Abstract
Plant-pathogenic fungi and their hosts engage in chemical warfare, attacking each other with toxic products of secondary metabolism and defending themselves via an arsenal of xenobiotic metabolizing enzymes. One such enzyme is homologous to arylamine N-acetyltransferase (NAT) and has been identified in Fusarium infecting cereal plants as responsible for detoxification of host defence compound 2-benzoxazolinone. Here we investigate functional diversification of NAT enzymes in crop-compromising species of Fusarium and Aspergillus, identifying three groups of homologues: Isoenzymes of the first group are found in all species and catalyse reactions with acetyl-CoA or propionyl-CoA. The second group is restricted to the plant pathogens and is active with malonyl-CoA in Fusarium species infecting cereals. The third group generates minimal activity with acyl-CoA compounds that bind non-selectively to the proteins. We propose that fungal NAT isoenzymes may have evolved to perform diverse functions, potentially relevant to pathogen fitness, acetyl-CoA/propionyl-CoA intracellular balance and secondary metabolism.
Plant-pathogenic fungi have developed diverse mechanisms to penetrate and colonize hosts, overcoming their physical barriers, chemical defences and complex cellular signalling responses1. Plants attack fungi with toxic products of secondary metabolism that can be broadly classified as phytoalexins or phytoanticipins2. Invading fungi, in turn, are often capable of overcoming the antimicrobial effects of such chemicals, by employing their xenobiotic metabolizing enzymes, including cytochrome P450, glucosyltransferases and others3,4.
Two such enzyme activities, essential for the detoxification of benzoxazinoids (antimicrobials produced by maize, wheat and rye), have been mapped at genetic loci FDB1 and FDB2 of the endophytic filamentous ascomycete Fusarium verticillioides5. Benzoxazinoids, like 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3-one (DIMBOA) and 2,4-dihydroxy-2H-1,4-benzoxazin-3-one (DIBOA), are phytoanticipins generated via a well-characterized biosynthetic pathway6,7, released as aglycones that rapidly degrade into the corresponding benzoxazolinones 6-methoxy-2-benzoxazolinone (MBOA) and 2-benzoxazolinone (BOA). These, too, exert toxic effects to unwelcome microorganisms, herbivores or weeds by reacting with nucleophilic groups (e.g. -NH2 and -SH) of biomolecules7. Endophytic fungi associated with cereal plants, like the maize pathogen F. verticillioides, have adapted their xenobiotic metabolism to effectively detoxify MBOA and BOA, by employing the metabolic activities encoded at the FDB1 and FDB2 loci5,8. The former activity mediates BOA hydrolysis to the intermediate compound 2-aminophenol (2AP), while the latter involves a N-malonyltransferase catalysing the conversion of 2AP to the non-toxic compound N-(2-hydroxyphenyl)malonamic acid (HPMA)5. Cloning of the N-malonyltransferase gene revealed a sequence highly homologous to arylamine N-acetyltransferase (NAT), while deletion of the FDB2 locus further demonstrated a secondary branch to the metabolic pathway, potentially involving another NAT homologue catalysing N-acetylation of 2AP to N-(2-hydroxyphenyl)acetamide (HPAA)9. Genomic analysis identified a total of four putative NAT loci in F. verticillioides, while screening of an additional 145 fungal genomes annotated multiple putative homologues in various ascomycetes within the subphylum of Pezizomycotina10,11.
NAT was originally identified by Nobel Laureate Fritz Lipmann and his co-workers12, as the hepatic enzymatic activity responsible for the acetyl-coenzyme A (CoA) dependent N-acetylation of arylamines (EC 2.3.1.5). It was subsequently demonstrated that the same enzyme entity also bears N-hydroxyarylamine O-acetyltransferase (EC 2.3.1.118) and arylhydroxamic acid N,O-acetyltransferase (EC 2.3.1.56) activities13. Pharmacological work demonstrated that human NAT is also active against aromatic hydrazines, such as the anti-tubercular drug isoniazid (INH), and polymorphic acetylation of INH and other common drugs has been the subject of intense pharmacogenetic research14. Polymorphic NAT activity has additionally been associated with xenobiotic-induced carcinogenicity of the bladder15. The mechanism of NAT-mediated N-acetylation has been studied in humans and laboratory mammals, as well as in prokaryotes, including mycobacteria16. Irrespectively of their kingdom of origin, all characterized NAT homologues appear to perform their enzyme function via a conserved “catalytic triad” (typically formed by residues cysteine-histidine-aspartate) similar to that of cysteine proteases17,18. Genomic surveys indicate NAT homologues in phylogenetically diverse organisms, although gaps in the evolution of the NAT gene family appear to exist, most notably in plants10,19. NATs are well-represented in fungi, particularly plant-associated ascomycetes which are predicted to harbour more than one putative paralogue in their genome10,11. Recent studies have also explored N-acetylation by fungal NATs as a potential route for detoxification of environmental pollutants20,21.
Our previous work on the BOA-detoxification pathway of F. verticillioides9, points towards possible functional diversification of NAT enzymes in plant-pathogenic fungi. Here, we characterize the NAT homologues of five representative species of Fusarium and Aspergillus, four of which (including F. verticillioides) compromise various crops. Our results support that different NAT isoenzymes of plant-associated ascomycetes are not just the product of genetic redundancy, but may have evolved to perform distinct functions.
Results
Characterization of NAT loci in plant-associated ascomycetes
Our previous survey10 predicted 16 putative NAT loci in the sequenced genomes of the maize pathogen Fusarium verticillioides (teleomorph Gibberella moniliformis; strain FGSC 7600; 4 NAT loci), the wheat pathogen Fusarium graminearum (teleomorph Gibberella zeae; strain PH-1; 3 NAT loci), the tomato pathogen Fusarium oxysporum f.sp. lycopersici (strain FOL 4287; 4 NAT loci), the grain contaminant Aspergillus flavus (strain NRRL 3357; 4 NAT loci) and the laboratory model fungus Aspergillus nidulans (teleomorph Emericella nidulans; strain FGSC A4; 1 NAT locus). To validate the computational annotations, we undertook PCR amplification and sequencing of those loci from genomic DNA and cDNA isolated from the five ascomycetes, followed by computational alignment of the identified sequences to determine the exon-intron structure of each NAT gene. Thirteen NAT loci were confirmed to generate transcripts with open reading frames (ORFs) that translate into peptide sequences with the characteristic conserved elements of NAT enzymes10,17,18 (Table 1, Supplementary Fig. S1). In contrast, amplification from cDNA with primers specific for the predicted NAT4 loci of F. verticillioides and A. flavus identified hypothetical ORFs compromised by multiple nonsense mutations, suggesting that those two loci may be transcribing pseudogenes. Moreover, the NAT1 locus of A. flavus is most likely not transcribed, under the standard culture conditions applied, since different combinations of primers generated specific amplification products from genomic DNA, but not from cDNA. The ORFs of fungal NAT genes, shown to generate protein-coding transcripts, were cloned and expressed in Escherichia coli, providing recombinant proteins with the expected size upon SDS-PAGE (Supplementary Fig. S2).
Table 1. Description of characterized NAT loci.
| Species (strain) | Taxon mnemonic1 | Taxon ID1 | Gene symbol | ORF (bp)2 | Protein (aa)2 | Exon span | Number of introns | Predicted locus tag3 | Nucleotide ID4 | |
|---|---|---|---|---|---|---|---|---|---|---|
| F. verticillioides (FGSC 7600) | GIBM7 | 334819 | ΝΑΤ1 | 1038 | 345 | 1 (1–1038) | 0 | FVEG_12636 | EU552489, FN687904 | |
| ΝΑΤ2 | 957 | 318 | 1 (1–957) | 0 | FVEG_03961 | FN687889, FN687905 | ||||
| ΝΑΤ3 | 978 | 325 | 1 (1–978) | 0 | FVEG_12062 | FN687890, FN687906 | ||||
| NAT45 | Transcribed pseudogene | 1 (1–298) 2 (461–1033) | 1 | FVEG_07425 | FN687891, LN829129 | |||||
| F. graminearum (PH-1) | GIBZE | 229533 | ΝΑΤ1 | 1032 | 343 | 1 (1–1032) | 0 | FGSG_00080 | FN687882, FN687897 | |
| ΝΑΤ2 | 957 | 318 | 1 (1–371) 2 (430–1015) | 1 | FGSG_09400 | FN687883, FN687898 | ||||
| ΝΑΤ3 | 960 | 319 | 1 (1–960) | 0 | FGSG_07888 | FN687884, FN687899 | ||||
| F. oxysporum f.sp. lycopersici (FOL 4287) | FUSO4 | 426428 | ΝΑΤ1 | 1053 | 350 | 1 (1–1053) | 0 | FOXG_15318 | FN687885, FN687900 | |
| ΝΑΤ2 | 957 | 318 | 1 (1–371) 2 (423–1008) | 1 | FOXG_06095 | FN687886, FN687901 | ||||
| ΝΑΤ3 | 999 | 332 | 1 (1–999) | 0 | FOXG_03795 | FN687887, FN687902 | ||||
| ΝΑΤ4 | 963 | 320 | 1 (1–365) 2 (548–1145) | 1 | FOXG_04301 | FN687888, FN687903 | ||||
| A. flavus (NRRL 3357) | ASPFN | 332952 | NAT15 | Elusive | Elusive | Elusive | Elusive | AFL2G_05055 | − | |
| ΝΑΤ2 | 981 | 326 | 1 (1–426) 2 (482–528) 3 (588–1095) | 2 | AFL2G_01915 | FN687893, FN687907 | ||||
| ΝΑΤ3 | 957 | 318 | 1 (1–395) 2 (449–1010) | 1 | AFL2G_11316 | FN687894, FN687908 | ||||
| ΝΑΤ45 | Transcribed pseudogene | 1 (1–91) 2 (143–692) | 1 | AFL2G_03311 | FN687895, FN687909 | |||||
| A. nidulans (FGSC A4) | EMENI | 227321 | ΝΑΤ1 | 960 | 319 | 1 (1–407) 2 (463–510) 3 (568–1072) | 2 | ANID_10723 | FN687881, FN687896 | |
1The taxon mnemonics and ID numbers are from the UniProt Taxonomy database (http://www.uniprot.org/taxonomy/). They correspond to sequenced strains of Fusarium verticillioides (teleomorph Gibberella moniliformis), Fusarium graminearum (teleomorph Gibberella zeae), Fusarium oxysporum f.sp. lycopersici, Aspergillus flavus and Aspergillus nidulans (teleomorph Emericella nidulans). According to current consensus nomenclature guidelines (Supplementary Methods and http://nat.mbg.duth.gr/), taxon mnemonics are attached to the symbols of NAT genes to identify their specific organism of origin.
2The sequences of open reading frames (ORF) in base pairs (bp), as well as of deduced proteins in amino acids (aa), were determined via alignment of amplification products generated from genomic DNA and cDNA.
3The locus tags represent annotations by the Broad Institute (http://www.broadinstitute.org/science/projects/fungal-genome-initiative/gene-finding-methods). F. verticillioides FVEG_07425 and A. flavus AFL2G_03311 tag genomic loci with sequences overlapping, but not coinciding, with the NAT sequences characterized experimentally in the present study.
4Two Nucleotide IDs were assigned to each fungal NAT, the first for the genomic and the second for the transcribed sequence of each locus.
5Annotation remains elusive for the NAT1 locus of A. flavus, as specific amplification from cDNA of the fungus has not been possible. The NAT4 loci of F. verticillioides and A. flavus appeared as transcribing pseudogenes with hypothetical ORFs that are disrupted by nonsense mutations.
Acyl-coenzyme A selectivity of fungal NAT enzymes
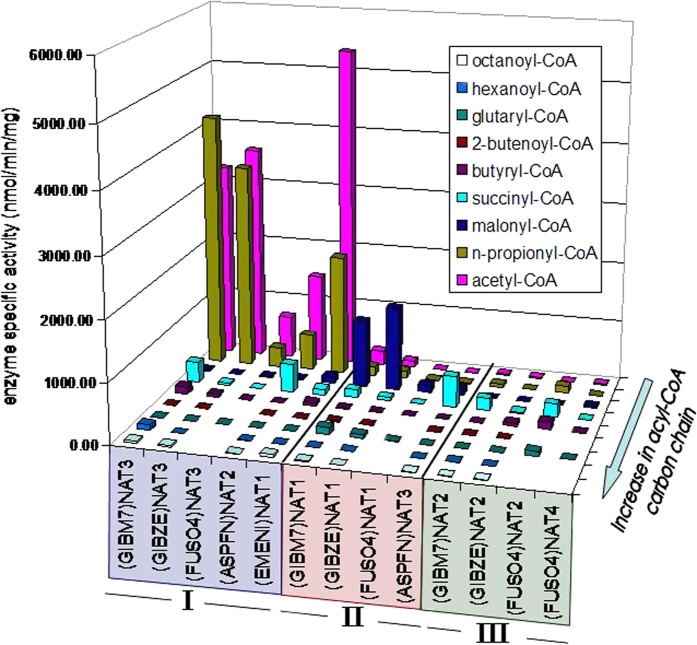
The N-malonyltransferase encoded by the FDB2 locus in F. verticillioides has been assigned symbol NAT19,10. Here, we show that this isoenzyme selectively employs malonyl-CoA to catalyze N-malonyl transfer to an arylamine, and that this enzymatic reaction is specific to the NAT1 homologues of F. verticillioides [(GIBM7)NAT1] and F. graminearum [(GIBZE)NAT1], the two fungi in our panel that are naturally exposed to BOA through their endophytic association with cereals. Homologous NAT isoenzymes are also predicted for the tomato pathogen F. oxysporum f.sp. lycopersici [(FUSO4)NAT1] and potentially for the non-endophytic maize pathogen A. flavus [(ASPFN)NAT3], although the N-malonyltransferase activity detected with those recombinant proteins was very low. No N-malonyltransferase homologue was found in A. nidulans, a fungus not associated with plants (Fig. 1 and Supplementary Fig. S3).
Figure 1. Activity of fungal NAT enzymes with different acyl-coenzyme A compounds.
Overview of the acyl-CoA selectivity pattern observed for the recombinant NAT isoenzymes of F. verticillioides (G. moniliformis-GIBM7), F. graminearum (G. zeae-GIBZE), F. oxysporum f.sp. lycopersici (FUSO4), A. flavus (ASPFN) and A. nidulans (E. nidulans-EMENI). Functional homologues are grouped together within coloured boxes labelled I-III. Each enzyme was assayed against a series of acyl-CoA compounds, used as acyl-group donors in reactions with 5-aminosalicylate as acceptor substrate. The results for each set of assays are presented in Supplementary Fig. S3.
The 13 fungal NAT proteins, expressed here in recombinant form, were subjected to enzymatic activity assays with a series of acyl-CoA compounds ranging in acyl-chain length from acetyl- to octanoyl-CoA. The screen identified three distinct groups (I-III) of NAT homologues in the five ascomycetes investigated (Fig. 1 and Supplementary Fig. S3). Each fungus has one NAT isoenzyme (group I) with typical N-acetyltransferase activity, and those homologues are also highly selective for n-propionyl-CoA. Group II comprises the N-malonyltransferase homologues described above, while the NAT proteins in group III show no specific preference for any of the acyl-CoAs tested. Malonyl-CoA was employed exclusively by the N-malonyltransferases, which also provided minimal activity with acetyl- and n-propionyl-CoA. Low levels of activity were also detected with succinyl-CoA for several NAT isoenzymes, but no specific selectivity pattern was evident among homologues. Other acyl-CoA compounds provided residual or no activity with fungal NATs (Fig. 1 and Supplementary Fig. S3).
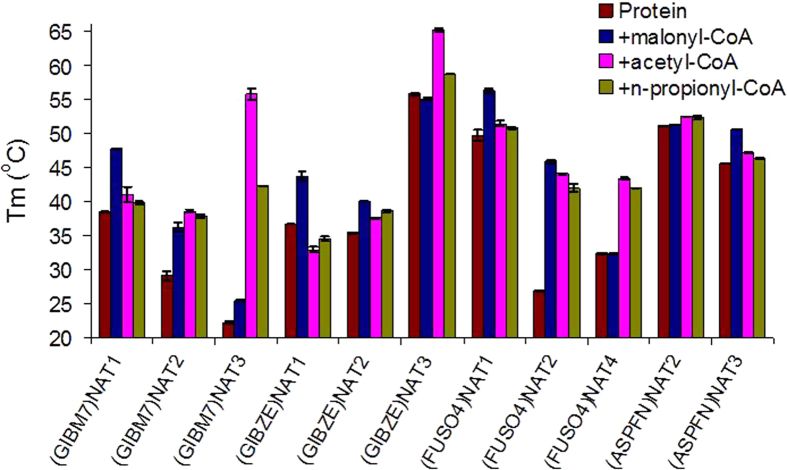
Binding of acyl-CoAs to fungal NATs was investigated using differential scanning fluorimetry (DSF), a technique that detects changes in the thermal stability of recombinant proteins upon interaction with their specific ligands22. The method was applicable with all recombinant NAT proteins in our panel, except the NAT3 of F. oxysporum [(FUSO4)NAT3] and the NAT1 of A. nidulans [(EMENI)NAT1] which were recovered at less optimal levels of yield and purity (Supplementary Fig. S2). The Tm values of different fungal NAT homologues varied considerably from around 20 to over 55 °C, with the NAT3 homologues of F. verticillioides [(GIBM7)NAT3] and F. graminearum [(GIBZE)NAT3] at opposite extremes (Fig. 2 and Supplementary Fig. S4). The addition of acyl-CoAs increased the Tm values of NAT proteins, with a pattern that reflected exactly the results of our activity assays for group I N-acetyl/N-propionyltransferases and group II N-malonyltransferases. In the case of group I homologues [(GIBM7)NAT3, (GIBZE)NAT3 and (ASPFN)NAT2], the shift in Tm was always greater with acetyl-CoA, intermediate with n-propionyl-CoA and minimal with malonyl-CoA. Conversely, for group II N-malonyltransferases [(GIBM7)NAT1, (GIBZE)NAT1, (FUSO4)NAT1 and (ASPFN)NAT3], the increase in Tm was substantial only with malonyl-CoA (Fig. 2 and Supplementary Fig. S4).
Figure 2. Effect of acyl-coenzyme A compounds on the Tm of fungal NAT proteins.
Overview of Tm values determined by differential scanning fluorimetry for recombinant NAT isoenzymes of F. verticillioides (G. moniliformis-GIBM7), F. graminearum (G. zeae-GIBZE), F. oxysporum f.sp. lycopersici (FUSO4) and A. flavus (ASPFN), in the absence or presence of various acyl-CoAs. Two replicate experiments were performed, generating overlapping curves for which the average Tm (± standard deviation) is shown. The results for each set of experiments are presented in Supplementary Fig. S4.
The results of the DSF analysis indicated that the NAT homologues of group III, appearing as functionally redundant in our activity assays with various acyl-CoAs (Fig. 1), are in fact capable of binding acetyl-, n-propionyl-, malonyl- and succinyl-CoA [(GIBM7)NAT2, (GIBZE)NAT2 and (FUSO4)NAT2]. The only deviation within group III was observed for the NAT4 isoenzyme of F. oxysporum [(FUSO4)NAT4], where the shift to Tm was evident with acetyl- and n-propionyl-CoA and absent with malonyl-CoA (Fig. 2 and Supplementary Fig. S4). It is possible that this particular isoenzyme of F. oxysporum is distinct from other fungal NAT homologues in our experimental panel.
Substrate selectivity of fungal NAT enzymes
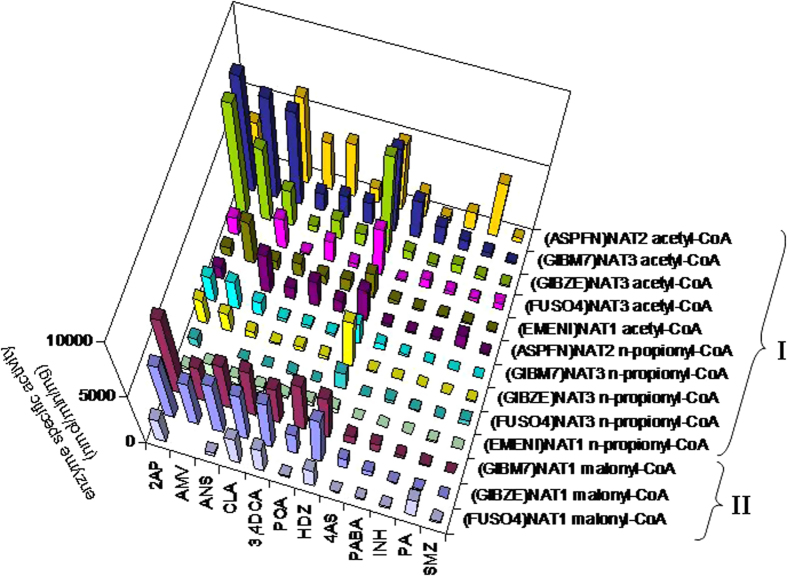
To assess the enzymatic activity of recombinant fungal NAT proteins towards different substrates, assays were performed with the preferred acyl-CoA of each isoenzyme (as identified by the enzymatic and DSF analyses of the previous section) against a representative panel of arylamine and arylhydrazine compounds23. Group I homologues provided higher enzyme activities with acetyl-CoA and lower with n-propionyl-CoA, but the overall specificity pattern with respect to acceptor substrate was similar with both acyl-group donor compounds (Fig. 3 and Supplementary Fig. S5).
Figure 3. Activity of fungal NAT enzymes with different acceptor substrates.
Overview of the acceptor substrate selectivity pattern observed for group I and II isoenzymes of F. verticillioides (G. moniliformis-GIBM7), F. graminearum (G. zeae-GIBZE), F. oxysporum f.sp. lycopersici (FUSO4), A. flavus (ASPFN) and A. nidulans (E. nidulans-EMENI). Recombinant NAT proteins were assayed with selective acyl-CoAs against a panel of arylamine and arylhydrazine substrates, and the results for each set of assays are presented in Supplementary Fig. S5. The full chemical names of compounds are: 2-aminophenol (2AP), 4-aminoveratrole (AMV), 4-anisidine (ANS), 4-chloroaniline (CLA), 3,4-dichloroaniline (3,4DCA), 4-phenoxyaniline (POA), hydralazine (HDZ), 4-aminosalicylate (4AS), 4-aminobenzoate (PABA), isoniazid (INH), procainamide (PA) and sulphamethazine (SMZ).
Our screening showed that 2AP, the substrate of NAT1-mediated N-malonyltransfer leading to BOA detoxification in F. verticillioides9, generates high levels of activity with several group I and group II homologues. Toxic substituted anilines, such as the haloanilines 4-chloroaniline (CLA) and 3,4-dichloroaniline (3,4DCA), and the alkoxyanilines 4-anisidine (ANS) and 4-aminoveratrole (AMV), were also effective substrates of fungal NAT enzymes. On the other hand, 4-aminosalicylate (4AS), 4-aminobenzoate (PABA), procainamide (PA), sulphamethazine (SMZ) and INH, all well-characterized pharmaceutical compounds metabolized by mammalian NAT isoenzymes, were demonstrated to be poorer substrates of fungal NATs. Of the two arylhydrazines used, the antihypertensive hydralazine (HDZ) generated considerably higher levels of NAT enzymatic activity than the anti-tubercular INH (Fig. 3 and Supplementary Fig. S5).
Endogenous NAT activities of fungi and effects of xenobiotics
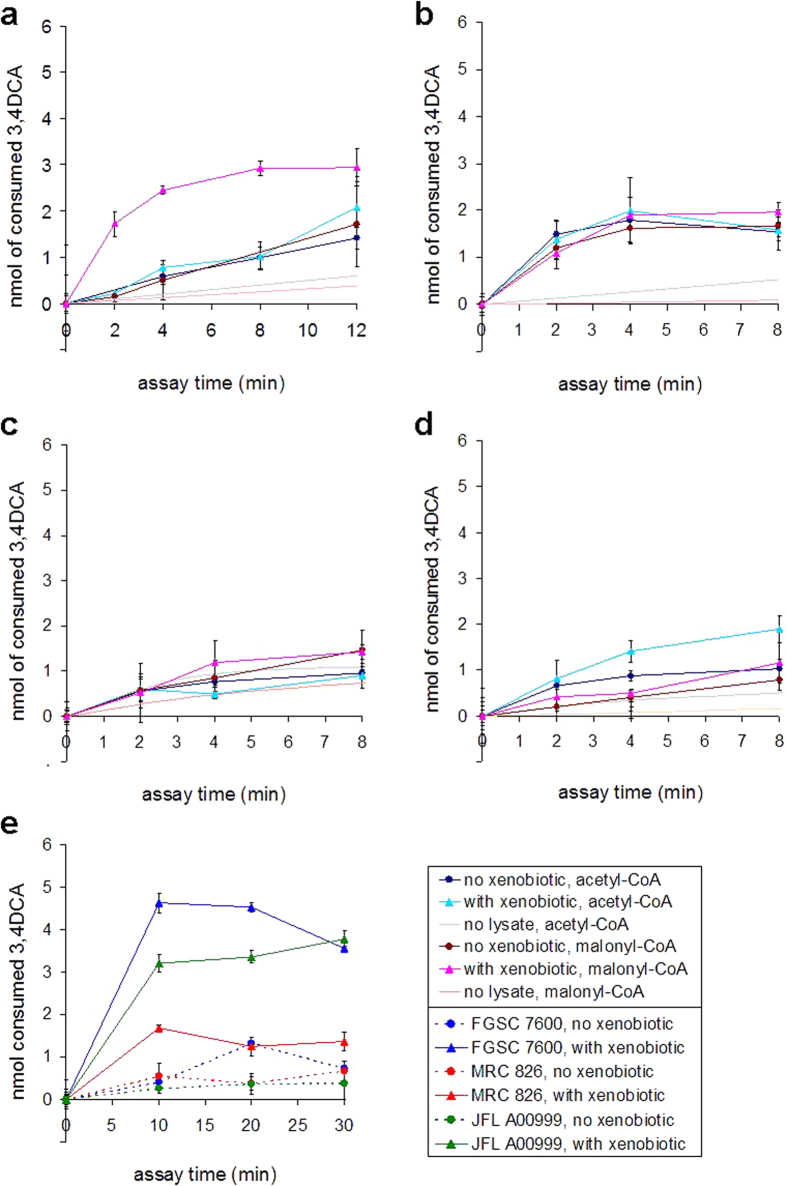
Cellular N-acetyltransferase and N-malonyltransferase activities were measured with 3,4DCA in soluble extracts of F. verticillioides, F. graminearum, F. oxysporum f.sp. lycopersici and A. flavus, grown in liquid media with or without xenobiotics. The fungi were challenged with a mixture of BOA and 3,4DCA, in order to achieve induction of the BOA detoxification pathway9 and potentially other cellular NAT enzymes. In A. nidulans, the yield of total soluble protein was reduced by over 2.5-fold (4.4 vs. 11.7 mg) in extracts from cultures with vs. without xenobiotics, suggesting that the fungus is unable to tolerate exposure. Xenobiotic challenge had only minimal effect (up to 1.5-fold increase) on the N-acetyltransferase activity of the four other fungi (Fig. 4).
Figure 4. NAT enzymatic activities in fungal cell extracts upon xenobiotic exposure.
Erlich’s reagent was used to measure NAT activity in fungal soluble extracts, following enzyme assays with 3,4-dichloroaniline (3,4DCA) and either acetyl- or malonyl-CoA. The graphs show comparison of NAT enzymatic activities measured in cell extracts from cultures challenged for 2 h with xenobiotics (mixture of 2-benzoxazolinone and 3,4DCA, each at 25 μg/ml), relative to extracts prepared from cultures grown in standard medium. Control assays, without cell extract, are also shown. Each data point is the average value of three replicates ± standard deviation. Results are shown for assays performed with cell extracts from F. verticillioides (G. moniliformis) strain FGSC 7600 (a), F. graminearum (G. zeae) strain PH-1 (b), F. oxysporum f.sp. lycopersici strain FOL 4287 (c) and A. flavus strain NRRL 3357 (d). The effects of xenobiotics on NAT enzymatic activity measured with malonyl-CoA in cell extracts from F. verticillioides strains FGSC 7600, MRC 826 and JFL A00999 are also compared (e).
In F. verticillioides strain FGSC 7600, the N-malonyltransferase activity was relatively low (1 nmol/min/mg), but increased sharply (by 8-fold) upon challenge with xenobiotics (Fig. 4a). Induction was also evident with F. verticillioides strains MRC 826 and JFL A00999 (Fig. 4e). In F. graminearum, N-malonyltransferase activity was higher (5.3 nmol/min/mg), but xenobiotic exposure had no apparent effect. N-malonyltransferase activity was also present in F. oxysporum (2.3 nmol/min/mg) and was not affected by the xenobiotics. In A. flavus, the N-malonyltransferase activity was low (0.8 nmol/min/mg), but xenobiotics caused a moderate (2.5-fold) increase (Fig. 4).
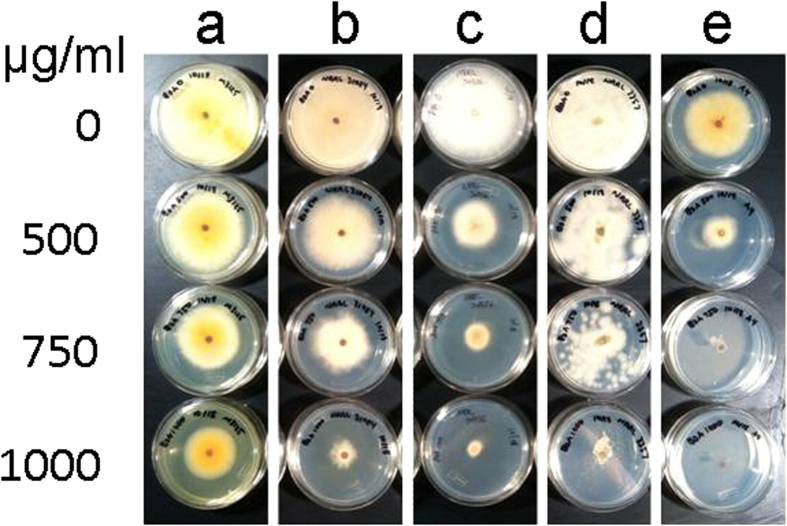
The results of the enzymatic assays measuring endogenous N-malonyltransferase activity are reflected in the ability of the ascomycetes to tolerate BOA in solid culture media. F. verticillioides grew effectively on media with up to 1 mg/ml BOA, a toxic concentration for other fungi. F. graminearum also tolerated BOA, but radial growth was slower and toxicity was evident at 1 mg/ml concentration of the compound. F. oxysporum and A. flavus, on the other hand, were sensitive to BOA, with delayed growth evident on media with lower concentrations of the compound. Consistent with our observations above, growth of N-malonyltransferase deficient A. nidulans was much reduced at the lowest concentration of BOA, and the higher concentrations completely inhibited growth (Fig. 5).
Figure 5. Fungal tolerance of 2-benzoxazolinone.
F. verticillioides (G. moniliformis) strain FGSC 7600 (a), F. graminearum (G. zeae) strain PH-1 (b), F. oxysporum f.sp. lycopersici strain FOL 4287 (c), A. flavus strain NRRL 3357 (d) and A. nidulans (E. nidulans) strain FGSC A4 (e) were grown on standard agar medium supplemented with up to 1000 μg/ml of 2-benzoxazolinone. Cultures are shown after 5 days of incubation.
Discussion
We have investigated 16 NAT loci from five filamentous ascomycetes, including four plant pathogens and three mycotoxigenic fungi of agricultural importance. Three of those loci are apparently non-functional, while the remaining 13 loci were demonstrated to comprise ORFs generating recombinant proteins with size and deduced amino acid sequences characteristic of prokaryotic and eukaryotic NAT homologues. Earlier studies of eukaryotic NAT genes have been confined mainly to humans and model mammals, where NAT ORFs are always contained in a single exon, although upstream non-coding exons have been experimentally characterized and alternative splicing in the 5′-untranslated region has been observed24,25. Segmented NAT ORFs, demonstrated experimentally here, have been computationally predicted previously for certain fungi, protists and lower chordates10,11.
Specificity of our 13 fungal NAT proteins was elucidated by enzymatic activity assays with nine different acyl-CoAs as donor substrates, followed by screens against a panel of arylamine and arylhydrazine acceptor substrates. To our knowledge, this dataset represents the largest assemblage of comparative specific activity analyses ever presented for NAT enzymes, revealing unique specificity profiles and supported by DSF experiments. We propose that fungal NATs may be divided into at least three biochemically distinct groups (I-III) of homologues, depending on their acyl-CoA specificity and postulated function. Such classification is exactly reflected in our previous phylogenetic analysis10, which indicated distinct, well-supported lineages of group I, II and III homologues. The group I homologues of A. flavus and A. nidulans formed a strongly supported lineage with other Aspergillus orthologues that is paraphyletic to the group I homologues of the three Fusarium species. Particularly interesting was the lineage of group II homologues, which appeared to include maize and wheat pathogens additional to Fusarium. Consistent with the biochemical analysis, the protein sequence phylogeny suggested that F. oxysporum NAT4 has diverged from the other Fusarium group III homologues. Lastly, although A. flavus NAT3 demonstrated biochemical characteristics of group II homologues, it appeared phylogenetically more related to group III. Continued investigation into these deviating homologues may enable a more accurate classification of their particular functions.
We demonstrate that the group II homologues of F. verticillioides and F. graminearum are N-malonyltransferases selectively utilizing malonyl-CoA to acylate arylamines, including the 2AP intermediate formed during hydrolysis and detoxification of BOA. To our knowledge, these are the first NAT isoenzymes conclusively shown to have specificity for malonyl-CoA instead of the typical acetyl-CoA. F. oxysporum and A. flavus also appear to possess a NAT homologue preferentially binding malonyl-CoA, but N-malonyltransferase activity was low with arylamine in preparations of both recombinant proteins and cell lysates. Upon xenobiotic exposure, F. verticillioides demonstrated substantial induction of arylamine N-malonyltransferase activity, reflecting the ability of this fungus to readily overcome BOA concentrations which are toxic to other fungi. F. graminearum, on the other hand, appeared to rely on its substantial, albeit non-inducible, arylamine N-malonyltransferase activity to manage delayed growth on BOA. In contrast, F. oxysporum and A. flavus were both more sensitive to BOA, presumably due to their low endogenous arylamine N-malonyltransferase activity, although other possible dysfunctions in their postulated BOA detoxification pathway cannot be ruled out. A. nidulans lacks a group II NAT homologue and was more sensitive to BOA than the other fungi. Experiments designed to accurately quantify the effects of xenobiotics on NAT gene transcriptional expression are currently underway, expanding the present biochemical analysis.
From this and our previous studies9, it is evident that group II NAT homologues have diverged to serve the purpose of Fusarium endophytic survival in the BOA-enriched environment of cereal plant tissues. Other ascomycetes, with less specific host associations, exhibited lower N-malonyltransferase activity (as in the case of non-endophytic pathogens F. oxysporum and A. flavus) or completely lacked the corresponding NAT gene (as in the case of A. nidulans). Interference with the BOA detoxification pathway, either via deletion of the NAT1 gene9,26 or co-culture with antagonistic Bacillus bacteria27, has been shown to compromise the BOA tolerance of endophytic Fusaria infecting cereal plants. Furthermore, a recent study26 demonstrates reduction in virulence of the wheat pathogen F. pseudograminearum, upon deletion of its NAT1 orthologue. NAT1 inhibition might thus represent a novel strategy to control maize and wheat pathogens, and available panels of NAT inhibitors16 could be relevant.
All five fungi studied were shown to possess one group I homologue with preference for acetyl-CoA, as expected by an archetypal NAT enzyme. Consistent with previous observations for Podospora anserina20, these homologues exhibited relatively low activity with established pharmaceutical substrates of NAT enzymes, but were highly active towards toxic substituted anilines, including 3,4DCA, a derivative compound of commonly applied agrochemicals and the subject of several bioremediation studies28,29. In cell lysates of our fungi, particularly F. graminearum, N-acetyltransferase activity was readily detectable with arylamine, but was apparently non-inducible upon challenge with BOA and 3,4DCA. Collectively, these data imply a role for group I N-acetyltransferases distinct from group II N-malonyltransferases. Moreover, the apparent constitutive expression of arylamine N-acetyltransferase activity in both challenged and unchallenged cultures suggests that group I NAT homologues may facilitate endogenous metabolism in a manner independent of xenobiotic acylation.
There are various hypotheses as to the possible endogenous roles of NAT enzymes. In human and transgenic mice, certain NAT homologues have been investigated for possible involvement in folate metabolism30,31. In actinobacteria, some NAT genes have been localized within operons regulating cholesterol catabolism to propionyl-CoA32,33, an acyl-group donor substrate of NATs, as shown by this and previous studies23,34. In tuberculous mycobacteria, the genes co-localizing with NAT are essential for survival of the pathogen on cholesterol and in host macrophage35, while NAT gene deletion in Mycobacterium bovis BCG compromises mycolic acid biosynthesis and cell wall integrity36. Here, we show that propionyl-CoA is employed as acyl-group donor substrate exclusively by group I NAT homologues of all five fungi examined. In contrast, group II and III homologues were essentially inactive towards propionyl-CoA, although group III homologues appeared to non-selectively bind the compound.
Propionyl-CoA is generated in microbial cells through oxidation of odd-chain fatty acids or catabolism of valine, isoleucine, and methionine. Uptake of propionate from environmental sources can also produce propionyl-CoA via thioesterification. Excess accumulation of propionyl-CoA is toxic to cells, due to inhibition of several primary metabolic enzymes, and propionate is commonly used as a food preservative to suppress microbial growth37. Additionally, propionyl-CoA accumulation in fungi has been shown to inhibit the biosynthesis of polyketide-derived secondary metabolites, such as ochratoxin, sterigmatocystin, and spore pigment37,38,39,40. Inhibition of polyketide formation is likely due to an intracellular imbalance of the ratio of propionyl-CoA to acetyl-CoA. For degradation of propionyl-CoA, fungi utilize the methylcitrate cycle, in which methylcitrate synthase condenses propionyl-CoA with oxaloacetate to generate methylcitrate and, ultimately, compounds used in primary metabolic pathways41. As speculated42, plant pathogenic fungi likely encounter carbon sources during plant infection that result in propionyl-CoA generation. Metabolic profiling of soybean roots inoculated with Fusarium tucumaniae showed pathogen-induced accumulation of valine and other amino acids, at early time points in the infection43. These data are consistent with the quantification of dipeptides (composed of branched-chain amino acids, predominantly leucine and isoleucine) in Arabidopsis roots, where they were abundant with significantly higher amounts in the epidermis and endodermis compared with other root cell types44. Degradation of these dipeptides would provide an abundance of isoleucine and valine amino acids, which could result in surplus accumulation of propionyl-CoA in a fungal pathogen infecting the roots. Given their dual preference for acetyl-CoA and propionyl-CoA, group I NAT homologues could function to alleviate toxicity when propionyl-CoA is in excess. This hypothesis could be tested using the methylcitrate synthase deficient A. nidulans mutant developed by Brock and colleagues41. This strain suffers from elevated intracellular levels of propionyl-CoA, resulting in toxicity and reduced mycotoxin production38,39. Loss of group I NAT homologues might have similar effects. We are currently pursuing development of genetically engineered strains of F. verticillioides, to investigate the effects of NAT gene inactivation or overexpression on cell viability, physiology and secondary metabolism. We speculate that group I NAT enzymes may act to maintain a balanced propionyl-CoA to acetyl-CoA ratio within cells.
Our screen of recombinant NAT proteins suggested a third group of homologues in plant-associated fungi, with essentially no activity towards any of the acyl-CoAs tested. However, these group III homologues were readily expressed and purified in substantial yields as soluble proteins in E. coli. Moreover, DSF analysis demonstrated their ability to non-selectively bind acetyl-, malonyl-, propionyl- and succinyl-CoA, the only deviation being the NAT4 of F. oxysporum which exhibited group I-like acyl-CoA binding specificity but no enzyme activity. Transcription of the corresponding genes was also verified by PCR from fungal cDNA. We propose that group III NAT homologues may have an as yet uncharacterized function.
Functional divergence of NATs has been reported before in the filamentous actinomycete Amycolatopsis mediterranei, where one NAT homologue (encoded by RifF locus of the rifamycin biosynthetic gene cluster) performs the final cyclization step of rifamycin and its consequent release from the polyketide synthase (PKS). This is an unexpected reaction for NAT, both in terms of excessive size of accommodated substrate and apparent acyl-CoA independence of the enzyme45. Prokaryotic filamentous actinomycetes demonstrate immense potential for secondary metabolism46 and our database searches suggest that many of their sequenced representatives (particularly Streptomycetes) harbour multiple NAT genes in their genome. This is very similar to the pattern observed for eukaryotic filamentous ascomycetes, where NAT genes are well-represented in sequenced genomes of Pezizomycotina (particularly endophytic and soil fungi with diverse metabolic capabilities), but absent in Saccharomycotina yeasts10,11. A bioinformatics analysis comparing Pezizomycotina vs. Saccharomycotina genomes indicates that genes present in the former and absent in the latter group of ascomycetes are primarily relevant to plant biomass decomposition, secondary metabolism and amino acid (particularly valine, leucine and isoleucine) degradation47. This potentially implicates Pezizomycotina NATs in xenobiotic detoxification, acetyl-CoA/propionyl-CoA homeostasis and/or secondary metabolism.
There is no solid evidence whether NAT genes might be part of polyketide biosynthetic gene clusters in the five ascomycetes examined. One recent study has placed NAT3 of A. flavus in a putative biosynthetic gene cluster predicted to be responsible for production of a polyketide that is some type of pigment48. The cluster has been experimentally studied in A. oryzae, but the role of NAT3 (aoiL locus in the study) is not clear49. The most similar orthologue of the PKS gene defining the postulated NAT3-containing cluster in Aspergillus is found on the gene cluster responsible for production of the red pigment bikaverin in Fusarium50. The bikaverin cluster does not bear any NAT orthologue; however, recent bioinformatics analyses suggest that certain NAT genes in Fusarium are contained within subtelomeric chromosomal regions which are known to be highly variable and harbour genes involved in secondary metabolism and adaptation of fungi to environmental stimuli51. Biosynthesis of polyketide-type secondary metabolites proceeds via iterative condensation and chain elongation, based on carbon units typically delivered by acetyl-CoA and malonyl-CoA52, again suggesting a possible role of NATs as mediators of intracellular acyl-CoA homeostasis.
This investigation provides insight into the distribution and function of NAT enzymes in mycotoxigenic fungi of global concern for agriculture and the food industry53. Recent comparative genomic analyses indicate that the full functional diversity and potential of many microbial proteins has not yet been demonstrated, as laboratory culture conditions provide limited opportunity for discovery of complex biochemical pathways related to secondary metabolism and cell adaptation to variable environmental stresses or sources of nutrients54. We believe such investigations may be relevant to fungal NAT enzymes and propose research directions stemming from comparative investigations of different groups of homologues.
Methods
Fungal strains
Fungi used in the study are described below, along with the strains examined (synonymous strain identifiers in parentheses), host organisms and geographical location of origin8. Fusarium verticillioides (teleomorph Gibberella moniliformis), sequenced strain FGSC 7600 (JFL A00149/FRC M3125/NRRL 20956), maize, California, USA55; strain JFL A00999 (FGSC 7603/ATCC 201261/FRC M3703/NRRL 20984), maize, Indiana, USA; strain MRC 826 (FRC M1325/NRRL 13447), maize, South Africa. Fusarium graminearum (teleomorph Gibberella zeae), sequenced strain PH-1 (NRRL 31084/ATCC MYA-4620/FGSC 9075), wheat, Michigan, USA56. Fusarium oxysporum f.sp. lycopersici, sequenced strain FOL 4287 (NRRL 34936/CBS 123668/FGSC 9935), tomato, Spain55. Aspergillus flavus, sequenced strain NRRL 3357 (ATCC 200026/FGSC A1120/JCM 12722/SRRC 167), maize and peanut, USA57. Aspergillus nidulans (teleomorph Emericella nidulans), sequenced strain FGSC A4 (ATCC 38163/CBS 112.46/NRRL 194/M139), Glasgow soil sample58.
Culture conditions and preparation of fungal cell extracts
Cultures (50 ml) were initiated from frozen stocks (−80 oC, 15% v/v glycerol), and the fungi were grown in media and at temperatures generally regarded as standard conditions for the respective genera. Starter cultures of Fusarium were grown in potato dextrose broth (Fluka) for 3 days at 25 oC in the dark (200 rpm). Starter cultures of Aspergillus were grown in complete medium (Fluka) for 24 h at 37 oC in the dark (200 rpm). Twenty flasks (each with 50 ml of appropriate medium), per experiment, were subsequently inoculated with 1 ml (Fusarium) or 6–7 ball-like clumps of hyphae (Aspergillus) from the starter cultures, and the new cultures were incubated under the same conditions. Half of the flasks then received a mixture of xenobiotics (fresh preparations of BOA and 3,4DCA, each compound added to the liquid culture at 25 μg/ml final concentration) and were incubated for 2 h, with cultures then placed on ice for immediate harvest. Cultures without xenobiotics were treated in exactly the same way for use as controls. Both xenobiotic-amended and control cultures contained 1% v/v ethanol. The multiple flasks of fungal growth for each treatment were combined together, centrifuged (6,000 xg, 10 min, 4 oC) to remove most of the medium, and the fungal pellet was filtered (Whatman 41 ashless filter paper) under vacuum. The recovered cell paste was flash-frozen and ground in liquid nitrogen, using mortar and pestle. Cell extracts were used to isolate DNA, total RNA or total soluble protein, as described in the sections that follow. Fusarium was also grown on potato dextrose agar solid medium at 25 oC, and Aspergillus on agar minimal medium at 37 oC (both media from Fluka), with or without xenobiotic (0–1 mg/ml BOA). For short-term induction experiments, the time of exposure (2 h) and the concentration of xenobiotics (50 μg/ml total) were empirically determined, so that the endogenous NAT activities could be quantified without toxic effects on fungal cells. In contrast, for agar-based tolerance assays, higher doses (up to 1 mg/ml) of xenobiotic were justified, to the level of toxicity, in order to distinguish tolerant from sensitive fungi.
Characterization of fungal NAT genes
The sequences of putative fungal NAT genes were retrieved from genomic databases and computationally annotated as described previously10. Isolation of nucleic acids was carried out from cell extracts, using the DNeasy (for DNA) or RNeasy (for RNA) Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA isolation procedure included on-column treatment with deoxyribonuclease I to eliminate DNA. The preparations were assessed on a Nanodrop ND-1000 spectrophotometer (Thermo Scientific) and by microfluidic analysis of RNA integrity (RNA integrity numbers of 6–8) on an Agilent 2100 Bioanalyzer (Agilent Technologies). Complementary DNA was generated from 1 μg/reaction of RNA, using AMV reverse transcriptase (Roche) and oligo [dT]16 primer. Mock reactions without reverse transcriptase were performed to confirm that RNA preparations were devoid of genomic DNA. High-fidelity Pfu-DNA polymerase (Finnzymes) and combinations of primers shown in Supplementary Table 1 were used to PCR-amplify fungal NAT genes from genomic DNA or cDNA template, followed by direct sequencing (GATC Biotech, Germany) of the products with the same primers. Validated genomic and transcribed NAT sequences were aligned with the BioEdit Sequence Alignment Editor 7.0.5.3, in order to determine the exon-intron structure of each gene. All fungal NAT sequences were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena). Gene symbols are according to the recommendations of the Arylamine N-acetyltransferase Gene Nomenclature Committee59, with specific details for naming fungal NATs provided in the Supplementary Methods.
Cloning of fungal NAT genes
Amplified products of the complete ORFs of fungal NAT genes (generated from cDNA or, in the case of intronless NAT genes, from genomic DNA) were A-tailed at the 3′-ends with Taq-DNA polymerase, then ligated to EcoRV-digested and T-tailed pGEM-5zf(+) cloning vector. Following transformation into E. coli JM109 competent cells, colonies were PCR-screened for insert orientation with vector-specific forward primer (T7-promoter) and the appropriate gene-specific reverse primer (Supplementary Table 1). Clones passing this test were sequenced from both ends with vector-specific primers (T7-promoter and SP6-promoter). For recombinant protein expression, the intronless ORFs of fungal NAT genes were recovered from validated pGEM-5zf(+) constructs, by cutting the vector at restriction sites flanking the beginning (SacII) and the end (NotI) of each insert. The excised fragments were then ligated to a NdeI-SacII adaptor (Supplementary Table 1), providing the NdeI-compatible end required for in-frame incorporation of the SacII/NotI-digested inserts into NdeI/NotI-digested pET28b(+) recombinant expression vector (Novagen). The adaptor allowed expression of NAT proteins with N-terminal hexa-histidine tags. For fungal NAT ORFs without NdeI sites, a more direct approach was used to transfer each insert from the pGEM-5zf(+) to the pET28b(+) vector, involving PCR with primers (Supplementary Table 1) designed to incorporate NdeI and NotI restriction sites at the beginning and the end of each amplified product, respectively. Ligation products with pET28b(+) vector were initially transformed into the JM109 strain and recombinant colonies were validated by sequencing with vector specific primers (T7-promoter and T7-terminator). The constructs were then transformed into E. coli BL21(DE3)pLysS competent cells and clones used for recombinant protein expression were again verified by sequencing with the same primers. Unless otherwise stated, all cloning reagents were from Promega. Sequencing of plasmid DNA was carried out by GATC Biotech or the USDA-ARS Eastern Regional Research Center Integrated Biomolecular Resources (Wyndmoor, PA, USA).
Recombinant expression-purification of fungal NAT proteins
Recombinant expression of fungal NAT proteins was carried out by modification of the protocol described for human NAT218 [ http://www.thesgc.org/structures/2pfr], and the details are provided in the Supplementary Methods.
Enzyme activity assays with recombinant proteins
Measurement of enzymatic release of CoA during NAT-catalyzed reactions was performed as described60 and the specific details are provided in the Supplementary Methods.
Enzyme activity assays with cell extracts
After grinding of filtered culture pellets, fungal cell extracts were prepared in buffer (1 ml per 50 ml culture) consisting of 20 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol (added fresh) and 1x protease inhibitors (Thermo/Pearce). Homogenous suspensions were generated through vigorous shaking, and the insoluble and soluble fractions were separated by centrifugation at 40,000 xg (30 min, 4 oC), followed by a second centrifugation (20,000 xg, 40 min, 4 oC) of the supernatant, to eliminate any remaining insoluble material. All manipulations were performed on ice and the supernatants were stored in −80 oC in aliquots that were allowed to thaw only once for immediate use. Quantification of total soluble protein in the preparations was performed against a series of bovine serum albumin standards (0–1 mg/ml), using the Bradford reagent (Sigma-Aldrich). Measurement of enzymatic conversion of arylamine during NAT-catalyzed reactions was subsequently performed as previously described60, and the specific details are provided in the Supplementary Methods.
Differential scanning fluorimetry
DSF was performed essentially as described60, each 20 μl reaction containing 3 μg of glycerol-free purified recombinant fungal NAT protein, or protein with 0.4 mM acyl-CoA (acetyl-, n-propionyl-, malonyl- or succinyl-CoA). Reactions were performed in duplicate, with SyproOrange (Invitrogen) in 20 mM Tris-HCl (pH 7.5), 0.5% v/v dimethyl sulphoxide (DMSO). Fluorescence was monitored on a 7500 real-time thermocycler (Applied Biosystems), and the generated sigmoid curves were fitted to the Boltzmann equation to accurately calculate Tm values. Tm peaks of proteins were determined via calculation of the derivative of generated thermal profiles for each set of conditions. Data analysis was performed using software OriginPro 8 SR0 (OriginLab).
Compounds used in enzyme assays
All compounds were purchased from Sigma-Aldrich. The following acyl-CoA compounds (5 mM in water) were tested in NAT enzymatic activity assays: acetyl-CoA sodium salt, n-propionyl-CoA lithium salt, malonyl-CoA lithium salt, succinyl-CoA sodium salt, butyryl-CoA lithium salt hydrate, 2-butenoyl-CoA lithium salt, glutaryl-CoA lithium salt, hexanoyl-CoA trilithium salt hydrate and octanoyl-CoA lithium salt hydrate. The tested arylamines and arylhydrazines (100 mM in DMSO, except hydralazine which is water-soluble) are described with abbreviations and PubChem ID numbers in the Supplementary Methods.
Additional Information
How to cite this article: Karagianni, E. P. et al. Homologues of xenobiotic metabolizing N-acetyltransferases in plant-associated fungi: Novel functions for an old enzyme family. Sci. Rep. 5, 12900; doi: 10.1038/srep12900 (2015).
Supplementary Material
Acknowledgments
This work was supported by the Bodossaki Foundation, Greece. For work at Oxford (U.K.), S.B. and E.P.K. were supported by a 2008 UNESCO-L’Oréal National Award for Women in Science and an EU-FP7 Coordination & Support Action “Capacities” REGPOT-2008-1 programme (acronym “BioStrength”). For work at Athens (GA, U.S.A.), S.B. was funded by a 2012 Fulbright-Schuman Research Scholarship. Work of E.K. and B.K. was funded by a Democritus University Internship Programme (Ministry of Education Operational Programme “Education and Lifelong Learning”), co-funded by the European Union (European Social Fund) and the Greek State (National Strategic Reference Framework 2007–2013). T.T. is recipient of graduate scholarships from the Onassis Foundation and the State Scholarships Foundation of Greece. We thank Dr. Ali Ryan, Professor Robert Sim and Dr. Areej Abuhammad (University of Oxford, U.K.) for helpful advice on protein expression/purification and enzyme activity assays. We are also grateful to Dr. Bogos Agianian and Dr. Giannoulis Fakis (Democritus University of Thrace, Greece) for useful discussions during the project and key comments on the manuscript.
Footnotes
Author Contributions S.B. and A.E.G. designed the study and wrote the paper; E.P.K. and S.B. characterized and cloned the genes, and optimized conditions for recombinant protein expression/purification; E.K. and S.B. expressed and purified proteins to perform enzymatic activity assays (including data analysis); B.K. and S.B. expressed and purified proteins to perform differential scanning fluorimetry (including data analysis); S.B., B.D. and A.E.G. optimized and implemented the enzyme assays with fungal cell extracts and analyzed the data; B.D. and A.E.G. grew cultures and provided nucleic acid preparations; T.T. provided assistance with gene cloning, protein expression/purification, enzyme activity assays and differential scanning fluorimetry; V.G. provided assistance with enzyme activity assays; E.S. provided important intellectual input and resources during optimization of recombinant protein expression/purification procedures and enzyme activity assays.
References
- Van De Wouw A. P. & Howlett B. J. Fungal pathogenicity genes in the age of ‘omics’. Mol Plant Pathol 12, 507–514, 10.1111/j.1364-3703.2010.00680.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A. Natural products and plant disease resistance. Nature 411, 843–847, 10.1038/35081178 (2001). [DOI] [PubMed] [Google Scholar]
- Lah L. et al. The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification. Mol Microbiol 81, 1374–1389, 10.1111/j.1365-2958.2011.07772.x (2011). [DOI] [PubMed] [Google Scholar]
- Sexton A. C., Minic Z., Cozijnsen A. J., Pedras M. S. & Howlett B. J. Cloning, purification and characterisation of brassinin glucosyltransferase, a phytoalexin-detoxifying enzyme from the plant pathogen Sclerotinia sclerotiorum. Fungal Genet Biol 46, 201–209, 10.1016/j.fgb.2008.10.014 (2009). [DOI] [PubMed] [Google Scholar]
- Glenn A. E., Gold S. E. & Bacon C. W. Fdb1 and Fdb2, Fusarium verticillioides loci necessary for detoxification of preformed antimicrobials from corn. Mol Plant Microbe Interact 15, 91–101, 10.1094/mpmi.2002.15.2.91 (2002). [DOI] [PubMed] [Google Scholar]
- Frey M. et al. Analysis of a chemical plant defense mechanism in grasses. Science 277, 696–699 (1997). [DOI] [PubMed] [Google Scholar]
- Frey M., Schullehner K., Dick R., Fiesselmann A. & Gierl A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70, 1645–1651, 10.1016/j.phytochem.2009.05.012 (2009). [DOI] [PubMed] [Google Scholar]
- Glenn A. E., Hinton D. M., Yates I. E. & Bacon C. W. Detoxification of corn antimicrobial compounds as the basis for isolating Fusarium verticillioides and some other Fusarium species from corn. Appl Environ Microbiol 67, 2973–2981, 10.1128/aem.67.7.2973-2981.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn A. E. & Bacon C. W. FDB2 encodes a member of the arylamine N-acetyltransferase family and is necessary for biotransformation of benzoxazolinones by Fusarium verticillioides. J Appl Microbiol 107, 657–671, 10.1111/j.1365-2672.2009.04246.x (2009). [DOI] [PubMed] [Google Scholar]
- Glenn A. E., Karagianni E. P., Ulndreaj A. & Boukouvala S. Comparative genomic and phylogenetic investigation of the xenobiotic metabolizing arylamine N-acetyltransferase enzyme family. FEBS Lett 584, 3158–3164, 10.1016/j.febslet.2010.05.063 (2010). [DOI] [PubMed] [Google Scholar]
- Martins M., Dairou J., Rodrigues-Lima F., Dupret J. M. & Silar P. Insights into the phylogeny or arylamine N-acetyltransferases in fungi. J Mol Evol 71, 141–152, 10.1007/s00239-010-9371-x (2010). [DOI] [PubMed] [Google Scholar]
- Chou T. C. & Lipmann F. Separation of acetyl transfer enzymes in pigeon liver extract. J Biol Chem 196, 89–103 (1952). [PubMed] [Google Scholar]
- Saito K., Shinohara A., Kamataki T. & Kato R. N-hydroxyarylamine O-acetyltransferase in hamster liver: identity with arylhydroxamic acid N,O-acetyltransferase and arylamine N-acetyltransferase. J Biochem 99, 1689–1697 (1986). [DOI] [PubMed] [Google Scholar]
- McDonagh E. M. et al. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genomics 24, 409–425, 10.1097/fpc.0000000000000062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein D. W. N-acetyltransferase 2 genetic polymorphism: effects of carcinogen and haplotype on urinary bladder cancer risk. Oncogene 25, 1649–1658, 10.1038/sj.onc.1209374 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim E., Fakis G., Laurieri N. & Boukouvala S. Arylamine N-acetyltransferases—from drug metabolism and pharmacogenetics to identification of novel targets for pharmacological intervention. Adv Pharmacol 63, 169–205, 10.1016/b978-0-12-398339-8.00005-7 (2012). [DOI] [PubMed] [Google Scholar]
- Sinclair J. C., Sandy J., Delgoda R., Sim E. & Noble M. E. Structure of arylamine N-acetyltransferase reveals a catalytic triad. Nat Struct Biol 7, 560–564, 10.1038/76783 (2000). [DOI] [PubMed] [Google Scholar]
- Wu H. et al. Structural basis of substrate-binding specificity of human arylamine N-acetyltransferases. J Biol Chem 282, 30189–30197, 10.1074/jbc.M704138200 (2007). [DOI] [PubMed] [Google Scholar]
- Vagena E., Fakis G. & Boukouvala S. Arylamine N-acetyltransferases in prokaryotic and eukaryotic genomes: a survey of public databases. Curr Drug Metab 9, 628–660 (2008). [DOI] [PubMed] [Google Scholar]
- Martins M. et al. An acetyltransferase conferring tolerance to toxic aromatic amine chemicals: molecular and functional studies. J Biol Chem 284, 18726–18733, 10.1074/jbc.M109.015230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocaign A. et al. Biotransformation of Trichoderma spp. and their tolerance to aromatic amines, a major class of pollutants. Appl Environ Microbiol 79, 4719–4726, 10.1128/aem.00989-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen F. H., Berglund H. & Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2, 2212–2221, 10.1038/nprot.2007.321 (2007). [DOI] [PubMed] [Google Scholar]
- Kawamura A. et al. Eukaryotic arylamine N-acetyltransferase. Investigation of substrate specificity by high-throughput screening. Biochem Pharmacol 69, 347–359, 10.1016/j.bcp.2004.09.014 (2005). [DOI] [PubMed] [Google Scholar]
- Boukouvala S., Price N., Plant K. E. & Sim E. Structure and transcriptional regulation of the Nat2 gene encoding for the drug-metabolizing enzyme arylamine N-acetyltransferase type 2 in mice. Biochem J 375, 593–602, 10.1042/bj20030812 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouvala S. & Sim E. Structural analysis of the genes for human arylamine N-acetyltransferases and characterisation of alternative transcripts. Basic Clin Pharmacol Toxicol 96, 343–351, 10.1111/j.1742-7843.2005.pto_02.x (2005). [DOI] [PubMed] [Google Scholar]
- Kettle A. J. et al. Degradation of the benzoxazolinone class of phytoalexins is important for virulence of Fusarium pseudograminearum towards wheat. Mol Plant Pathol, 10.1111/mpp.12250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C. W., Hinton D. M., Glenn A. E., Macias F. A. & Marin D. Interactions of Bacillus mojavensis and Fusarium verticillioides with a benzoxazolinone (BOA) and its transformation product, APO. J Chem Ecol 33, 1885–1897, 10.1007/s10886-007-9347-5 (2007). [DOI] [PubMed] [Google Scholar]
- Carvalho G. et al. Biological treatment of propanil and 3,4-dichloroaniline: kinetic and microbiological characterisation. Water Res 44, 4980–4991, 10.1016/j.watres.2010.08.006 (2010). [DOI] [PubMed] [Google Scholar]
- Castillo J. M., Nogales R. & Romero E. Biodegradation of 3,4 dichloroaniline by fungal isolated from the preconditioning phase of winery wastes subjected to vermicomposting. J Hazard Mater 267, 119–127, 10.1016/j.jhazmat.2013.12.052 (2014). [DOI] [PubMed] [Google Scholar]
- Laurieri N. et al. From arylamine N-acetyltransferase to folate-dependent acetyl CoA hydrolase: impact of folic acid on the activity of (HUMAN)NAT1 and its homologue (MOUSE)NAT2. PLoS One 9, e96370, 10.1371/journal.pone.0096370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher N. J. & Minchin R. F. Arylamine N-acetyltransferase 1: a novel drug target in cancer development. Pharmacol Rev 64, 147–165, 10.1124/pr.110.004275 (2012). [DOI] [PubMed] [Google Scholar]
- Anderton M. C. et al. Characterization of the putative operon containing arylamine N-acetyltransferase (nat) in Mycobacterium bovis BCG. Mol Microbiol 59, 181–192, 10.1111/j.1365-2958.2005.04945.x (2006). [DOI] [PubMed] [Google Scholar]
- Van der Geize R. et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA 104, 1947–1952, 10.1073/pnas.0605728104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack N. A. et al. Temperature stability of proteins essential for the intracellular survival of Mycobacterium tuberculosis. Biochem J 418, 369–378, 10.1042/bj20082011 (2009). [DOI] [PubMed] [Google Scholar]
- Ouellet H., Johnston J. B. & De Montellano P. R. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol 19, 530–539, 10.1016/j.tim.2011.07.009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakta S. et al. Arylamine N-acetyltransferase is required for synthesis of mycolic acids and complex lipids in Mycobacterium bovis BCG and represents a novel drug target. J Exp Med 199, 1191–1199, 10.1084/jem.20031956 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M. & Buckel W. On the mechanism of action of the antifungal agent propionate. Eur J Biochem 271, 3227–3241, 10.1111/j.1432-1033.2004.04255.x (2004). [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Brock M. & Keller N. P. Connection of propionyl-CoA metabolism to polyketide biosynthesis in Aspergillus nidulans. Genetics 168, 785–794, 10.1534/genetics.104.027540 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Q. & Keller N. P. Blockage of methylcitrate cycle inhibits polyketide production in Aspergillus nidulans. Mol Microbiol 52, 541–550, 10.1111/j.1365-2958.2004.03994.x (2004). [DOI] [PubMed] [Google Scholar]
- Maerker C., Rohde M., Brakhage A. A. & Brock M. Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J 272, 3615–3630, 10.1111/j.1742-4658.2005.04784.x (2005). [DOI] [PubMed] [Google Scholar]
- Brock M., Fischer R., Linder D. & Buckel W. Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol Microbiol 35, 961–973 (2000). [DOI] [PubMed] [Google Scholar]
- Domin N., Wilson D. & Brock M. Methylcitrate cycle activation during adaptation of Fusarium solani and Fusarium verticillioides to propionyl-CoA-generating carbon sources. Microbiology 155, 3903–3912, 10.1099/mic.0.031781-0 (2009). [DOI] [PubMed] [Google Scholar]
- Scandiani M. M. et al. Metabolic profiles of soybean roots during early stages of Fusarium tucumaniae infection. J Exp Bot 66, 391–402, 10.1093/jxb/eru432 (2015). [DOI] [PubMed] [Google Scholar]
- Moussaieff A. et al. High-resolution metabolic mapping of cell types in plant roots. Proc Natl Acad Sci USA 110, E1232–1241, 10.1073/pnas.1302019110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss H. G. & Yu T. W. Lessons from the rifamycin biosynthetic gene cluster. Curr Opin Chem Biol 3, 592–597 (1999). [DOI] [PubMed] [Google Scholar]
- Liu G., Chater K. F., Chandra G., Niu G. & Tan H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol Mol Biol Rev 77, 112–143, 10.1128/mmbr.00054-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvas M. et al. Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina. BMC Genomics 8, 325, 10.1186/1471-2164-8-325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K. C. & Mack B. M. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins (Basel) 6, 1916–1928, 10.3390/toxins6061916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T. et al. Overexpressing transcriptional regulator in Aspergillus oryzae activates a silent biosynthetic pathway to produce a novel polyketide. Chembiochem 13, 855–861, 10.1002/cbic.201200107 (2012). [DOI] [PubMed] [Google Scholar]
- Wiemann P. et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog 9, e1003475, 10.1371/journal.ppat.1003475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Waalwijk C., De Wit P. J., Tang D. & Van Der Lee T. Relocation of genes generates non-conserved chromosomal segments in Fusarium graminearum that show distinct and co-regulated gene expression patterns. BMC Genomics 15, 191, 10.1186/1471-2164-15-191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A. & Sherman D. H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet 24, 37–66, 10.1146/annurev.ge.24.120190.000345 (1990). [DOI] [PubMed] [Google Scholar]
- Wu F. Mycotoxin risk assessment for the purpose of setting international regulatory standards. Environ Sci Technol 38, 4049–4055 (2004). [DOI] [PubMed] [Google Scholar]
- Brakhage A. A. & Schroeckh V. Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet Biol 48, 15–22, 10.1016/j.fgb.2010.04.004 (2011). [DOI] [PubMed] [Google Scholar]
- Ma L. J. et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373, 10.1038/nature08850 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo C. A. et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317, 1400–1402, 10.1126/science.1143708 (2007). [DOI] [PubMed] [Google Scholar]
- Rokas A. et al. What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud Mycol 59, 11–17, 10.3114/sim.2007.59.02 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E. et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115, 10.1038/nature04341 (2005). [DOI] [PubMed] [Google Scholar]
- Hein D. W., Boukouvala S., Grant D. M., Minchin R. F. & Sim E. Changes in consensus arylamine N-acetyltransferase gene nomenclature. Pharmacogenet Genomics 18, 367–368, 10.1097/FPC.0b013e3282f60db0 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirka T., Boukouvala S., Agianian B. & Fakis G. Polymorphism p.Val231Ile alters substrate selectivity of drug-metabolizing arylamine N-acetyltransferase 2 (NAT2) isoenzyme of rhesus macaque and human. Gene 536, 65–73, 10.1016/j.gene.2013.11.085 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.