Abstract
A novel Streptomyces, strain MUSC 149T was isolated from mangrove soil. A polyphasic approach was used to study the taxonomy of MUSC 149T, which shows a range of phylogenetic and chemotaxonomic properties consistent with those of the members of the genus Streptomyces. The diamino acid of the cell wall peptidoglycan was LL-diaminopimelic acid. The predominant menaquinones were identified as MK9(H8) and MK9(H6). Phylogenetic analysis indicated that closely related strains include Streptomyces rhizophilus NBRC 108885T (99.2% sequence similarity), S. gramineus NBRC 107863T (98.7%) and S. graminisoli NBRC 108883T (98.5%). The DNA–DNA relatedness values between MUSC 149T and closely related type strains ranged from 12.4 ± 3.3% to 27.3 ± 1.9%. The DNA G + C content was determined to be 72.7 mol%. The extract of MUSC 149T exhibited strong antioxidant activity and chemical analysis reported identification of an antioxidant agent, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-. These data showed that metabolites of MUSC 149T shall be useful as preventive agent against free-radical associated diseases. Based on the polyphasic study of MUSC 149T, the strain merits assignment to a novel species, for which the name S. mangrovisoli sp. nov. is proposed. The type strain is MUSC 149T (=MCCC 1K00699T=DSM 100438T).
Keywords: Streptomyces mangrovisoli, novel taxa, antioxidant, DPPH, mangrove
Introduction
Oxidative stress has been implicated in physiological aging which may contribute to the development of chronic diseases. The disequilibrium of oxidation status has been associated with development of neurodegenerative diseases which includes Parkinson’s disease and Alzheimer’s disease (Floyd and Hensley, 2002; Farooqui and Farooqui, 2009). In fact, oxidative stress is recognized to play a critical role in carcinogenesis as well. It is plausible that the accumulation of free radicals results in various modifications or damages to biological macromolecules such as protein, lipid, and DNA (Reuter et al., 2010). These unwanted, harmful effects then expedite DNA mutation and increase cancer risks. Therefore, the discovery of the antioxidants from natural resources has always sparked great interest of researchers (Lee et al., 1998).
The mangrove is an exclusive woody plant area of intertidal coasts in tropical and subtropical coastal regions. This ecosystem is among the world’s most prolific environments and produces commercial forest products, protects coastlines and supports coastal fisheries. Mangrove ecosystems are habitats of various flora and fauna of marine, freshwater and terrestrial species (Jennerjahn and Ittekkot, 2002). Recently, there has been increasing interest in exploitation of mangrove microorganism resources as the constant changes in factors such as salinity and tidal gradient in the mangrove ecosystems are consideration to be driving forces for metabolic pathway adaptations that could direct to the production of valuable metabolites (Hong et al., 2009; Lee et al., 2014a). Lately, numerous studies have discovered novel actinobacteria from the different mangrove environments globally, such as the isolation of Streptomyces avicenniae (Xiao et al., 2009), S. xiamenensis (Xu et al., 2009), S. sanyensis (Sui et al., 2011), S. qinglanensis (Hu et al., 2012), S. pluripotens (Lee et al., 2014b), and S. gilvigriseus (Ser et al., 2015).
Waksman and Henrici (1943) had proposed the genus Streptomyces; the genus Streptomyces is comprised of ca. 600 species with validly published names (http://www.bacterio.cict.fr/) at the time of writing (May 2015). Many members of this genus have made vital contributions to mankind due to their capabilities to produce various natural products (Berdy, 2005). These Streptomyces-derived secondary metabolites have attracted much attention from the community as they possess diverse bioactivities such as antibacterial, antifungal, antitumor, and antioxidant (Kaneko et al., 1989; Kim et al., 2008; Olano et al., 2009a; Saurav and Kannabiran, 2012; Thenmozhi and Kannabiran, 2012; Wang et al., 2013; Kumar et al., 2014; Khieu et al., 2015). Notably, some of the bioactivities described were associated with production of cyclic compounds such as cyclomarins and pyrrolizidines (Renner et al., 1999; Karanja et al., 2010; Fu and MacMillan, 2015).
In this study, this particular strain of Streptomyces was isolated from a mangrove soil located from the Tanjung Lumpur mangrove forest located in east coast of Peninsular Malaysia. With the polyphasic approach, it is revealed that MUSC 149T represents a novel species of the Streptomyces genus, for which the name S. mangrovisoli sp. nov. is proposed. In our very initial attempt to explore the potential biological activity possessed by MUSC149T, antioxidant activity was examined. The result indicated that MUSC149T extract exhibited a significant antioxidant property. To the best of our knowledge, the antioxidant activity of MUSC149T has hitherto not been reported. The chemical analysis was then conducted to identify the chemical constituents present in the extract of MUSC149T. The outcomes derived from this research have provided a strong foundation for further in depth biological studies to be performed particularly focusing on free-radical associated diseases.
Materials and Methods
Isolation and Maintenance of Isolate
Strain MUSC 149T was isolated from a soil sample collected at site MUSC-TLS1 (3° 48′ 3.2′′ N 103° 20′ 11.0′′ E), located in the mangrove forest of Tanjung Lumpur in the state of Pahang, Peninsular Malaysia, in December 2012. Topsoil samples of the upper 20-cm layer (after removing the top 2–3 cm) were collected and sampled into sterile plastic bags using an aseptic metal trowel, and stored at –20°C. Air-dried soil samples were ground with a mortar and pestle. Selective pretreatment of soil samples was performed using wet heat in sterilized water (15 min at 50°C; Takahashi et al., 1996). Five grams of the pretreated air-dried soil was mixed with 45 ml sterilized water and mill ground, spread onto the isolation medium ISP 2 (Shirling and Gottlieb, 1966) supplemented with cycloheximide (25 μg ml-1) and nystatin (10 μg ml-1), and incubated at 28°C for 14 days. Pure cultures of strain MUSC 149T were isolated and maintained on slants of ISP 2 agar at 28°C and as glycerol suspensions (20%, v/v) at –20°C for long term preservation.
Genomic and Phylogenetic Analyses
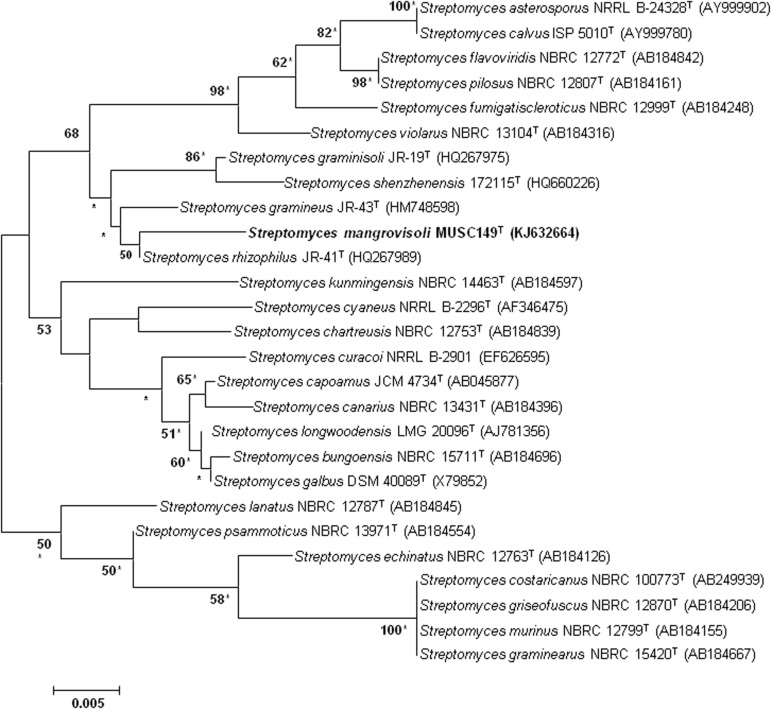
The extraction of genomic DNA for PCR was performed as described by Hong et al. (2009). In short, approximately 0.5 g of each culture was suspended in TE buffer (0.5 ml) and ribolised for 30 s at a speed of 5.5 m/s following the addition of sterile glass beads (0.5 g, 100 mesh). The resultant preparations were extracted with an equal volume of chloroform: iso-amyl alcohol (24:1, v/v) and centrifuged at 15,000 g for 5 min at 4°C. The upper aqueous layers, which contained the DNA, were transferred to fresh tubes and used as template DNA. The amplification of 16S rRNA gene was performed according to Lee et al. (2014b). Briefly the PCR reactions were performed in a final volume of 50 μl according to protocol of SolGentTM 2X Taq PLUS PCR Smart mix using the Kyratex PCR Supercycler (Kyratec, Australia) with the following cycling conditions: (i) 95°C for 5 min, (ii) 35 cycles of 94°C for 50 s, 55°C for 1 min and 72°C for 1 min 30 s; and (iii) 72°C for 8 min. The 16S rRNA gene sequence of strain MUSC 149T was aligned with representative sequences of related type strains of the genus Streptomyces retrieved from the GenBank/EMBL/DDBJ databases using CLUSTAL-X software (Thompson et al., 1997). The alignment was verified manually and then used to generate phylogenetic tree. Phylogenetic trees were constructed with the maximum-likelihood (Felsenstein, 1981) (Figure 1) and neighbor-joining (Saitou and Nei, 1987) (Supplementary Figure S1) algorithms using MEGA version 5.2 (Tamura et al., 2011). Evolutionary distances for the neighbor-joining algorithm were computed using Kimura’s two-parameter model (Kimura, 1980). The EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/; Kim et al., 2012) was used for calculations of sequence similarity. The stability of the resultant trees topologies were evaluated by using the bootstrap based on 1000 resampling method of Felsenstein (1985).
FIGURE 1.
Maximum-likelihood phylogenetic tree based on 1487 nucleotides of 16S rRNA gene sequence showing the relationship between strain MUSC 149T and representatives of related taxa. Numbers at nodes indicate percentages of 1000 bootstrap re-samplings, only values above 50% are shown. Bar, 0.005 substitutions per site. Asterisks indicate that the corresponding nodes were also recovered using the neighbor-joining tree-making algorithm.
BOX-PCR fingerprint analysis was used to characterize strain MUSC 149T and the closely related strains using the primer BOX-A1R (5′-CTACGGCAAGGCGACGCTGACG-3′) (Versalovic et al., 1991; Lee et al., 2014c). The BOX-PCR cycling parameters were 5 min at 94°C for pre-denaturation, 35 cycles each of 30 s at 94°C for denaturation, 30 s at 53°C for annealing, 7 min at 65°C for extension and a final extension at 65°C for 8 min (Lee et al., 2014d). The PCR products were visualized by 2% agarose gel electrophoresis.
The protocol of Cashion et al. (1977) was used for the extraction of genomic DNA for DNA-DNA hybridization of strain MUSC 149T, S. graminisoli NBRC 108883T, S. gramineus NBRC 107863T and S. rhizophilus NBRC 108885T. DNA–DNA hybridization was carried out by the Identification Service of the DSMZ, Braunschweig, Germany following the protocol of De Ley et al. (1970) under consideration of the modifications described by Huss et al. (1983). The G + C content of strain MUSC 149T was determined by HPLC (Mesbah et al., 1989).
Phenotypic Characteristics
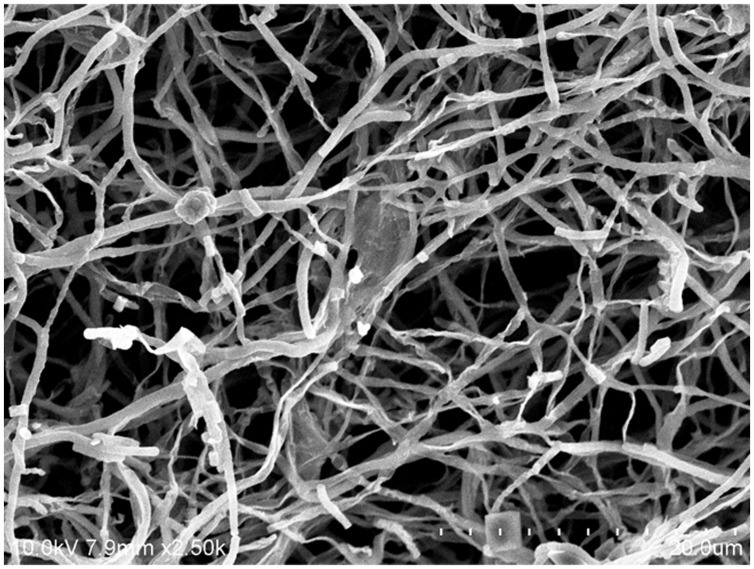
The cultural characteristics of strain MUSC 149T were determined following growth on ISP 2, ISP 3, ISP 4, ISP 5, ISP 6, ISP 7 (Shirling and Gottlieb, 1966), actinomycetes isolation agar (AIA; Atlas, 1993), Streptomyces agar (SA; Atlas, 1993), starch casein agar (SCA; Küster and Williams, 1964), and nutrient agar (Macfaddin, 2000) for 14 days at 28°C. Light microscopy (80i, Nikon) and scanning electron microscopy (JEOL-JSM 6400) were used to observe the morphology of the strain after incubation on ISP 2 agar at 28°C for 7–14 days (Figure 2). The designation of colony color was determined by using the ISCC-NBS color charts (Kelly, 1964). Gram staining was performed by standard Gram reaction and confirmed by using KOH lysis (Cerny, 1978). The growth temperature range was tested at 4-40 °C at intervals of 4 °C on ISP 2 agar. The pH range for growth was tested in tryptic soy broth (TSB) between pH 2.0 and 10.0 at intervals of 1 pH unit. The NaCl tolerance was tested in TSB and salt concentrations ranging from 0 to 10% (w/v) at intervals of 2%. The responses to temperature, pH and NaCl were observed for 14 days. Catalase activity and production of melanoid pigments were determined following protocols described by Lee et al. (2014e). The production of melanoid pigments was examined using ISP 7 medium. Hemolytic activity was assessed on blood agar medium containing 5% (w/v) peptone, 3% (w/v) yeast extract, 5% (w/v) NaCl, and 5% (v/v) horse blood (Carrillo et al., 1996). The plates were examined for hemolysis after incubation at 28°C for 7–14 days. Amylolytic, cellulase, chitinase, lipase, protease, and xylanase activities were determined by growing cells on ISP 2 agar and following protocols as described by Meena et al. (2013). The presence of clear zones around the colonies was taken to indicate the potential of isolates for surfactant production. Antibiotic susceptibility tests were performed by the disk diffusion method as described by Shieh et al. (2003). Antimicrobials used and their concentrations per disk (Oxoid, Basingstoke, UK) were as follows: ampicillin (10 μg), ampicillin sulbactam (30 μg), cefotaxime (30 μg), cefuroxime (30 μg), cephalosporin (30 μg), chloramphenicol (30 μg), ciprofloxacin (10 μg), erythromycin (15 μg), gentamicin (20 μg), nalidixic acid (30 μg), Penicillin G (10 μg), streptomycin (10 μg), tetracycline (30 μg), and vancomycin (30 μg). Carbon-source utilization and chemical sensitivity assays were determined using Biolog GenIII MicroPlates (Biolog, USA) according to the manufacturer’s instructions. All of the phenotypic assays mentioned were performed concurrently for strain MUSC 149T, S. graminisoli NBRC 108883T, S. gramineus NBRC 107863T, and S. rhizophilus NBRC 108885T.
FIGURE 2.
Scanning electron microscope of Streptomyces mangrovisoli MUSC 149T.
Chemotaxonomic Characteristics
The analyses of peptidoglycan amino acid composition and sugars of strain MUSC 149T were carried out by the Identification Service of the DSMZ using protocols of Schumann (2011). Major diagnostic cell wall sugars of strain MUSC 149T were obtained as described by Whiton et al. (1985) and analyzed by TLC on cellulose plates (Staneck and Roberts, 1974). Analysis of respiratory quinones, polar lipids (Kates, 1986) and fatty acids (Sasser, 1990) were carried out by the Identification Service of the DSMZ.
Extract preparation of MUSC 149T
MUSC 149T was grown in TSB for 14 days prior to fermentation process. The fermentation medium used was FM3 (Hong et al., 2009; Lee et al., 2012a). The medium was autoclaved at 121°C for 15 min prior to experiment. Fermentation was carried out in test tubes (30 mm × 200mm) containing 20 mL of FM3, at an angle of 45° for 7–10 days at 28°C. The resulting FM3 medium was recovered by centrifugation at 12000 g for 15 min. The supernatant was filtered and subjected to freeze dry process. Upon freeze-drying, the sample was extracted with methanol for 72 h and the methanol-containing extract was filtered and collected. The residue was re-extracted under the same condition twice at 24 h interval. Subsequently, the methanol-containing extract was evaporated using rotary vacuum evaporator at 40°C. The extract of MUSC 149T was collected and suspended in dimethyl sulphoxide (DMSO) as vehicle reagent prior to assay.
Determination of Antioxidant Activity of MUSC 149T Extract using 2,2-diphenyl-1- picrylhydrazyl (DPPH) Radical Scavenging Method
The stable radical 2,2-diphenyl-1-picrylhydrazyl (DPPH; Sigma–Aldrich) was used to examine antioxidant activity by measuring its hydrogen donating or radical scavenging ability. Scavenging activity on DPPH free radicals by MUSC 149T extract was accessed following previous method with minor modification (Ling et al., 2009). The decrease in radical is measured as decrease in the absorbance of 515 nm. Volume of 195 μL of 0.016% DPPH ethanolic solution was added to 5 μL of extract solution to make up final volume of 200 μL. Gallic acid was included as positive control. Reactions were carried out at room temperature in dark for 20 min before measurement with spectrophotometer at 515 nm. DPPH scavenging activity was calculated as follows:
Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
Gas chromatography–mass spectrometry (GC–MS) analysis was performed in accordance with our previous developed method with slight modification (Supriady et al., 2015). The machine used was Agilent Technologies 6980N (GC) equipped with 5979 Mass Selective Detector (MS), HP-5MS (5% phenyl methyl siloxane) capillary column of dimensions 30.0 m × 250 μm × 0.25 μm and used helium as carrier gas at 1 mL/min. The column temperature was programmed initially at 40°C for 10 min, followed by an increase of 3°C/min to 250°C and was kept isothermally for 5 min. The MS was operating at 70 eV. The constituents were identified by comparison of their mass spectral data with those from NIST 05 Spectral Library.
Results and Discussion
Phenotypic, Phylogenetic, and Genomic Analyses
Strain MUSC 149T was observed to grow well on ISP 2, ISP 3, ISP 5, ISP 6, ISP 7 agar, actinomycetes isolation agar, starch casein agar, and nutrient agar after 7–14 days at 28°C, and to grow poorly on Streptomyces agar, and did not grow on ISP 4 medium. The colors of the aerial and substrate mycelium were media-dependent (Supplementary Table S1). The morphological observation of a 15-day-old culture grown on ISP 2 agar revealed a smooth spore surface and abundant growth of both aerial and vegetative hyphae, which were well developed and not fragmented. These morphological features are consistent with grouping of the strain to the genus Streptomyces (Williams et al., 1989). Growth occurred at pH 5.0–8.0 (optimum pH 6.0–7.0), with 0–4% NaCl tolerance (optimum 0–2%) and at 24–36°C (optimum 28–32°C). Cells were found to be positive for catalase but negative for both melanoid pigment production and hemolytic activity. Hydrolysis of carboxymethylcellulose was found to be positive, but negative for hydrolysis of casein, chitin, soluble starch, tributyrin (lipase), and xylan. Strain MUSC 149T can be differentiated from closely related members of the genus Streptomyces using a range of phenotypic properties (Table 1). In chemical sensitivity assays, cells are resistant to aztreonam, D-serine, fusidic acid, guanine HCl, lincomycin, lithium chloride, minocycline, nalidixic acid, niaproof 4, potassium tellurite, rifamycin RV, sodium bromate, sodium butyrate, 1% sodium lactate, tetrazolium blue, tetrazolium violet, troleandomycin, and vancomycin.
Table 1.
Differentiation characteristics of strain MUSC 149T and type strains of phylogenetically closely related species of the genus Streptomyces.
| Characteristics | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Morphology (on ISP 2) | ||||
| Color of aerial mycelium | Pale yellow | Grayish yellow | Yellowish white | Light greenish yellow |
| Color of substrate mycelium | Grayish yellow | Grayish yellow | Pale orange yellow | Grayish yellow |
| Growth at | ||||
| 24°C | (+) | + | + | (+) |
| 36°C | (+) | (+) | (+) | + |
| pH 5 | (+) | (+) | (+)γ | (+) |
| pH 8 | (+) | + | (+)γ | (+) |
| 4% NaCl | (+) | + | (+)γ | (+) |
| Catalase | + | + | + | - |
| Hemolytic | - | - | - | - |
| Hydrolysis of | ||||
| Casein (protease) | - | - | + | - |
| Tributyrin (lipase) | - | - | + | + |
| Starch (amylolytic) | - | + | +γ | + |
| Carboxymethylcellulose (cellulase) | + | + | - | + |
| Xylan (xylanase) | - | - | - | + |
| Carbon source utilization | ||||
| D-trehalose | + | - | + | - |
| D-cellobiose | + | - | + | + |
| α-D-lactose | + | - | + | - |
| β-methyl-D-glucoside | + | - | - | + |
| N-acetyl-β-D-mannosamine | + | - | - | - |
| N-acetyl-D-galactosamine | + | - | - | - |
| N-acetyl-neuraminic acid | + | - | - | - |
| D-mannose | - | + | + | + |
| 3-methyl glucose | - | + | - | - |
| Inosine | + | - | + | - |
| D-mannitol | + | - | + | + |
| D-serine | - | + | - | - |
| Glycyl-L-proline | - | + | + | + |
| L-alanine | + | - | + | + |
| L-arginine | - | + | + | + |
| L-pyroglutamic acid | - | + | + | + |
| D-gluconic acid | - | + | + | + |
| Mucic acid | + | - | + | - |
| Quinic acid | - | - | + | + |
| D-saccharic acid | - | + | + | - |
| D-lactic acid methyl ester | - | + | + | + |
| D-malic acid | + | - | + | + |
| Chemical sensitivity assays | ||||
| Troleandomycin | + | + | - | - |
| Lithium chloride | + | + | - | - |
Strains: 1, S. mangrovisoli sp. nov. MUSC 149T; 2, S. rhizophilus NBRC 108885T; 3, S. gramineus NBRC 107863T; 4, S. graminisoli NBRC 108883T. All data were obtained concurrently in this study. +, Positive; –, negative; (+), weak. All strains are positive for utilization of Dextrin, D-maltose, gentiobiose, D-melibiose, α-D-glucose, D-fructose, D-galactose, L-fucose, L-rhamnose, gelatine, L-serine, pectin, oxp-hydroxy-phenylacetic acid, methyl pyruvate, L-malic acid, bromo-succinic acid, tween 40, oxγ-amino-butyric acid, α- hydroxy-butyric acid, oxβ-hydroxy-D,L-butyric acid and oxα-keto-butyric acid. S. graminisoli NBRC 108883T. γResults in accordance with that published for S. gramineus NBRC 107863T by Lee et al. (2012b).
The nearly complete 16S rRNA gene sequence was obtained for strain MUSC 149T (1487 bp; GenBank/EMBL/DDBJ accession number KJ632664) and phylogenetic trees were reconstructed to determine the phylogenetic position of this strain (Figure 1; Supplementary Figure S1). Phylogenetic analysis exhibited that strain MUSC 149T is closely related to S. rhizophilus JR-41T, as they formed a distinct clade (Figure 1; Supplementary Figure S1). The type strain S. rhizophilus JR-41T was isolated from a bamboo (Sasa borealis) rhizosphere soil (Lee and Whang, 2014). The 16S rRNA gene sequence analysis of strain MUSC 149T showed the highest similarity to that of S. rhizophilus NBRC 108885T (99.2% sequence similarity), followed by S. gramineus NBRC 107863T (98.7%) and S. graminisoli NBRC 108883T (98.5%); sequences similarities of less than 98.3% were obtained with the type strains of other species of the genus Streptomyces. The DNA–DNA hybridization values between strain MUSC 149T and S. rhizophilus NBRC 108885T (12.4 ± 3.3%), followed by S. gramineus NBRC 107863T (13.7 ± 0.5%) and S. graminisoli NBRC 108883T (27.3 ± 1.9%) were significantly below 70%, the threshold value for the delineation of bacterial species (Wayne et al., 1987). The BOX-PCR results indicated that strain MUSC 149T yielded a unique BOX-PCR fingerprint compared with the closely related type strains (Supplementary Figure S2). These results are in agreement with results of DNA–DNA hybridizations, which indicate that strain MUSC 149T represents a novel species.
Chemotaxonomic Analyses
Chemotaxonomic analyses showed that the cell wall of strain MUSC 149T is of cell-wall type I (Lechevalier and Lechevalier, 1970) as it contains LL-diaminopimelic. The presence of LL-diaminopimelic has been observed in many other species of the genus Streptomyces (Lee et al., 2005, 2014b; Xu et al., 2009; Hu et al., 2012; Ser et al., 2015). The predominant menaquinones of strain MUSC 149T were identified as MK-9(H8) (59%) and MK-9(H6) (15%). This is in agreement with Kim et al. (2003) that the predominant menaquinones of members of the genus Streptomyces are MK-9(H6) and MK-9(H8). The cell wall sugars detected were glucose, mannose and ribose. Strain MUSC149T shared the same sugar profile with S. gilvigriseus (Ser et al., 2015). Furthermore the sugars glucose and ribose were detected in other members of the genus Streptomyces such as S. rhizophilus JR-41T, S. graminisoli JR-19T (Lee and Whang, 2014), S. gramineus JR-43T (Lee et al., 2012b), S. shenzhenensis 172115T (Hu et al., 2011), and S. pluripotens (Lee et al., 2014b). The G + C content of strain MUSC 149T was determined to be 72.7 mol%; this is within the range of 67.0–78.0 mol% described for species of the genus Streptomyces (Kim et al., 2003).
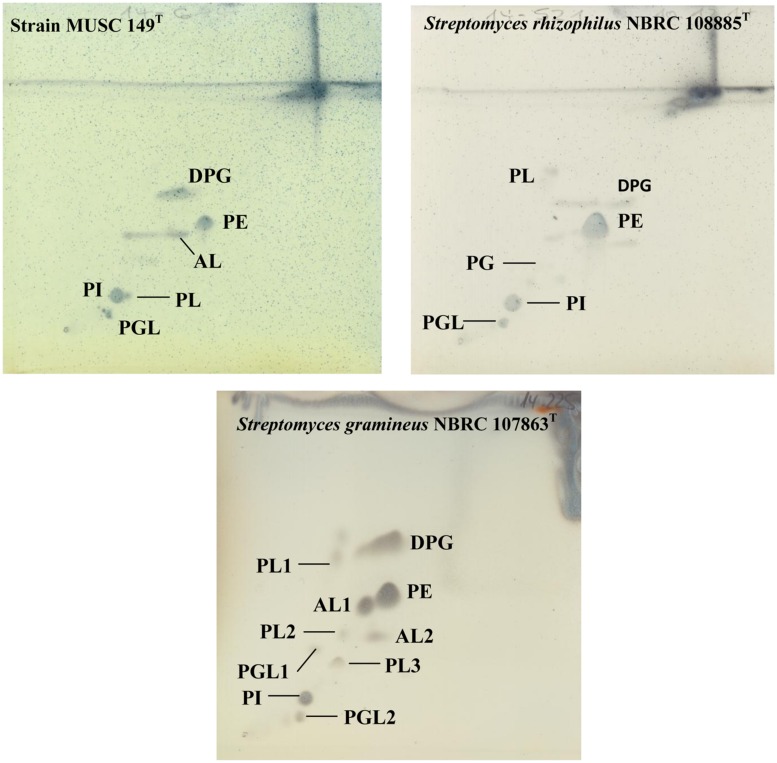
The polar lipid analysis showed the presence of aminolipid, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphoglycolipid, and phospholipid (Figure 3). Differences in polar lipid profiles indicated that MUSC 149T is different from related type strains (Figure 3); for example, strain MUSC 149T was found to contain aminolipid, lipid that was not detected in S. rhizophilus NBRC 108885T (Figure 3). The fatty acids profiles of strain MUSC 149T and closely related type strains are given shown in Table 2.
FIGURE 3.
Two dimensional total lipid profile of strain MUSC 149T, S. rhizophilus NBRC 108885T and S. gramineus NBRC 107863T. AL, Aminolipid; DPG, Diphosphatidylglycerol; PL, Phospholipid; PI, Phosphatidylinositol; PE, Phosphatidylethanolamine; PG, Phosphatidylglycerol; PGL, Phosphoglycolipid.
Table 2.
Cellular fatty acid composition of strain MUSC 149T and its closely related Streptomyces species.
| Fatty acid | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| iso-C12:0 | 0.1 | – | – | 0.1 |
| C12:0 | 0.1 | – | – | 0.1 |
| iso-C13:0 | 0.4 | 0.6 | 0.2 | 0.4 |
| anteiso-C13:0 | 0.5 | 0.3 | 0.2 | 0.3 |
| iso-C14:0 | 2.9 | 3.5 | 4.3 | 5.7 |
| C14:0 | 0.6 | 0.5 | 0.2 | 1.1 |
| iso-C15:0 | 17.7 | 18.3 | 19.1 | 12.5 |
| anteiso-C15:0 | 26.2 | 26.5 | 21.4 | 17.5 |
| C15:0w6c | – | – | – | 0.3 |
| C15:0 | 1.8 | 1.5 | – | 2.1 |
| iso-C16:1 H | 1.9 | – | 1.3 | 1.7 |
| iso-C16:0 | 16.0 | 15.4 | 19.2 | 25.1 |
| C16:1Cis9 | 2.8 | – | – | – |
| C16:0 | 4.0 | 9.4 | 4.3 | 7.9 |
| iso-C17:1w9c | – | 1.3 | 5.0 | 3.1 |
| anteiso-C17:1w9c | 2.4 | 0.6 | 1.9 | 1.9 |
| iso-C17:0 | 6.1 | 10.3 | 10.4 | 5.0 |
| anteiso-C17:0 | 11.3 | 10.7 | 9.6 | 9.2 |
| C17:1w8c | – | – | 0.4 | 0.4 |
| C17:0 CYCLO | 0.4 | – | 0.5 | 0.6 |
| C17:0 | 0.3 | 0.7 | 0.7 | 0.5 |
Strains: 1, S. mangrovisoli sp. nov. MUSC 149T; 2, S. rhizophilus NBRC 108885T; 3, S. gramineus NBRC 107863T; 4, S. graminisoli NBRC 108883T. –, <0.1% or not detected. All data are obtained concurrently from this study.
The major cellular fatty acids in MUSC 149T were identified as anteiso-C15: 0 (26.2%), iso-C15:0 (17.7%), iso-C16:0 (16.0%) and anteiso-C17:0 (11.3%). The fatty acids profile of MUSC 149T is consistent with those of closely related phylogenetic neighbors such as S. rhizophilus NBRC 108885T, S. gramineus NBRC 107863T, and S. graminisoli NBRC 108883T, which contain anteiso-C15:0 (26.5–17.5%), iso-C16:0 (25.1–15.4%), and iso-C15:0 (18.3–12.5%) as their major fatty acids (Table 2). However, the fatty acid profile of MUSC 149T was quantitatively different from those of these type strains; for example, although anteiso-C15:0 (26.2%) was found to be predominant in strain MUSC 149T, the amount of anteiso-C15:0 was significantly lesser (17.5%) in S. graminisoli NBRC 108883T (Table 2).
Based on the results of DNA-DNA hybridization, phylogenetic analysis, chemotaxonomic, phenotypic and DNA fingerprinting, strain MUSC 149T merits assignment to a novel species in the genus Streptomyces, for which the name S. mangrovisoli sp. nov. is proposed.
Antioxidant Activity of MUSC 149T Extract
The antioxidant evaluation assay DPPH is based upon the reduction of DPPH free radical. It is widely used to determine free radical scavenging capacity of the tested samples (Blois, 1958; Molyneux, 2004). As a free radical, DPPH is observed as purple solution when dissolved in appropriate solvent. It is known to exhibit a high absorption at 515 nm when measured with visible spectroscopy. In the presence of free radical-scavenging agent(s) or hydrogen donor(s), the odd electron of DPPH will be paired off, it will subsequently result in discoloration of solution to become either yellowish or colorless. The strength of the radical scavenging or anti-oxidant activity can then be quantified by the difference of absorbance obtained with the samples when is comparing to control.
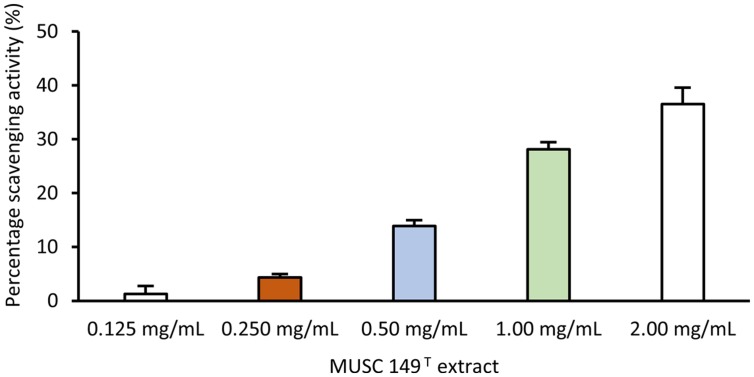
The DPPH scavenging assay was employed to examine the antioxidant activity of MUSC 149T extract. The extract was tested for a dose-response study with five different concentrations (0.125, 0.25, 0.5, 1.0, and 2.0 mg/mL). Based on the results obtained, the extract of MUSC 149T displayed a dose-dependent manner of antioxidant activity. It was inferred by a gradual increase in scavenging activity of MUSC 149T extract with a low concentration of extract at 0.125 mg/mL to the highest concentration at 2.0 mg/mL. The scavenging activity of lowest concentration at 0.125 mg/mL and the highest concentration at 2.0 mg/mL was recorded at 1.1 ± 1.4% and 36.5 ± 3.0%, respectively (Figure 4). The ability of MUSC 149T extract to scavenge DPPH free radicals indicates the possible presence of antioxidant agent(s) in the tested MUSC 149T extract.
FIGURE 4.
Antioxidant activity of MUSC 149T methanolic extract. Antioxidant activity of MUSC 149T was evaluated at different concentration and values are SEM of four replicates.
GC–MS Analysis of MUSC 149T Methanolic Extract
Growing evidence implies that the accumulation of free radicals may contribute to pathogenesis of chronic diseases including Parkinson’s disease and various types of cancers (Floyd and Hensley, 2002; Farooqui and Farooqui, 2009; Goldkorn et al., 2014; Mahalingaiah and Singh, 2014). Synthetic antioxidants may be able to scavenge these notorious free radicals, however, currently available antioxidants display low solubility and may promote negative health impacts (Barlow, 1990; Panicker et al., 2014). With this in mind, the search of the antioxidants from natural resources has always been one of the major focuses for many researchers (Lee et al., 1998; Harvey et al., 2015). In order to explore this premise, we examined the antioxidant activity of the extract of MUSC 149T. The results obtained demonstrated that MUSC 149T extract was posing significant antioxidant activity. This has prompted the necessities to further examine the chemical constituents which present in the extract of MUSC 149T.
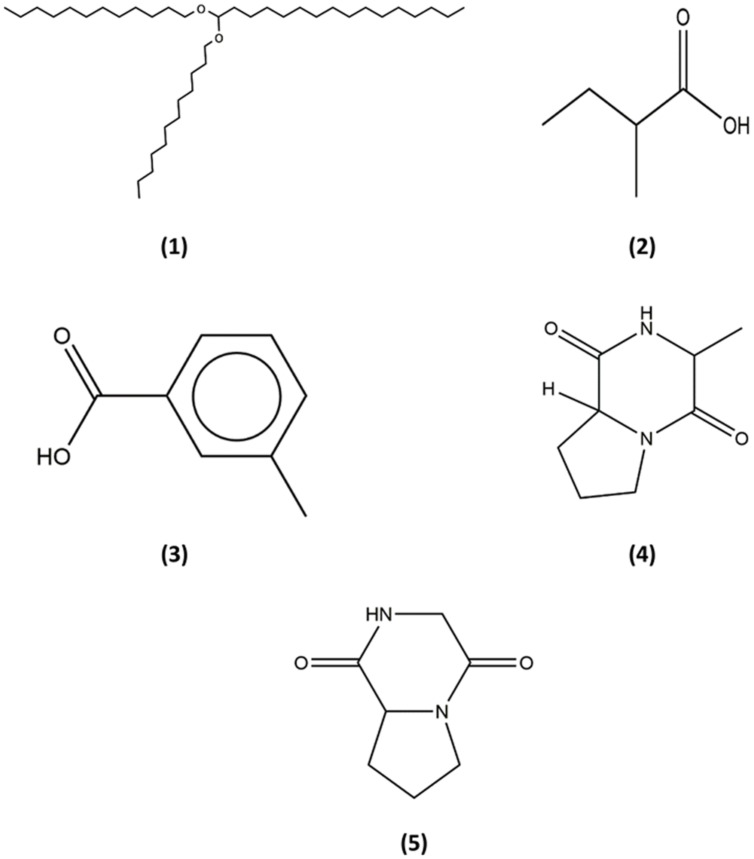
As Streptomyces are known to produce various secondary metabolites with diverse biological activity, numerous studies have incorporated powerful analytical techniques such as GC–MS to assist with the chemical analysis (Pollak and Berger, 1996; Karanja et al., 2010; Sudha and Masilamani, 2012; Ara et al., 2014; Jog et al., 2014). This robust technique produces reliable results as it combines separation power of GC and detection power of MS by generating characteristic mass spectral fragmentation patterns for each compounds present in mixture (Hites, 1997). For instance, recent study by Kim et al. (2008) has described detection of the bioactive compound (protocatechualdehyde) present in the extract of S. lincolnensis M-20 by using the GC–MS. With this intention, GC–MS analysis was performed in this study to explore the chemical constituents present in the extract of MUSC 149T. Using this analytical technique, we have identified chemical constituents of the extract of MUSC 149T (Table 3) and the chemical structures (Figure 5) as Hexadecane, 1,1-bis(dodecyloxy) (1), Butanoic acid, 2-methyl- (2), Benzoic acid, 3-methyl- (3) (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo[1,2-a]pyrazine-1,4-dione (4), and Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- (5).
Table 3.
Compounds identified from MUSC 149T extract through Gas chromatography–mass spectrometry (GC–MS).
| No. | Retention time | Compound | Formula | Molecular weight | Similarity (%) |
|---|---|---|---|---|---|
| 1 | 5.913 | Hexadecane, 1,1-bis(dodecyloxy) | C40H82O2 | 595 | 64 |
| 2 | 9.753 | Butanoic acid, 2-methyl- | C5H10O2 | 102 | 74 |
| 3 | 33.499 | Benzoic acid, 3-methyl- | C8H8O2 | 136 | 90 |
| 4 | 51.535 | (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo [1,2-a]pyrazine-1,4-dione | C8H12N2O2 | 168 | 90 |
| 5 | 52.994 | Pyrrolo [1,2-a]pyrazine-1,4-dione, hexahydro- | C7H10N2O2 | 154 | 90 |
FIGURE 5.
Chemical structures of the identified compounds from MUSC 149T. (1) Hexadecane, 1,1-bis(dodecyloxy); (2) Butanoic acid, 2-methyl-; (3) Benzoic acid, 3-methyl-; (4) (3R,8aS)-3-methyl-1,2,3,4,6,7,8,8a-octahydropyrrolo[1,2-a]pyrazine-1,4-dione; (5) Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-.
The detection of heterocyclic organic compound in extract is deemed as one of the most important findings in current study. Pyrrolizidines are widely present or synthesized in several marine Streptomyces species (Olano et al., 2009b; Robertson and Stevens, 2014). Furthermore, pyrrolizidines are known to exhibit a wide range of bioactivities which including antitumor, anti-angiogenesis, and antioxidant activities. For instance, the detection of the compound known as pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- (Table 3; Figure 5) in the extract has suggested the antioxidant activity could be contributed by this compound. Furthermore, other recent findings conducted on this compound suggested strong antioxidant activities as well (Gopi et al., 2014; Balakrishnan et al., 2015). These findings have demonstrated that pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- was able to scavenge or reduce amount of free radicals as evaluated by using reducing power assay. In short, an antioxidant is likely to play important roles in prevention and treatment of chronic diseases (Morales-González, 2013). The strong free radical scavenging effect possessed by the extract of MUSC 149T warrants the future investigations into different type of biological activities.
Description of S. mangrovisoli sp. nov.
Streptomyces mangrovisoli sp. nov. (man.gro.vi.so′li. N.L. n. mangrovum, mangrove; L. gen. n. soli, of soil; N.L. gen. n. mangrovisoli, of mangrove soil, referring to the source of the inoculum).
Cells stain Gram-positive and form pale yellow aerial and grayish yellow substrate mycelium on ISP 2 agar. The colors of the aerial and substrate mycelium are media-dependent (Supplementary Table S1). Grows well on ISP 2, ISP 3, ISP 5, ISP 6, ISP 7 agar, actinomycetes isolation agar, starch casein agar, and nutrient agar after 1–2 weeks at 28°C; and to grow poorly on Streptomyces agar, whereas no growth on ISP 4 medium. Grows occur at pH 5.0–8.0 (optimum pH 6.0–7.0), with 0–4% NaCl tolerance (optimum 0–2%) and at 24–36°C (optimum 28–32°C). Cells are positive for catalase but negative for both melanoid pigment production and hemolytic activity. Carboxymethylcellulose is hydrolysed but negative for hydrolysis of casein, chitin, soluble starch, tributyrin (lipase), and xylan. The following compounds are utilized as sole carbon sources: acetic acid, acetoacetic acid, α-D-glucose, α-D-lactose, α-hydroxy-butyric acid, α-keto-butyric acid, α-keto-glutaric acid, β-hydroxyl-D,L-butyric acid, β-methyl-D-glucoside, bromo-succinic acid, citric acid, D-cellobiose, Dextrin, D-fructose, D-fructose-6-phosphate, D-fucose, D-galactose, D-galacturonic acid, D-gluconic acid, D-glucose-6-phosphate, D-glucuronic acid, D-lactic acid methyl ester, D-malic acid, D-maltose, D-mannitol, D-melibiose, D-raffinose, D-saccharic acid, D-sorbitol, D-trehalose, D-turanose, formic acid, gelatin, gentiobiose, glucuronamide, inosine, L-fucose, L-galactonic acid lactone, L-lactic acid, L-malic acid, L-rhamnose, methyl pyruvate, mucic acid, N-acetyl-β-D-mannosamine, N-acetyl-D-galactosamine, N-acetyl-D-glucosamine, N-acetyl-neuraminic acid, pectin, p-hydroxyl-phenylacetic acid, propionic acid, quinic acid, stachyose, sucrose, Tween 40, and γ-amino-butyric acid. The following compounds are not utilized as sole carbon sources: D-salicin, D-mannose, D-arabitol, myo-inositol, glycerol, D-aspartic acid, D-serine, glycyl-L-proline, and 3-methyl glucose. L-alanine, L-histidine, and L-serine are utilized as sole nitrogen sources. L-arginine, L-aspartic acid, L-glutamic acid, and L-pyroglutamic acid are not utilized as sole nitrogen sources. Extract of the type strain exhibits strong antioxidant activity in a dose-dependent manner. The G + C content of the genomic DNA of the type strain is 72.7 mol%.
The type strain is MUSC 149T (=MCCC 1K00699T=DSM 100438T), isolated from mangrove soil collected from the Tanjung Lumpur mangrove forest located in the state of Pahang, Peninsular Malaysia. The 16S rRNA gene sequence of strain MUSC 149T has been deposited in GenBank/EMBL/DDBJ under the accession number KJ632664.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027 and No. A000001-50001) awarded to K.-G. C. and External Industry Grants from Biotek Abadi Sdn Bhd (vote no. GBA-808138 and GBA-808813) awarded to L.-H. L. The authors are thankful to Professor Bernhard Schink for the support in the Latin etymology of the new species name.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00854
References
- Ara I., Bukhari N. A., Aref N., Shinwari M. M., Bakir M. (2014). Antiviral activities of Streptomycetes against tobacco mosaic virus (TMV) in Datura plant: evaluation of different organic compounds in their metabolites. Afr. J. Biotech. 11 2130–2138. [Google Scholar]
- Atlas R. M. (1993). Handbook of Microbiological Media ed. Parks L. C. Boca Raton, FL: CRC Press. [Google Scholar]
- Balakrishnan D., Bibiana A., Vijayakumar A., Santhosh R., Dhevendaran K., Nithyanand P. (2015). Antioxidant activity of bacteria associated with the Marine Sponge Tedania anhelans. Ind. J. Microbiol. 55 13–18. 10.1007/s12088-014-0490-8 [DOI] [Google Scholar]
- Barlow S. M. (1990). Toxicological Aspects of Antioxidants Used as Food Additives. Food Antioxidants. Amsterdam: Springer; 253–307. [Google Scholar]
- Berdy J. (2005). Bioactive microbial metabolites. J. Antibiot. 58 1–26. 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Blois M. S. (1958). Antioxidant determinations by the use of a stable free radical. Nature 181 1199–1200. 10.1038/1811199a0 [DOI] [Google Scholar]
- Carrillo P., Mardaraz C., Pitta-Alvarez S., Giulietti A. (1996). Isolation and selection of biosurfactant-producing bacteria. World J. Microbiol. Biotech. 12 82–84. 10.1007/BF00327807 [DOI] [PubMed] [Google Scholar]
- Cashion P., Holder-Franklin M. A., McCully J., Frankiln M. (1977). A rapid method for the base ratio determination of bacterial DNA. Anal. Biochem. 81 461–466. 10.1016/0003-2697(77)90720-5 [DOI] [PubMed] [Google Scholar]
- Cerny G. (1978). Studies on the aminopeptidase test for the distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotech. 5 113–122. 10.1007/BF00498805 [DOI] [Google Scholar]
- De Ley J., Cattoir H., Reynaerts A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12 133–142. 10.1111/j.1432-1033.1970.tb00830.x [DOI] [PubMed] [Google Scholar]
- Farooqui T., Farooqui A. A. (2009). Aging: an important factor for the pathogenesis of neurodegenerative diseases. Mech. Ageing Dev. 130 203–215. 10.1016/j.mad.2008.11.006 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17 368–376. 10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–789. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Floyd R. A., Hensley K. (2002). Oxidative stress in brain aging: Implications for therapeutics of neurodegenerative diseases. Neurobiol. Aging 23 795–807. 10.1016/S0197-4580(02)00019-2 [DOI] [PubMed] [Google Scholar]
- Fu P., MacMillan J. B. (2015). Spithioneines A and B, two new bohemamine derivatives possessing ergothioneine moiety from a marine-derived Streptomyces spinoverrucosus. Organic Lett. 17 3046–3049. 10.1021/acs.orglett.5b01328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldkorn T., Filosto S., Chung S. (2014). Lung injury and Lung cancer caused by cigarette smoke-induced oxidative stress: molecular mechanisms and therapeutic opportunities involving the ceramide-generating machinery and epidermal growth factor receptor. Antioxid. Redox signal. 21 2149–2174. 10.1089/ars.2013.5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopi M., Dhayanithi N. B., Devi K. N., Kumar T. T. A. (2014). Marine natural product, Pyrrolo [-a] pyrazine–dione, hexahydro-(C7H10N2O2) of antioxidant properties from Bacillus species at Lakshadweep archipelago. J. Coastal Life Med. 2 632–637. [Google Scholar]
- Harvey A. L., Edrada-Ebel R., Quinn R. J. (2015). The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Dis. 14 111–129. 10.1038/nrd4510 [DOI] [PubMed] [Google Scholar]
- Hites R. A. (1997). “Gas chromatography mass spectrometry,” in Handbook of Instrumental Techniques for Analytical Chemistry ed. Settle F. (Upper Saddle River, NJ: Prentice Hall; ) 609–626. [Google Scholar]
- Hong K., Gao A.-H., Xie Q.-Y., Gao H. G., Zhuang L., Lin H.-P., et al. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7 24–44. 10.3390/md7010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Lin H.-P., Xie Q., Li L., Xie X.-Q., Hong K. (2012). Streptomyces qinglanensis sp. nov., isolated from mangrove sediment. Intl. J. Syst. Evol. Microbiol. 62 596–600. 10.1099/ijs.0.032201-0 [DOI] [PubMed] [Google Scholar]
- Hu H., Lin H.-P., Xie Q., Li L., Xie X.-Q., Sun M., et al. (2011). Streptomyces shenzhenensis sp. nov., a novel actinomycete isolated from mangrove sediment. Antonie Van Leeuwenhoek 100 631–637. 10.1007/s10482-011-9618-6 [DOI] [PubMed] [Google Scholar]
- Huss V. A. R., Festl H., Schleifer K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4 184–192. 10.1016/S0723-2020(83)80048-4 [DOI] [PubMed] [Google Scholar]
- Jennerjahn T. C., Ittekkot V. (2002). Relevance of mangroves for the production and deposition of organic matter along tropical continental margins. Naturwissenschaften 89 23–30. 10.1007/s00114-001-0283-x [DOI] [PubMed] [Google Scholar]
- Jog R., Pandya M., Nareshkumar G., Rajkumar S. (2014). Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiology 160 778–788. 10.1099/mic.0.074146-0 [DOI] [PubMed] [Google Scholar]
- Kaneko I., Katuo K., Shuji T. (1989). Complestain, a potent anti-complement substance produced by Streptomyces lavendulae. I. Fermentation, isolation and biological characterization. J. Antibiotics 42 236–241. [DOI] [PubMed] [Google Scholar]
- Karanja E., Boga H., Muigai A., Wamunyokoli F., Kinyua J., Nonoh J. (2010). “Growth characteristics and production of secondary metabolites from selected novel Streptomyces species isolated from selected Kenyan national parks,” in Proceedings of the 5th JKUAT Scientific, Technological and Industrialization Conference (Juja: Jomo Kenyatta University of Agriculture and Technology; ) 51–80. [Google Scholar]
- Kates M. (1986). Lipid Extraction Procedures. Techniques of Lipidology. Amsterdam: Elsevier; 100–111. [Google Scholar]
- Kelly K. L. (1964). Inter-Society Color Council-National Bureau of Standards Color Name Charts Illustrated with Centroid Colors. Washington, DC: U.S. Government Printing Office. [Google Scholar]
- Khieu T.-N., Liu M.-J., Nimaichand S., Quach N.-T., Chu-Ky S., Phi Q.-T., et al. (2015). Characterization and evaluation of antimicrobial and cytotoxic effects of Streptomyces sp. HUST012 isolated from medicinal plant Dracaena cochinchinensis Lour. Front. Microbiol. 6:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-J., Kim M.-A., Jung J.-H. (2008). Antitumor and antioxidant activity of protocatechualdehyde produced from Streptomyces lincolnensis M-20. Arch. Pharmacal. Res. 31 1572–1577. 10.1007/s12272-001-2153-7 [DOI] [PubMed] [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Intl. J. Syst. Evol. Microbiol. 62 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Kim S. B., Lonsdale J., Seong C. N., Goodfellow M. (2003). Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waskman and Gencici 1943AL) emend. Rainey etal. 1997. Antonie Van Leeuwenhoek 83 107–116 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kumar V., Naik B., Gusain O., Bisht G. S. (2014). An actinomycete isolate from solitary wasp mud nest having strong antibacterial activity and kills the Candida cells due to the shrinkage and the cytosolic loss. Front. Microbiol. 5:446 10.3389/fmicb.2014.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küster E., Williams S. (1964). Media for the isolation of Streptomycetes: starch casein medium. Nature 202 928–929. 10.1038/202928a0 [DOI] [PubMed] [Google Scholar]
- Lechevalier M. P., Lechevalier H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Intl. J. Syst. Evol. Microbiol. 20 435–443. [Google Scholar]
- Lee H.-J., Whang K.-S. (2014). Streptomyces graminisoli sp. nov. and Streptomyces rhizophilus sp. nov., isolated from bamboo (Sasa borealis) rhizosphere soil. Intl. J. Syst. Evol. Microbiol. 64 1546–1551. 10.1099/ijs.0.055210-0 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Lee J. Y., Jung H. W., Hwang B. K. (2005). Streptomyces koyangensis sp. nov., a novel actinomycete that produces 4-phenyl-3-butenoic acid. Intl. J. Syst. Evol. Microbiol. 55 257–262. 10.1099/ijs.0.63168-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Cheah Y.-K., Sidik S. M., Ab Mutalib N.-S., Tang Y.-L. Lin H.-P. et al. (2012a). Molecular characterization of Antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotech. 28 2125–2137. 10.1007/s11274-012-1018-1 [DOI] [PubMed] [Google Scholar]
- Lee H.-J., Han S.-I., Whang K.-S. (2012b). Streptomyces gramineus sp. nov., an antibiotic-producing actinobacterium isolated from bamboo (Sasa borealis) rhizosphere soil. Intl. J. Syst. Evol. Microbiol. 62 856–859. 10.1099/ijs.0.030163-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Zainal N., Azman A.-S., Eng S.-K., Goh B.-H., Yin W.-F., et al. (2014a). Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014 14 10.1155/2014/698178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.-H., Zainal N., Azman A.-S., Eng S.-K., Ab Mutalib N.-S. Yin W.-F. et al. (2014b). Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Intl. J. Syst. Evol. Microbiol. 64 3297–3306. 10.1099/ijs.0.065045-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Azman A.-S., Zainal N., Eng S.-K., Fang C.-M., Hong K., et al. (2014c). Novosphingobium malaysiense sp. nov. isolated from mangrove sediment. Intl. J. Syst. Evol. Microbiol. 64 1194–1201. 10.1099/ijs.0.059014-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Azman A.-S., Zainal N., Eng S.-K., Ab Mutalib N.-S., Yin W.-F., et al. (2014d). Microbacterium mangrovi sp. nov., an amylotytic actinobacterium isolated from mangrove forest soil. Intl. J. Syst. Evol. Microbiol. 64 3513–3519. 10.1099/ijs.0.062414-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Zainal N., Azman A.-S., Ab Mutalib N.-S., Hong K., Chan K.-G. (2014e). Mumia flava gen. nov., sp. nov., an actinobacterium of the family Nocardioidaceae. Intl. J. Syst. Evol. Microbiol. 64 1461–1467. 10.1099/ijs.0.058701-0 [DOI] [PubMed] [Google Scholar]
- Lee S. K., Mbwambo Z., Chung H., Luyengi L., Gamez E., Mehta R., et al. (1998). Evaluation of the antioxidant potential of natural products. Comb. Chem. High Throughput Screen. 1 35–46. [PubMed] [Google Scholar]
- Ling L. T., Yap S.-A., Radhakrishnan A. K., Subramaniam T., Cheng H. M., Palanisamy U. D. (2009). Standardised Mangifera indica extract is an ideal antioxidant. Food Chem. 113 1154–1159. 10.1016/j.foodchem.2008.09.004 [DOI] [Google Scholar]
- Macfaddin J. (2000). Biochemical Tests for Identification of Medical Bacteria. Philadelphia, PA: Lippincott Williams and Wilkins. [Google Scholar]
- Mahalingaiah P. K. S., Singh K. P. (2014). Chronic oxidative stress increases growth and tumorigenic potential of mcf-7 breast cancer cells. PLoS ONE 9:e87371 10.1371/journal.pone.0087371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena B., Rajan L. A., Vinithkumar N. V., Kirubagaran R. (2013). Novel marine actinobacteria from emerald Andaman & Nicobar Islands: a prospective source for industrial and pharmaceutical byproducts. BMC Microbiol. 13:145 10.1186/1471-2180-13-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah M., Premachandran U., Whitman W. B. (1989). Precise measurement of the G+ C content of deoxyribonucleic acid by high-performance liquid chromatography. Intl. J. Syst. Evol. Microbiol. 39 159–167. [Google Scholar]
- Molyneux P. (2004). The use of the stable free radical diphenylpicrylhydrazyl (DPPH) for estimating antioxidant activity. Songklanakarin. J. Sci. Technol .26 211–219. [Google Scholar]
- Morales-González J. A. (ed.). (2013). Oxidative Stress and Chronic Degenerative Diseases-A Role for Antioxidants. Rijeka: InTech, 500; 10.5772/45722 [DOI] [Google Scholar]
- Olano C., Mendez C., Salas J. A. (2009a). Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat. Prod. Rep. 26 628–660. 10.1039/b822528a [DOI] [PubMed] [Google Scholar]
- Olano C., Méndez C., Salas J. A. (2009b). Antitumor compounds from marine actinomycetes. Mar. Drugs 7 210–248. 10.3390/md7020210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker V. P., George S., Krishna D. (2014). Toxicity study of butylated hydroxyl toluene (BHT) in rats. World J. Pharm. Pharmaceut. Sci. 3 758–763. [Google Scholar]
- Pollak F. C., Berger R. G. (1996). Geosmin and related volatiles in bioreactor-cultured Streptomyces citreus CBS 109.60. Appl. Environ. Microbiol. 62 1295–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M. K., Shen Y.-C., Cheng X.-C., Jensen P. R., Frankmoelle W., Kauffman C. A., et al. (1999). Cyclomarins AC, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J. Amer. Chem. Soc. 121 11273–11276. 10.1021/ja992482o [DOI] [Google Scholar]
- Reuter S., Gupta S. C., Chaturvedi M. M., Aggarwal B. B. (2010). Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 49 1603–1616. 10.1016/j.freeradbiomed.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J., Stevens K. (2014). Pyrrolizidine alkaloids. Nat. Prod. Rep. 31 1721–1788. 10.1039/c4np00055b [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sasser M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101 Newark DEMIDI Inc. Schmieder R, Edwards R (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurav K., Kannabiran K. (2012). Cytotoxicity and antioxidant activity of 5-(2,4-dimethylbenzyl)pyrrolidin-2-one extracted from marine Streptomyces VITSVK5 spp. Saudi J. Biol. Sci 19 81–86. 10.1016/j.sjbs.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann P. (2011). Peptidoglycan structure. Met. Microbiol. 38 101–129. 10.1016/B978-0-12-387730-7.00005-X [DOI] [Google Scholar]
- Ser H.-L., Zainal N., Palanisamy U. D., Goh B.-H., Yin W.-F., Chan K.-G., et al. (2015). Streptomyces gilvigriseus sp. nov., a novel actinobacterium isolated from mangrove forest soil. Antonie Van Leeuwenhoek 107 1369–1378. 10.1007/s10482-015-0431-5 [DOI] [PubMed] [Google Scholar]
- Shieh W. Y., Chen Y.-W., Chaw S.-M., Chiu H.-H. (2003). Vibrio ruber sp. nov., a red, facultatively anaerobic, marine bacterium isolated from sea water. Intl. J. Syst. Evol. Microbiol. 53 479–484. 10.1099/ijs.0.02307-0 [DOI] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Intl. J. Syst. Evol. Microbiol 16 313–340. 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Staneck J. L., Roberts G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudha S., Masilamani S. M. (2012). Characterization of cytotoxic compound from marine sediment derived actinomycete Streptomyces avidinii strain SU4. Asian Pac. J. Trop. Biomed. 2 770–773. 10.1016/S2221-1691(12)60227-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J.-L., Xu X.-X., Qu Z., Wang H.-L., Lin H.-P., Xie Q.-Y., et al. (2011). Streptomyces sanyensis sp. nov., isolated from mangrove sediment. Intl. J. Syst. Evol. Microbiol. 61 1632–1637. 10.1099/ijs.0.023515-0 [DOI] [PubMed] [Google Scholar]
- Supriady H., Kamarudin M. N. A., Chan C. K., Goh B. H., Kadir H. A. (2015). SMEAF attenuates the production of pro-inflammatory mediators through the inactivation of Akt-dependent NF-κB, p38 and ERK1/2 pathways in LPS-stimulated BV-2 microglial cells. J. Funct. Foods 17 434–448. 10.1016/j.jff.2015.05.042 [DOI] [Google Scholar]
- Takahashi Y., Matsumoto A., Seino A., Iwai Y., Omura S. (1996). Rare actinomycetes isolated from desert soils. Actinomycetologica 10 91–97. 10.3209/saj.10_91 [DOI] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thenmozhi M., Kannabiran K. (2012). Antimicrobial and antioxidant properties of marine actinomycetes Streptomyces sp VITSTK7. Oxid. Antioxid. Med. Sci. 1 51–57. [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman S. A., Henrici A. T. (1943). The nomenclature and classification of the actinomycetes. J. Bacteriol. 46 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang Z., Qiao X., Li Z., Li F., Chen M., et al. (2013). Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol. Lett. 341 45–51. 10.1111/1574-6968.12088 [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Brenner D. J., Colwell R. R., Grimont P. A. D., Kandler O., Krichevsky M. I., et al. (1987). Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37 463–464. 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- Whiton R. S., Lau P., Morgan S. L., Gilbart J., Fox A. (1985). Modifications in the alditol acetate method for analysis of muramic acid and other neutral and amino sugars by capillary gas chromatography-mass spectrometry with selected ion monitoring. J. Chromatogr. A 347 109–120. 10.1016/S0021-9673(01)95474-3 [DOI] [PubMed] [Google Scholar]
- Williams S. T., Goodfellow M., Alderson G. (1989). “Genus Streptomyces Waksman and Henrici 1943 339AL,” in Bergey’s Manual of Systematic Bacteriology eds Williams S. T., Sharpe M. E., Holt J. G. (Baltimore: Williams & Wilkins; ) 2452–2492. [Google Scholar]
- Xiao J., Wang Y., Luo Y., Xie S.-J., Ruan J.-S., Xu J. (2009). Streptomyces avicenniae sp. nov., a novel actinomycete isolated from the rhizosphere of the mangrove plant Avicennia mariana. Intl. J. Syst. Evol. Microbiol. 59 2624–2628. 10.1099/ijs.0.009357-0 [DOI] [PubMed] [Google Scholar]
- Xu J., Wang Y., Xie S.-J., Xu J., Xiao J., Ruan J.-S. (2009). Streptomyces xiamenensis sp. nov., isolated from mangrove sediment. Intl. J. Syst. Evol. Microbiol. 59 472–476. 10.1099/ijs.0.000497-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.