Abstract
Influenza B virus causes significant disease but remains understudied in tropical regions. We sequenced 72 influenza B viruses collected in Kuala Lumpur, Malaysia, from 1995 to 2008. The predominant circulating lineage (Victoria or Yamagata) changed every 1 to 3 years, and these shifts were associated with increased incidence of influenza B. We also found poor lineage matches with recommended influenza virus vaccine strains. While most influenza B virus lineages in Malaysia were short-lived, one circulated for 3 to 4 years.
TEXT
Influenza B virus causes seasonal epidemics and a significant disease burden in humans, with reported rates of up to 82% of total influenza cases around the world and 22 to 44% of influenza-related child deaths in the United States (1). Since the mid-1970s, influenza B virus has formed two antigenically distinct lineages, “B/Victoria/2/87-like” and “B/Yamagata/16/88-like,” referred to as the Victoria and Yamagata lineages, respectively (2). Although the two lineages cocirculate in humans, Vijaykrishna et al. (3) recently demonstrated significant evolutionary and epidemiological differences between the two lineages. The Victoria lineage is subject to stronger seasonal bottlenecks, higher transmission rates, greater antigenic variation, and stronger positive selection and infects mostly younger age groups. In contrast, the Yamagata lineage experiences less severe bottlenecks and infects older people (3). In tropical Kuala Lumpur in Malaysia, there is year-round influenza activity, with biannual epidemics in May to July and November to January (4). A retrospective epidemiological study of hospitalized children in Kuala Lumpur from 1982 to 2008 found that 297 of 2,708 (11%) patients with confirmed respiratory virus infection were positive for influenza virus, making it the third most common respiratory virus affecting children aged <5 years, with higher rates of infection in children aged 1 to 5 years than in children under 12 months old (5). While epidemiological data in Malaysia have been well documented (4–7), no genetic study has been conducted on influenza B virus from Malaysia.
We obtained a total of 338 laboratory-confirmed influenza cases from children and adults (1 month to 49 years old) admitted to the University Malaya Medical Centre in Kuala Lumpur during 1995 to 2008. All patients resided in Kuala Lumpur or the surrounding conurbation. Of these, 88 cases (26.0%) were diagnosed with influenza B viruses, whereas 250 cases (74.0%) were infected with influenza A viruses. Isolates of influenza B virus were cultured in Madin-Darby canine kidney (MDCK) cells and passaged up to three times prior to extraction of viral RNA as described previously (8). Complete genomes of 72 influenza B viruses and partial genomes of 5 influenza B viruses were sequenced (8–10) and deposited in GenBank (see Table S1 in the supplemental material). These sequences represent all full-length hemagglutinin (HA) or complete genome sequences from Malaysia (from 1995 to 2008) available in public databases at the time of writing. Phylogenetic analyses were performed for all eight gene segments (PB2, PB1, PA, HA, NP, NA, MP, and NS) using a combination of Malaysian and global sequences (see Table S1). Temporal phylogenies were reconstructed using the coalescent-based Gaussian Markov random field (GMRF) method with the time-aware smoothing parameter (11) in BEAST v.1.8.1 (12). For all analyses, the uncorrelated log-normal relaxed molecular clock and the SRD06 codon position model (13) with the HKY85+Γ substitution model were used. Three independent analyses of 100 million generations, with sampling every 10,000 generations, were performed.
Our study indicates that 75.4% of positive influenza B cases were in young children aged <5 years, with a median age of 2.0 years (see Table S2 in the supplemental material). The incidence of influenza B virus infection in our study group was generally lower than that of influenza A virus infection; however, a higher incidence of influenza B virus (>30% of influenza cases) was observed in 2000, 2004, 2005, and 2007 (Fig. 1A), coinciding with shifts in the predominant lineage. In most years, only a single influenza B virus lineage was detected, with Yamagata viruses detected exclusively in 6 of the 14 study years, while only Victoria strains were detected in just 3 years (Fig. 1B). Cocirculation of both lineages was detected only in 2004 and 2007 in our study, in contrast to a recent study that showed cocirculation in Australia and New Zealand in most years (3). Although our study obtained a small number of sequences in most years, with no influenza virus detected in three of the years, the predominant influenza B virus lineages circulating in Malaysia as a whole from 2005 to 2008 (6), identified by hemagglutination inhibition, were similar to our data from Kuala Lumpur.
FIG 1.

Influenza virus activity in Kuala Lumpur, Malaysia, between 1995 and 2008. (A) The numbers of laboratory-confirmed human cases of influenza A and B viruses are shown for each year. The vertical bars represent the yearly proportion (percentage) of influenza B virus cases against the total number of positive influenza virus cases recorded. (B) Bars represent the prevalence (number of cases) of Victoria and Yamagata lineages. Cases of undetermined lineage refer to samples that tested positive for influenza B virus by immunofluorescence but were not successfully cultured for sequencing.
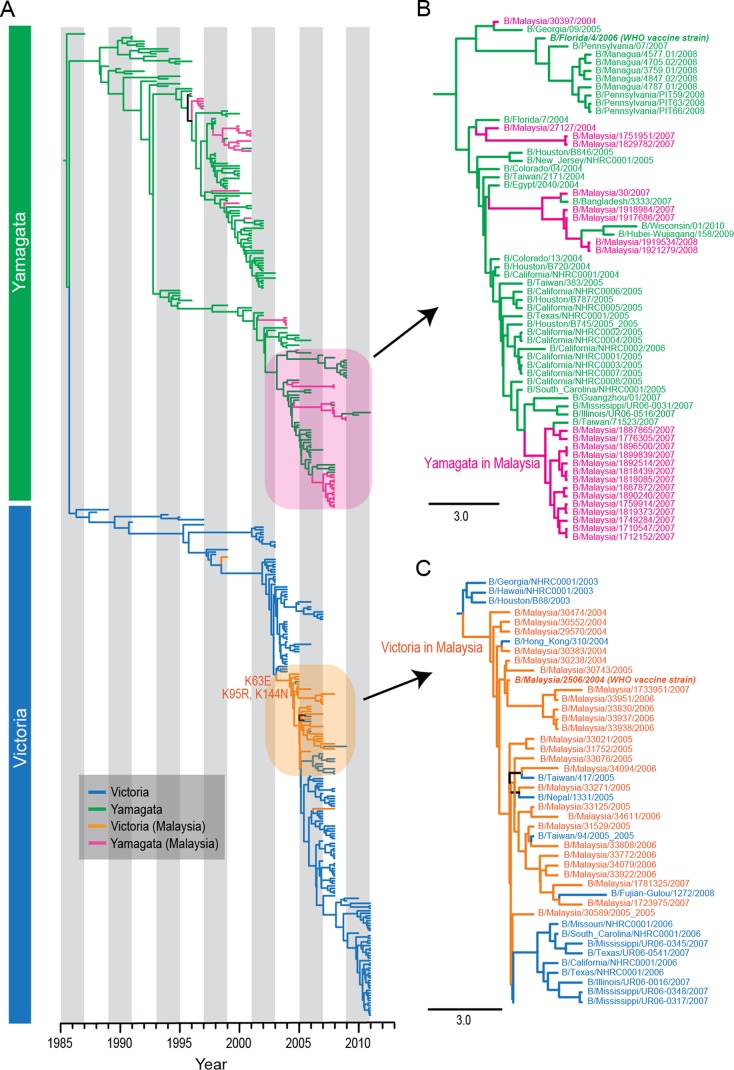
The HA phylogeny indicated that there had been multiple independent introductions of the two influenza B virus lineages into Malaysia that occurred sporadically throughout the study period (Fig. 2A). The markedly greater influenza B virus activity observed in 2007 (Fig. 1) was largely due to multiple introductions of Yamagata viruses (denoted by red branches in Fig. 2A), including a monophyletic clade that may indicate that a point source outbreak occurred in Kuala Lumpur (Fig. 2B). Genetic analysis also showed that the Victoria lineage viruses, which we found were more prevalent from 2004 to 2007 than in previous years (Fig. 1B), were B/Malaysia/2506/2004-like viruses (Fig. 2A). This strain was the WHO-recommended influenza B virus vaccine component in the Northern and Southern Hemispheres from 2006 to 2007 (14). These viruses appear to have circulated in Malaysia continuously from 2004 to 2007, and the Malaysian strains fall in a position basal to all other B/Malaysia/2506/2004-like viruses (Fig. 2C). Mapping of amino acid substitutions at the node leading to the B/Malaysia/2506/2004-like viruses showed two HA mutations, K95R and K144N (Fig. 2C). Residue 144 is a known antigenic site (15), and the K144N mutation may have led to antigenic drift of these viruses. Interestingly, these basal B/Malaysia/2506/2004-like viruses were isolated from pediatric patients aged 2 to 10 months old (see Table S2 in the supplemental material), although this may be a sampling artifact due to the undersampling of adult and elderly patients in our study. Given that B/Malaysia/2506/2004-like viruses made up 23/25 (92%) influenza B viruses collected between 2004 and 2006 from our study (see Fig. S4 in the supplemental material) and over half of the 165 influenza B viruses collected in 2005 and 2006 from around Malaysia (6), we speculate that these B/Malaysia/2506/2004-like viruses may have caused an epidemic in Malaysia before spreading to other countries.
FIG 2.
(A) Phylogeny of the HA gene of influenza B viruses, stratified by date. Victoria and Yamagata lineages are represented by blue and green branches, respectively, whereas Malaysian isolates of the Victoria and Yamagata lineages are represented by orange and pink branches, respectively. (B) Magnification of Yamagata lineages shown in the pink box in panel A. (C) Magnification of Victoria lineages shown in the orange box in panel A. The virus strain B/Malaysia/2506/2004 was previously selected as a WHO vaccine strain. The horizontal scale bar represents the number of substitutions per site.
Cocirculation of the two lineages in humans may lead to reassortment between Yamagata and Victoria viruses (3, 16). While the NP and MP phylogenies were consistent with the HA tree, phylogenies of the other internal genes showed that 6 of the Malaysian strains were reassortants (see arrows in Fig. S1 to S8 in the supplemental material). Despite frequent interlineage reassortments, the distinction between the PB1–PB2–HA gene complexes of the Yamagata and Victoria lineages was maintained (16).
The World Health Organization recommends trivalent influenza virus vaccine composition on a semiannual basis. Influenza virus vaccination control in Malaysia follows the Southern Hemisphere recommendations (6), but we found mismatches in 3 of 8 years (37.5% mismatch rate) for which sequence data were available in Kuala Lumpur (Fig. 1B), compared with global vaccine mismatch rates of 38 to 54% for influenza B virus (17, 18). However, influenza virus vaccination rates are low in Southeast Asian countries, including Malaysia (19). If increased vaccine use were to be considered in Malaysia, influenza B vaccine mismatch rates could be improved either by considering rapid adoption of the Northern and Southern Hemisphere recommended formulations or by using the quadrivalent vaccine that includes influenza B virus strains from both lineages.
Malaysia's influenza surveillance system is based on two major components, disease-based and laboratory-based surveillance. Disease-based surveillance collects influenza-like illness data from sentinel sites, i.e., health clinics and general practices, but these data are not publicly available. Our data from Kuala Lumpur consist of laboratory-confirmed influenza, and we show that influenza B virus accounts for 26% of all influenza cases, suggesting a considerable national burden of disease, which is consistent with previously reported data indicating that influenza B virus accounted for 9 to 48% of confirmed influenza cases in Malaysia from 2006 to 2014 (4, 6, 20). Our study also showed an overwhelming burden of influenza B virus cases in young children, consistent with previous studies globally (3, 21–24).
Active coordination of surveillance efforts in sentinel clinics and hospitals, including the standardization of patient and epidemiological data collected, is needed to effectively monitor influenza disease dynamics in Malaysia. This study highlights the necessity of establishing systematic and ongoing disease surveillance programs in Malaysia in order to provide accurate information on circulating influenza virus strains that may be used to improve vaccination strategies in the country.
Nucleotide sequence accession numbers.
The sequences generated in this study were deposited in GenBank under accession numbers CY118283 to CY118417, CY119546 to CY120017, and CY120029 to CY120046.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by contracts HHSN272200900007C and HHSN272201400006C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, and University Malaya (High Impact Research grant number UM.C/625/1/HIR/MOHE/MED/42).
The data for preparation of the manuscript were generated while D.E.W. was employed at the J. Craig Venter Institute. The opinions expressed in this article are the authors' own and do not reflect the views of the Centers for Disease Control, the Department of Health and Human Services, or the United States government.
I.-C.S. has received research funding and consultancy fees from Sanofi-Pasteur.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00708-15.
REFERENCES
- 1.Glezen WP, Schmier JK, Kuehn CM, Ryan KJ, Oxford J. 2013. The burden of influenza B: a structured literature review. Am J Public Health 103:e43–e51. doi: 10.2105/AJPH.2012.301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R, Holmes EC. 2008. The evolutionary dynamics of human influenza B virus. J Mol Evol 66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijaykrishna D, Holmes EC, Joseph U, Fourment M, Su YC, Halpin R, Lee RT, Deng YM, Gunalan V, Lin X, Stockwell TB, Fedorova NB, Zhou B, Spirason N, Kühnert D, Boskova V, Stadler T, Costa AM, Dwyer DE, Huang QS, Jennings LC, Rawlinson W, Sullivan SG, Hurt AC, Maurer-Stroh S, Wentworth DE, Smith GJ, Barr I. 2015. The contrasting phylodynamics of human influenza B viruses. eLife 4:e05055. doi: 10.7554/eLife.05055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sam IC, Abdul-Murad A, Karunakaran R, Rampal S, Chan YF, Nathan AM, Ariffin H. 2010. Clinical features of Malaysian children hospitalized with community-acquired seasonal influenza. Int J Infect Dis 14(Suppl 3):e36–e40. doi: 10.1016/j.ijid.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Khor CS, Sam IC, Hooi PS, Quek KF, Chan YF. 2012. Epidemiology and seasonality of respiratory viral infections in hospitalized children in Kuala Lumpur, Malaysia: a retrospective study of 27 years. BMC Pediatr 12:32. doi: 10.1186/1471-2431-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saat Z, Rashid TRTA, Yusof MA, Kassim FM, Thayan R, Kuen KS, Othman KA, Saraswathy TS. 2010. Seasonal influenza virus strains circulating in Malaysia from 2005 to 2009. Southeast Asia J Trop Med Public Health 41:1368–1373. http://www.tm.mahidol.ac.th/seameo/2010-41-6/14-4858.pdf. [PubMed] [Google Scholar]
- 7.Sam IC, Shaw R, Chan YF, Hooi PS, Hurt AC, Barr IG. 2013. Seroprevalence of seasonal and pandemic influenza A in Kuala Lumpur, Malaysia in 2008-2010. J Med Virol 85:1420–1425. doi: 10.1002/jmv.23622. [DOI] [PubMed] [Google Scholar]
- 8.Li K, Brownley A, Stockwell TB, Beeson K, McIntosh TC, Busam D, Ferriera S, Murphy S, Levy S. 2008. Novel computational methods for increasing PCR primer design effectiveness in directed sequencing. BMC Bioinformatics 9:191. doi: 10.1186/1471-2105-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, Dernovoy D, Tatusova T, Bao Y, St George K, Taylor J, Lipman DJ, Fraser CM, Taubenberger JK, Salzberg SL. 2005. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza a viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. 2006. Recommendations for influenza vaccine composition. Southern hemisphere: 2006. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/vaccines/vaccinerecommendations1/en/index8.html. [Google Scholar]
- 15.Eshaghi A, Duvvuri VR, Li A, Patel SN, Bastien N, Li Y, Low D, Gubbay JB. 2014. Genetic characterization of seasonal influenza A (H3N2) viruses in Ontario during 2010-2011 influenza season: high prevalence of mutations at antigenic sites. Influenza Other Respir Viruses 8:250–257. doi: 10.1111/irv.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudas G, Bedford T, Lycett S, Rambaut A. 2015. Reassortment between influenza B lineages and the emergence of a coadapted PB1–PB2–HA gene complex. Mol Biol Evol 32:162–172. doi: 10.1093/molbev/msu287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard SA, Viboud C, Miller MA. 2010. Evaluation of Southern Hemisphere influenza vaccine recommendations. Vaccine 28:2693–2699. doi: 10.1016/j.vaccine.2010.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heikkinen T, Ikonen N, Ziegler T. 2014. Impact of influenza B lineage-level mismatch between trivalent seasonal influenza vaccines and circulating viruses, 1999-2012. Clin Infect Dis 59:1519–1524. doi: 10.1093/cid/ciu664. [DOI] [PubMed] [Google Scholar]
- 19.Gupta V, Dawood FS, Muangchana C, Lan PT, Xeuatvongsa A, Sovann L, Olveda R, Cutter J, Oo KY, Ratih TS, Kheong CC, Kapella BK, Kitsutani P, Corwin A, Olsen SJ. 2012. Influenza vaccination guidelines and vaccine sales in Southeast Asia: 2008-2011. PLoS One 7:e52842. doi: 10.1371/journal.pone.0052842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. 15 Jun 2015. World Health Organization global health atlas. World Health Organization, Geneva, Switzerland: http://apps.who.int/globalatlas/dataQuery/default.asp. [Google Scholar]
- 21.Yu H, Huang J, Huai Yang Guan X, Klena J, Liu S, Peng Y, Yang H, Luo J, Zheng J, Chen M, Peng Z, Xiang N, Huo X, Xiao L, Jiang H, Chen H, Zhang Y, Xing X, Xu Z, Feng Z, Zhan F, Yang W, Uyeki TM, Wang Y, Varma JK. 2014. The substantial hospitalization burden of influenza in central China: surveillance of severe, acute respiratory infection, and influenza viruses, 2010-2012. Influenza Other Respir Viruses 8:53–65. doi: 10.1111/irv.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cromer D, van Hoek AJ, Jit M, Edmunds WJ, Fleming D, Miller E. 2014. The burden of influenza in England by age and clinical risk group: a statistical analysis to inform vaccine policy. J Infect 68:363–371. doi: 10.1016/j.jinf.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Silva ML, Perrier L, Späth H-M, Grog I, Mosnier A, Havet N, Cohen JM, IBGP Team. 2014. Economic burden of seasonal influenza B in France during winter 2010-2011. BMC Public Health 14:56. doi: 10.1186/1471-2458-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvala H, Smith D, Salvatierra K, Gunson R, von Wissmann B, Reynolds A, Frew C, MacLean A, Hunt A, Yirrell D, Simmonds P, McMenamin J, Templeton K. 2014. Burden of influenza B virus infections in Scotland in 2012/13 and epidemiological investigations between 2000 and 2012. Euro Surveill 19:pii=20903 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.