflugheber J, Sumpter R Jr, Sodora DL, Hui D, Sen GC, Gale M Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol 77:3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saikia P, Fensterl V, Sen GC. 2010. The inhibitory action of P56 on select functions of E1 mediates interferon's effect on human papillomavirus DNA replication. J Virol 84:13036–13039. doi: 10.1128/JVI.01194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terenzi F, Saikia P, Sen GC. 2008. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J 27:3311–3321. doi: 10.1038/emboj.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura T, Katoh H, Kayama H, Saiga H, Okuyama M, Okamoto T, Umemoto E, Matsuura Y, Yamamoto M, Takeda K. 2013. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′-capped 2′-O unmethylated RNA. J Virol 87:9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SH, Dong H, Li XF, Xie X, Zhao H, Deng YQ, Wang XY, Ye Q, Zhu SY, Wang HJ, Zhang B, Leng QB, Zuest R, Qin ED, Qin CF, Shi PY. 2013. Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J Virol 87:5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M Jr, Virgin HW, Klein RS, Sen GC, Diamond MS. 2012. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog 8:e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zust R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, Toh YX, Jiang T, Li SH, Deng YQ, Ellis BR, Ellis EM, Poidinger M, Zolezzi F, Qin CF, Shi PY, Fink K. 2013. Rational design of a live attenuated dengue vaccine: 2′-O-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog 9:e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil-Cruz C, Ziebuhr J, Thiel V, Pichlmair A. 2013. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog 9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Sweeney TR, Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. 2014. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res 42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C, Tonelli M, Paessler S, Takeda K, Klimstra WB, Amarasinghe GK, Diamond MS. 2014. A viral RNA structural element alters host recognition of nonself RNA. Science 343:783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbas YM, Pichlmair A, Gorna MW, Superti-Furga G, Nagar B. 2013. Structural basis for viral 5′-ppp-RNA recognition by human IFIT proteins. Nature 494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol 12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 20.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog 8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyde JL, Diamond MS. 2015. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 479–480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale M Jr, Diamond MS. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol 80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beutner C, Roy K, Linnartz B, Napoli I, Neumann H. 2010. Generation of microglial cells from mouse embryonic stem cells. Nat Protoc 5:1481–1494. doi: 10.1038/nprot.2010.90. [DOI] [PubMed] [Google Scholar]
- 25.You Y, Richer EJ, Huang T, Brody SL. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- 26.Boon AC, deBeauchamp J, Krauss S, Rubrum A, Webb AD, Webster RG, McElhaney J, Webby RJ. 2010. Cross-reactive neutralizing antibodies directed against pandemic H1N1 2009 virus are protective in a highly sensitive DBA/2 mouse influenza model. J Virol 84:7662–7667. doi: 10.1128/JVI.02444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J Virol 81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. 1998. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 178:651–661. doi: 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- 29.Proenca-Modena JL, Sesti-Costa R, Pinto AK, Richner JM, Lazear HM, Lucas T, Hyde JL, Diamond MS. 2015. Oropouche virus infection and pathogenesis are restricted by MAVS, IRF-3, IRF-7, and type I interferon signaling pathways in nonmyeloid cells. J Virol 89:4720–4737. doi: 10.1128/JVI.00077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med 11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You Y, Brody SL. 2013. Culture and differentiation of mouse tracheal epithelial cells. Methods Mol Biol 945:123–143. doi: 10.1007/978-1-62703-125-7_9. [DOI] [PubMed] [Google Scholar]
- 32.Fensterl V, Sen GC. 2015. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol 89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson JL, Holloway B, Kolakofsky D. 1984. La Crosse virions contain a primer-stimulated RNA polymerase and a methylated cap-dependent endonuclease. J Virol 52:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson JL, Kolakofsky D. 1984. Characterization of La Crosse virus small-genome transcripts. J Virol 49:680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weik M, Modrof J, Klenk HD, Becker S, Muhlberger E. 2002. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J Virol 76:8532–8539. doi: 10.1128/JVI.76.17.8532-8539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Murti KG, Webster RG, Jones IM. 1988. Localization of RNA polymerases on influenza viral ribonucleoproteins by immunogold labeling. Virology 164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 39.Schreier E, Ladhoff AM, Stompor S, Michel S. 1988. Interaction between anti-influenza viral polymerase antibodies and RNP particles using the in vitro transcription process and an immunogold labelling technique. Acta Virol 32:403–408. [PubMed] [Google Scholar]
- 40.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A 84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 42.Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, Mirazimi A, Weber F. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menachery VD, Yount BL Jr, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho H, Shrestha B, Sen GC, Diamond MS. 2013. A role for Ifit2 in restricting West Nile virus infection in the brain. J Virol 87:8363–8371. doi: 10.1128/JVI.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]