Abstract
The response regulator OtpR is critical for the growth, morphology and virulence of Brucella melitensis. Compared to its wild type strain 16 M, B. melitensis 16 MΔotpR mutant has decreased tolerance to acid stress. To analyze the genes regulated by OtpR under acid stress, we performed RNA-seq whole transcriptome analysis of 16 MΔotpR and 16 M. In total, 501 differentially expressed genes were identified, including 390 down-regulated and 111 up-regulated genes. Among these genes, 209 were associated with bacterial metabolism, including 54 genes involving carbohydrate metabolism, 13 genes associated with nitrogen metabolism, and seven genes associated with iron metabolism. The 16 MΔotpR also decreased capacity to utilize different carbon sources and to tolerate iron limitation in culture experiments. Notably, OtpR regulated many Brucella virulence factors essential for B. melitensis intracellular survival. For instance, the virB operon encoding type IV secretion system was significantly down-regulated, and 36 known transcriptional regulators (e.g., vjbR and blxR) were differentially expressed in 16 MΔotpR. Selected RNA-seq results were experimentally confirmed by RT-PCR and RT-qPCR. Overall, these results deciphered differential phenomena associated with virulence, environmental stresses and cell morphology in 16 MΔotpR and 16 M, which provided important information for understanding the detailed OtpR-regulated interaction networks and Brucella pathogenesis.
Brucella spp. is a group of facultative intracellular bacteria1. The virulence of Brucella depends on their ability to survive in professional and non-professional phagocytes. The intracellular niche of phagocytes contains various harsh environments like nutrition deprivation, low-pH conditions, and low-oxygen tension2,3,4. Brucella use several transcription regulators, including OtpR5, NtrYX6, and Rsh7, to activate a series of interacting signal networks in response to these environmental stresses. Among these transcription regulators, our studies first found that Brucella OtpR (BMEI0066) regulates stress responses, cell growth and cell morphology of Brucella5,8. Brucella OtpR (BMEI0066) is a cytoplasmic protein that shows significant similarity to the OmpR subfamily with two conserved domains: a signal receiver domain with a phosphoacceptor site and an effector domain with DNA-binding activity9. Our previous experiments demonstrated that 16 MΔotpR, an otpR mutant of the virulent B. melitensis strain 16 M, was avirulent in mice and had a reduced capacity to invade phagocytic cells8. Compared with its parental strains, 16 MΔotpR displayed an unusual, irregular deformation of the cell envelope8. These findings suggested that OtpR could regulate the cell morphology and cell growth. The expression of some cell division–associated proteins, including FtsQ, is reduced in 16 MΔotpR as compared with the parental strain8. However, the breadth of genes and the gene networks that are regulated by OtpR are still unclear.
One major mechanism of Brucella pathogenesis is the capacity of virulent Brucella surviving in an acidic environment inside macrophages10,11. Porte et al. found that early acidification of phagosomes containing B. suis is essential for intracellular survival inside macrophages4. The pH in the phagosomes containing live B. suis decreased to values of pH 4.0 ± 0.5. At 1 h postinfection, the phagosome was already acidic and remained acidic for at least 5 h. An early neutralization of vacuolar pH in fact inhibits the survival of B. suis inside macrophages4. Brucella is able to resist well to an acidic condition of pH 3.2 for several hours inside macrophages12. Similarly, virulent B. abortus also survives well inside an acidic intracellular condition13. Boschiroli et al. found that transcription of Brucella virB operon, which encodes for the Type IV secretion system (T4SS), was induced specifically within macrophages, and the phagosome acidification is a key intracellular signal inducing virB expression14. B. melitensis was found to induce a specific set of proteins (e.g., DnaK) in response to acidic pH15. Similar findings have also been reported in other bacteria. For example, Salmonella typhimurium activates virulence gene transcription within acidified macrophages16. These studies indicate that low pH acts as an intracellular signal on the regulation genes involved in survival and multiplication within phagocytic cells4,16. However, the detailed mechanism of the regulatory gene network of Brucella under acidic environment is still unclear.
Our previous study has observed that acid stress induces an approximately 1.5-fold increase in OtpR expression, suggesting that OtpR is activated under acid stress. 16 MΔotpR has a significantly reduced capacity in response to acid stress5. As described above, OtpR is also critical to regulate cell morphology and cell growth and several proteins (e.g., FtsQ). In addition, many other proteins such as T4SS proteins14, DnaK15, VjbR17, and Hfq18, are also regulated at acidic conditions. Therefore, to further define the OtpR-regulated Brucella pathogenesis mechanism, we hypothesized that OtpR plays a major role as an important transcription regulator in regulating Brucella genes critical for intracellular survival under an acidic condition. Our previous study has observed that acid stress induces an approximately 1.5-fold increase in OtpR expression, suggesting that OtpR is activated under acid stress. 16 MΔotpR has a significantly reduced capacity in response to acid stress5. As described above, we also found that OtpR is critical to regulate cell morphology and cell growth and several proteins (e.g., FtsQ). In addition, many other proteins (e.g., T4SS proteins, DnaK) were also regulated at acidic conditions (see the description above). Therefore, to further define the OtpR-regulated Brucella pathogenesis mechanism, we hypothesized that OtpR play a major role as an important transcription regulator in regulating Brucella genes critical for intracellular survival under an acidic condition. To address this hypothesis, it would be ideal to use a high throughput technology to detect and compare the gene expression profiles of 16 MΔotpR and its parental strain 16 M under an acid stress. Through high efficient sequencing of complementary DNAs (cDNAs) that are reverse transcribed from RNAs, the RNA-seq technology has many advantages compared to the microarray technology in whole-genome gene expression analysis19,20. RNA-seq does not require probe sequences and also has a greater dynamic range for measuring very low or very high gene expression levels21. The power of RNA-seq has been demonstrated in the transcriptomics studies for Brucella22,23,24 and many other bacteria19,20,21. Therefore, we used RNA-seq in current study. The results provided fundamental gene-level evidence and detailed gene expression profiles regulated by OtpR in Brucella.
Materials and Methods
Bacteria strains
Bacterial strains used in the present study were B. melitensis 16 M, B. melitensis 16 MΔotpR, 16McΔotpR. Strain 16 M is a commonly used, virulent, wild type B. melitensis strain. 16 MΔotpR is the otpR mutant of 16 M that has an avirulent phenotype. 16McΔotpR is the virulent complementation strain of 16 MΔotpR. Both 16 MΔotpR and 16McΔotpR were generated in our laboratory and previously reported8.
Bacterial growth and RNA preparation
Brucella 16 M and 16 MΔotpR were grown with 100 mL of Tryptic Soy Broth (TSB; BD; final pH = 7.3) in a 500-mL water-bath shaker (180 rpm) at 37 °C until early-log phase (OD600 ≅ 0.6 − 0.7). The acid treatment experiment followed the same protocol as previously reported5. Under this protocol, the cells were treated with the same TSB medium but with an acidic condition (pH 3.4 − 4.4)5. After the treatment, the cell cultures were collected and centrifuged. After the centrifugation, the supernatants were removed, and the RNA protect Bacteria Reagent (Qiagen, Hilden, Germany) was added to the pellets to prevent RNA degradation.
The B. melitensis RNAs for Solexa/Illumina sequence were isolated and purified with RNeasy Mini System (Qiagen, Hilden, Germany). RNA was eluted from the column using RNase-free water. Total RNA was incubated with DNase (Ambion, Foster City, CA) and then purified using two phenol-chloroform extractions and one chloroform extraction. RNA was resuspended in RNAase free TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0; Ambion). The purity and integrity of RNA was assessed using the 2100 Bioanalzyer (Agilent Technologies, Palo Alto, CA, USA). B. melitensis mRNA was enriched by removal of 16 S and 23 S rRNA from two 5 μg aliquots of total RNA using a MicrobExpress Bacterial mRNA purification Kit (Ambion). As ≤ 5 μg total RNA was treated per reaction, a separate enrichment reaction was performed for each RNA sample to enrich the RNA volume for the subsequent experiments. The mRNA sample was assessed with the 2100 Bioanalyzer to confirm the reduction of 16 S and 23 S rRNAs prior to the preparation of cDNA fragment libraries.
cDNA library preparation and sequencing using the Illumina Genome Analyzer
The RNA was subjected to Solexa/Illumina sequencing at Beijing Genomics Institute. The cDNA library was constructed as previously25. Briefly, each mRNA sample was fragmented into short sequences with divalent cations and heat25. Using these short fragments as templates, the first-strand cDNA was synthesized with random hexamer primers and reverse transcriptase (Invitrogen, Carlsbad, CA). The second-strand DNA was synthesized using RNase H (Invitrogen) and DNA polymerase I (New England Biolabs, Beverly, MA, USA), respectively. The amplified fragments were purified with QiaQuick PCR Purification kit (Qiagen, Hilden, Germany) and resolved with EB buffer for the end preparation and poly (A) addition. Individual paired-end libraries for each sample were constructed and loaded onto independent flow cells. Sequencing was carried out by running 35 cycles on the Illumina HiSeq 2000 platform.
Raw 90-bp sequence data were generated using the Illumina Genome Analyzer II system. All sequences were examined for possible sequencing errors. The raw sequence data was filtered by removing reads that contained adaptor sequences, consisted of >5% ambiguous residues (Ns), or had the majority base quality of <5. The raw data have been submitted to the National Center for Biotechonology Information-Gene Expression Omnibus (NCBI GEO) database http://www.ncbi.nlm.nih.gov/geo/). The Accession ID is GSE48165.
RNA-seq alignment and identification of transcribed and annotated CDS
To increase the quality of the reads, the raw reads with the length of 90-bp each were trimmed to 75 bp after quality evaluation using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The trimmed reads were aligned with the B. melitensis 16 M genome (NC_003317 and NC_003318) and annotated gene sets obtained from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/) using the Short Oligonucleotide Analysis Package (SOAP)26. cDNAs with matches to the reference genome of >80% were retained for further analysis. Those sequencing reads that matched annotated genes in the B. melitensis 16 M genome reflect the genes transcribed under the given experimental conditions. Gene expression was quantified as Reads Per Kilobase of coding sequence per Million reads RPKM algorithm27. A gene was considered to be differentially expressed if the difference in RPKM values between the two samples (16 M and 16 MΔotpR) was ≥2.0-fold (i.e., log2 ratio >1.0) and the p-value was <0.0527.
COG category and Pathway analysis of the otpR-dependent genes
The next generation sequencing method resulted in the identification of the transcription levels of genes in 16 MΔotpR and 16 M under acid stress. All possible otpR-dependent genes were identified using statistical methods as performed in a previous study with modifications28. COG annotations for the chosen genes were obtained from NCBI COG database (http://www.ncbi.nlm.nih.gov/COG/). The program OntoCOG was used for the COG enrichment test as previously described29.
Reverse Transcriptase-polymerase Chain Reaction (RT-PCR) and Quantitative Real-time PCR (RT-qPCR) analyses
To confirm the RNA-Seq results, 28 up- or down-regulated genes from the RNA-Seq analysis were selected, and RT-PCR and RT-qPCR were carried out to confirm the gene expression changes on these 28 genes. PCR primers were designed using Primer 5.0 software (Primer-E Ltd., Plymouth, United Kingdom) and are listed in Supplemental Data 1. The same experimental protocols were used to culture both wild type 16 M and 16 MΔotpR and extract RNA samples. The immunofluorescence analysis was performed with SYBR Green Master Mix (Applied Biosystems, Foster City, CA) using the 7500 Real Time PCR System (Applied Biosystems) as previously described8. Relative gene expression was calculated by the 2–∆∆Ct method8. All reactions were carried out in triplicates.
Stress challenge assays
To monitor extracellular growth under limited nutrition, the minimal medium (0.5% lactic acid, 3% glycerol, 0.75% NaCl, 1% K2HO4, 0.01% Na2S2O3·5H2O, 10 μg/ml Mg++, 0.1 μg/ml Fe++, 0.1 μg/ml Mn++, 0.21 μg/ml thiamine·HCl, 0.2 μg/ml nicotinic acid, 0.04 μg/ml calcium pantothenate, 0.001 μg/ml biotin, 5 mg/ml glutamate; pH 6.8–7.0 with NaOH) was inoculated with 106 colony forming units (CFU)/ml of the 16 MΔotpR, 16 M, or 16McΔotpR strains. The cultures were incubated at 37 °C, and growth was monitored based on the OD600.
To assess the survival of 16 MΔotpR in iron-limited medium, the bacteria were grown in TSB medium with a range of concentrations of the Fe2+chelator 2,2`-dipyridyl (DIP; Sigma-Aldrich, Shanghai, China) with an initial density of 3.0 × 107 CFU/ml30. CFUs were determined at 24, 48, and 72 h after inoculation. All assays were performed in triplicates.
Statistical analysis
The differences between the means of gene expressions for the experimental and control groups were analyzed by the Student’s unpaired t-test (equal sample sizes, equal variance) using SPSS 18.0. For the RNA-seq study, the P-values with the FDR (False Discovery Rate) multi-test adjustment were used to determine the differential expressed genes in the experimental groups compared to the control groups. The FDR P-value ≤0.001 and the absolute value of log2Ratio ≥1 (i.e., 2-fold change) were used as the thresholds to identify the genes showing statistically significant gene expression changes. For the RT-PCR study, P-value <0.05 was considered as statistically significant. The Student’s unpaired t-test was also used to analyze the bacterial survival rates under a stress condition.
Results
OtpR differentially regulated 501 genes in B. melitensis
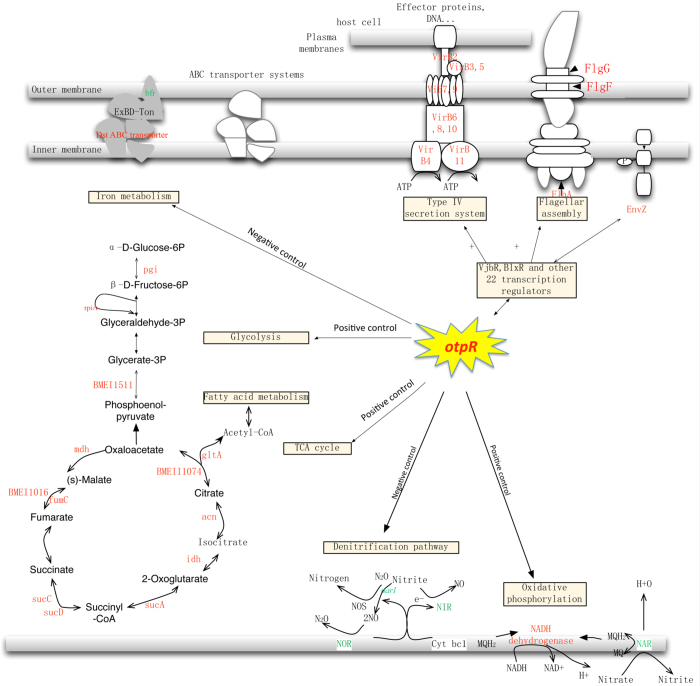
To detect all the possible genes regulated by OtpR during acid stress, the Next-Generation Sequencing (NGS) technology was used to sequence the whole transcriptomic profiles of 16 MΔotpR and its wild-type strain 16 M. The raw sequence output of the two strain transcriptomes included 150 million reads in total. Approximately 50% reads were perfectly matched to the reference genome B. melitensis 16 M. Based on the genomic alignment, our analysis determined the expression of 3,163 genes in each strain. In total, 501 genes in B. melitensis were identified to be differentially expressed in OtpR (Fig. 1 and Supplemental Data 2). Among these genes, 390 genes were down-regulated and 111 genes were up-regulated in 16 MΔotpR compared to the 16 M control. Most of these differentially expressed genes were associated with carbohydrate metabolism (10.78%), energy metabolism (7.39%), amino acid metabolism (6.19%), nucleotide metabolism (4.59%), lipid metabolism (1.80%), membrane transport (7.39%) and transcription (7.19%) (Fig. 1).
Figure 1. Functional categories of the differentially expressed genes in the otpR mutant as compared with the wild-type strain.
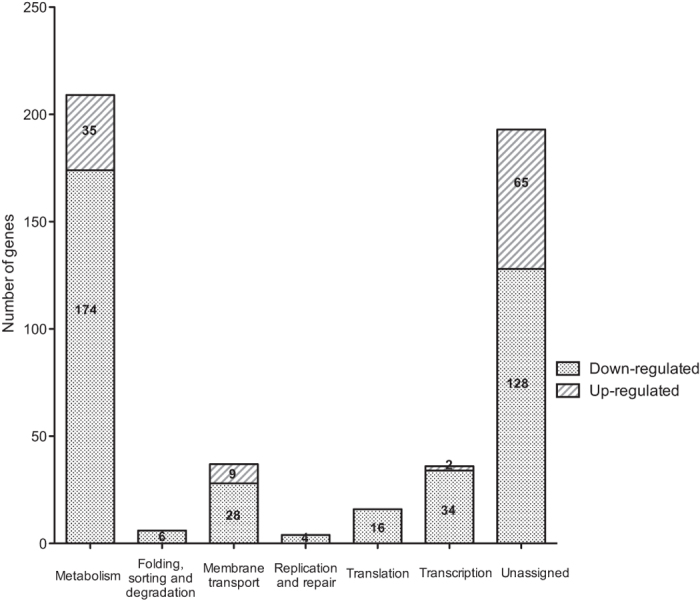
Only genes that were up- or down-regulated by ≥2.0-fold are shown.
It is noted that the traditional experiments typically determine relevant expression levels of the genes using internal housekeeping gene control (e.g., β-actin) to normalize the results31,32,33. However, the next-generation sequencing (NGS), including RNA-seq, counts the absolute numbers of sequence reads mapped to the genomes34,35. After the counting, the gene expression quantification was measured using the Reads Per Kilobase of coding sequence per Million reads (RPKM) algorithm27. With the RPKM algorithm, there is no need to have an internal control as typically seen in many traditional RT-PCR or microarray experiments. By comparing the expression values between the two samples (16 MΔotpR vs 16 M control), we were able to identify which genes were significantly regulated by OtpR under the same experimental condition.
More specific analysis results of these up- and down-regulated genes are described below.
OtpR regulates Brucella cell division and cell envelope generation
Our sequencing analysis found OtpR regulates many genes directly involved in the cell division cycle. For example, three filamentous temperature sensitive genes ftsK (BMEII0742), ftsQ (BMEI0582), and ftsZ (BMEI0585) were down-regulated in the otpR mutant 16 MΔotpR. These genes encode for three cell division proteins FtsK, FtsQ, and FtsZ36. FtsK acts as a bifunctional protein: its C-terminal domain facilitates segregation of chromosome dimers and its N-terminal may acts in the developing septum36. FtsQ (BMEI0582) is a highly conserved protein of the bacterial divisome, which is critical in linking the upstream and downstream cell division proteins to form the divisome37. The GTP-binding protein FtsZ is the key factor in the initiation of cell division by the formation of a ring-shaped structure38. In addition, our study also found that OtpR up-regulated intracellular septation protein BMEI0130.
Fatty acids participate in a number of cellular processes, most importantly in generating the cell envelope. Five genes for fatty-acid biosynthesis (BMEI1180; BMEI1473; fabG, BMEII0514; BMEI1521; BMEI1522; fadD, BMEI1632; cfa, BMEI1484) were down-regulated in the 16 MΔotpR, suggesting that OtpR up-regulates these five fatty-acid biosynthesis genes.
In addition to fatty-acid biosynthesis genes, OtpR regulates many other genes directly involving cell envelope protein generation, assembly, transport, and structure. In Gram-negative bacteria, lipoproteins are one of the most abundant proteins anchored to the outer membrane through the lipids, which regulates the bacteria-host interaction and intracellular survival39. Compared to strain 16 M, the lipoprotein oprf (BMEII0036) was 2.25-fold down-regulated in strain 16 MΔotpR. The down-regulated lipoprotein might lead to the modification of the cell surface proteins40. Two chaperone proteins GroES and GroEL were detected to be down-regulated in 16 MΔotpR. These chaperones mediated the protein folding and could stimulate an immune response of T cells41. The GroESL homologues belong to a family of selective stress proteins during the intracellular growth, which could be induced by many stress stimuli including acid shock, heat shock, or oxidative injury42,43. Other genes participating in cell envelope protein generation or transport, including an ABC transporter substrate binding protein (BMEI1954), apbE (BMEII1010), and bactoprenol glucosyl transferase (BMEII1101), were also down-regulated in 16 MΔotpR compared to its wild type control.
In 16 MΔotpR, eleven genes associated with ribosomal proteins were down-regulated. Ribosomal proteins are critical for protein production, cell replication, and bacterial growth.
OtpR regulates carbon, nitrogen, and energy metabolism in B. melitensis
The transcriptome analysis indicated that many genes associated with carbon and energy metabolism were significantly down-regulated in the otpR mutant under an acid stress. Most interestingly, these included twelve genes involved in the tricarboxylic acid (TCA) cycle (mdh, BMEI0137; sucD, BMEI0138; BMEI0139; sucA, BMEI0140; BMEI0791; gltA, BMEI0836; BMEI0855; BMEI0856; class I fumarate hydratase, BMEI1016; acn, BMEI1855; fumC, BMEII1051; and citrate lyase beta chain, BMEII1074; Fig. 2). The TCA cycle is critical for carbon metabolism and energy generation. The pyruvate metabolism supplies energy to living cells through the TCA cycle when oxygen is present (aerobic respiration), and alternatively through fermentation when oxygen is lacking44. Several genes relating the pyruvate metabolism were down-regulated in the otpR mutant, including mdh (BMEI0137), FAD-linked oxidase (BMEI0599), pdhB (BMEI0855), and aceF (BMEI0856). Furthermore, the entire NADH dehydrogenase operon was down-regulated in 16 MΔotpR. The genes encoding the cytochrome D ubiquinol oxidase subunits I, II, and III (BMEII0759, BMEII0760, BMEI1899, BMEI1900, BMEI1901) were all down-regulated in 16 MΔotpR. The NADH dehydrogenase operon and cytochrome D ubiquinol oxidase subunits participate in the oxidative phosphorylation, an important metabolic process for electron transport and energy release45.
Figure 2. Basic metabolism regulated by Brucella OtpR identified in transcriptome analysis.
Up-regulated genes in 16 MΔotpR are shown in green and down-regulated genes in 16 MΔotpR are shown in red.
We also found that the expression levels of five genes associated with nitrogen metabolism (npd, BMEII0460; narI, BMEII0953; BMEII0952; nirV, BMEII0987; norF, BMEII1000; and norE, BMEII1001) were altered in 16 MΔotpR under acid stress. Meanwhile, two genes (BMEII0952, BMEII0953) participating in the denitrification pathway were up-regulated (Supplemental Data 2). Brucella applies denitrification metabolism to generate energy at a low-oxygen condition in an intracellular niche inside host macrophages46.
To further investigate the importance of OtpR in regulating carbon and nitrogen metabolisms, we used a defined minimal medium that contains only carbon and nitrogen nutrients (without amino acids and growth factors). The minimal medium was used to separately culture parental strain 16 M, 16 MΔotpR, and the mutant complementing strain 16 McΔotpR, followed by the measuring of their dynamic growth profiles. All these three strains were able to grow in the minimal medium, indicating that the inorganic carbon and nitrogen resources provide sufficient nutrients for Brucella growth and replication. Compared to 16 M, the mutant 16MΔotpR showed reduced growth at the late log phase (Fig. 3). The phenomenon suggested that OtpR was important to sustain regular cell growth through the regulation of the carbon and nitrogen metabolism. The observation was further confirmed by the complementation of the gene in the mutant as shown by the full recovery of the cell growth in 16 McΔotpR (Fig. 3).
Figure 3. Growth curves of the parental B. melitensis strain 16 M, its otpR mutant 16 MΔotpR, and the mutant complementation strain 16 McΔotpR in a synthetic minimal medium.
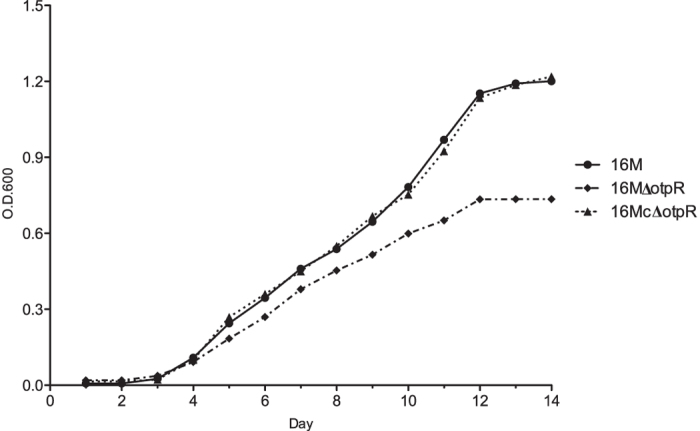
The minimal medium contains only carbon and nitrogen as the nutrient resources. Compared to the wild type strain, the otpR mutant had a decreased OD600 value. The gene complementation of the otpR mutant resumed the OD600 level. A curve of the optical density values at OD600 determined at several time points reflects the growth dynamics of a bacterial strain in the culture medium over the different time points.
OtpR regulates iron metabolism in B. melitensis
Our transcriptomics analysis also found that OtpR regulates many genes in iron metabolism (Supplemental Data 2). Compared to the 16 M, 16MΔotpR mutant presented down-regulation of two ABC transporter systems. One ABC transporter system includes an ATP-binding protein DstD (BMEII0604, ATP/GTP-binding site-containing protein A) and a permease DstE (BMEII0606, ferric anguibactin transport system permease protein). This Dst protein–dependent ABC transporter is responsible for the utilization of iron by B. melitensis in low-iron medium47. Both dstD and dstE were also down-regulated in 16 MΔotpR. The other ABC transporter system is the TonB-ExbB-ExbD complex that is critical to transport iron-siderophore complexes into bacterial cell. The TonB system is associated with 2, 3-dihydroxybenzoic acid assimilation in B. melitensis and allows adaptation to low-iron medium47. The expressions of both exbB (BMEI0365) and exbD (BMEI0366) were down-regulated in 16 MΔotpR. Several other OtpR-regulated iron-related genes include bfr (BMEII0704, bacterioferritin), BMEII0584 (iron-binding periplasmic protein), BMEII0607 (ferric anguibactin-binding protein), irrf2 (BMEII0707, RrF2 family protein), and fecD (BMEII0536, Fe3+dicitrate transport system permease protein fecd).
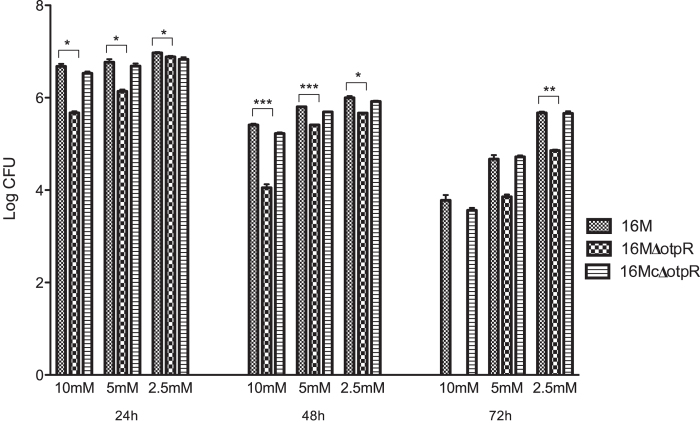
To confirm that OtpR regulates iron metabolism, the tolerance of 16 MΔotpR under an experimental condition of low iron was assessed after adding varying concentrations of the Fe2+ chelator DIP into the medium. In the presence of 2.5 mM, 5.0 mM, or 10 mM DIP, the survival capability of the mutant strain 16 MΔotpR was less than its parental strain 16 M (Fig. 4), suggesting that OtpR is critical to the utilization of iron in the low iron medium. The otpR gene complementation of 16 MΔotpR recovered the bacterial survival probably due to the recovered function of OtpR in the iron uptake. These results suggest that although the tolerance of 16 McΔotpR to low-iron medium was similar to that of 16 M, the otpR mutant appeared to affect longer-term survival in iron-limited medium.
Figure 4. Survival of B. melitensis 16 MΔotpR, 16 McΔotpR, and the wild-type strain under iron-limited conditions.
The survival of the strains was measured in the TSB medium containing 2.5 mM, 5.0 mM and 10.0 mM Fe2+ chelator 2,2`-dipyridyl. The measurements were taken at 24 h, 48 h and 72 h after inoculation. Experiments were performed in triplicates and a significant difference was observed. Statistical analysis was performed by comparing the CFUs of 16 M versus 16 MΔotpR bacteria at different time points using the Student’s t-test. p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***) represents different levels of significant differences. It is noted that in the presence of 10.0 mM DIP, the otpR mutant could be detected at 24 and 48 h after inoculation, but not at 72 h. When the CFU/mL was determined, 200 μL of culture sample was used in each condition, which equals to a detection limit of 5 CFU/ml (or ~0.7 LOG CFU). It is possible that there were still some viable otpR mutant cells survived in the iron-limited medium at 72 h; however, the level of survival was below the laboratory’s detection limit.
OtpR regulates the expression of many known Brucella virulence factors and regulators
Many Brucella virulence-related genes were differentially expressed in 16 MΔotpR under acid stress. All of the 12 Type IV secretion system genes in the virB operon were down-regulated in 16MΔotpR (fold change > 4; Fig. 2 and Supplemental Data 2). This system is critical for the translocation of Brucella effectors to the host for trafficking into macrophages. As compared with the wild type, three genes associated with flagellar assembly, flgG (coding for flagellar basal-body components in the distal portion of the rod), flhA (encoding a protein of the flagellar type III export apparatus), and flgF (coding for flagellar basal-body components), were down-regulated in the otpR mutant (Fig. 2).
Among the genes down-regulated in the otpR mutant under acid stress are 34 known transcriptional regulators including two quorum sensing regulators (Supplemental Data 2). The two quorum sensing regulators VjbR and BlxR may directly regulate specific biological processes in Brucella48,49,50. In addition, two other transcriptional regulators, PhoP and NorS, were up-regulated in the otpR mutant.
RT-qPCR validates the RNA-Seq results of selected B. melitensis genes
To validate the data generated from the RNA-seq experiment, we repeated the acid induction experiment and used RT-qPCR assays to detect transcript levels of 22 genes that were down-regulated in 16 MΔotpR and of 6 genes that were up-regulated in 16 MΔotpR. Out of the 501 differentially expressed genes detested by our RNA-Seq analysis, these 28 genes were selected based on three criteria: (i) Gene function. We selected one or two genes that were differentially expressed from each functional group, e.g., BMEI0655 belonging to ABC transporter system, BMEI1153 involved to oxidative phosphorylation, BMEI1325 belonging to a two-component system, and BMEII0704 associated with cell division. (ii) Virulence factor role. We purposely chose many important virulence-related genes out of the 501 gene list, such as BMEII0025 (virb1) and BMEII0035 (virb11) (T4SS components), and BMEII1116 (vjbR) (a quorum sensing-dependent transcriptional regulator). (iii) Gene position in the genome. These chosen 28 genes are located in different positions in the genome.
The mRNA levels of these 28 genes as determined by RT-qPCR were in good accordance with those from the RNA-Seq analysis (Table 1). Together, these results support the model that OtpR is critical in regulating Brucella virulence.
Table 1.
| B. melitensisORF | Gene name, predicted function | 2−ΔΔCt |
|---|---|---|
| BMEI0040 | gslc,glutamate synthase [nadph] large chain | 0.034 |
| BMEI0655 | ABC transporter ATP-binding protein | 0.014 |
| BMEI0747 | lsp,lsu ribosomal protein l10p | 0.022 |
| BMEI1016 | aerobic,fumarate hydratase class i | 0.012 |
| BMEI1153 | NADH-quinone oxidoreductase chain f | 0.019 |
| BMEI1325 | Sensory transduction protein kinase | 0.039 |
| BMEI1464 | pif, protoheme ix farnesyltransferase | 0.014 |
| BMEI1652 | Urease alpha subunit | 0.022 |
| BMEI1758 | blxR, transcriptional activator | 0.043 |
| BMEI1855 | aconitate hydratase | 0.014 |
| BMEII0025 | virb1, type IV secretion system | 0.011 |
| BMEII0035 | virb11, type IV secretion system | 0.026 |
| BMEII0071 | gntR, transcriptional regulator | 0.011 |
| BMEII0245 | usp, universal stress protein family | 0.043 |
| BMEII0409 | osmop, Osmotically inducible protein c | 0.016 |
| BMEII0531 | fusB,fusaric acid resistance protein | 0.018 |
| BMEII0704 | Bacterioferritin | 0.029 |
| BMEII0742 | ftsK, cell division protein | 0.015 |
| BMEII0953 | Respiratory nitrate reductase 2 gamma chain | 0.027 |
| BMEII1047 | groES,10 kDa chaperonin groES | 0.013 |
| BMEII1051 | Fumarate hydratase c | 0.046 |
| BMEII1114 | flhB, flagellar biosynthetic protein | 0.013 |
| BMEII1116 | vjbR, transcriptional activator | 0.023 |
| BMEI1355 | Hypothetical Cytosolic Protein | 10 |
| BMEI1751 | Two component response regulator | 4.62 |
| BMEII0477 | Uronate isomerase | 3.58 |
| BMEI1384 | Transcriptional regulator, arac family | 8.972 |
| BMEI1766 | Sulfite reductase (ferredoxin) | 2.223 |
Validation of twenty-eight otpR-independent genes identified by RNA-Seq analysis. A 2–∆∆Ct value >1 indicates that the gene was overexpressed in the otpR mutant, and a value of <1 indicates that the gene was expressed at a lower level in the mutant.
Discussion
Our RNA-seq study found that under acidic stress, OtpR regulated 501 genes associated with many important functions, including metabolism, membrane transport, transcription, regulation, translation, and DNA replication and repair51,52,53,54. Many environmental stresses, such as heat and oxygen limitation, may affect the expression of genes associated with these functions15. However, the specific mechanisms of the gene regulations on these functions are unclear. Our results provide evidence to show that OtpR is an important Brucella regulator that regulates metabolism processes and bacterial virulence under acidic stress.
The identification of the critical role of OtpR in regulating a large number of genes involving metabolic processes expands our understanding of the gene in Brucella and possibly other bacteria (Fig. 2). Our previous study shows that Brucella OtpR regulates cell growth and cell morphology8. The OtpR homologue in Caulobacter crescentus, CenR, is also found to be important in regulating bacterial growth and cell cycle progression55. However, previous studies did not show the generic regulatory mechanism of OtpR in regulating the bacterial cell growth and cell cycle progression8. This study found some OtpR-regulated genes associated with cell cycle progression, and the maintenance of cell morphology. The iron acquisition within the host cell influences the capacity of Brucella to survive in a host10,56. This study first showed that OtpR regulated the iron metabolism. Compared to the parental strain and the complementation strain of 16 MΔotpR, 16 MΔotpR had a reduced ability to survive in low-iron media. It suggests that OtpR plays an important role in B. melitensis to survive in low-iron media under acidic stress or in normal conditions.
Importantly, this study demonstrated that OtpR regulated many genes related to Brucella virulence. The entire virB operon and 34 transcriptional regulators were significantly down-regulated in the otpR mutant, suggests that OtpR positively regulated the expression of these genes. The virB operon can be induced by acid stimuli or phagosome acidification2. Its expression is directly regulated by VjbR, BvrR, IHF and HutC through promoter binding48,57,58,59. This study showed that the transcriptional regulators VjbR and BlxR were also markedly down-regulated in the otpR mutant, especially VjbR. VjbR and BlxR in Brucella are the two quorum sensing-related LuxR-type factors that regulate the transcription of other genes, including the virB operon and genes for flagellar and outer membrane components48,60,61. Our findings suggested that OtpR might indirectly regulate VirB genes through direct interaction between OtpR and VjbR. Considering that Brucella survives in an acidic environment inside macrophages10, it was likely that once inside macrophages, OtpR becomes activated and regulate these virulence factors.
Considering OtpR is important in Brucella metabolism regulation and virulence, further research is required to analyze the OtpR-mediated regulatory mechanisms. Structural analysis revealed that OtpR contains a phosphoacceptor site, which suggests that it might belong to a two-component regulator system. Our amino acid sequence analysis found that OtpR is highly homologous to that of CenR in Caulobacter crescentus. In C. Crescentus, CenR is the regulator of a two-component system CenR/CenS that senses and acts on various environmental stimuli55. The CenS is the sensor of the CenR/CenS two-component system. Although a genome sequence analysis identified a possible gene homologous to CenS, our studies found that the gene does not act like a sensor for OtpR. More investigation is still required to identify the sensor of the OtpR regulator. Since many genes identified to be regulated by OtpR, it might be possible to use bioinformatics and experimental methods to predict and identify the binding site(s) of OtpR. Another area of research is to identify how OtpR interacts with and regulates this large number of Brucella genes. Instead of up- or down-regulating a large number of genes simultaneously, it is more likely that OtpR regulates these many genes through one or more defined pathways in a time-dependent matter. In addition to the acidic stress condition, other experimental factors may also regulate the functions of OtpR. The eventual discovery of the detailed OtpR-regulated interaction networks will be critical to understand Brucella pathogenesis and will support the rational design of therapeutic drugs and preventive vaccines.
In conclusion, through comparative transcriptome analysis, differential expressions of many genes, involving the carbohydrate metabolism, nitrogen metabolism, and iron metabolism, were observed in 16 MΔotpR and its parental strain 16 M under acid stress. The results indicated that cell division proteins and iron metabolism could be regulated by OtpR, and several important virulence factors were also differentially expressed in 16 MΔotpR. For examples, virB operon was significantly down-regulated, and the genes encoding for 36 known transcriptional regulators, including quorum-sensing regulators VjbR and BlxR were also down-regulated. Selective RNA-seq results were experimentally verified, which further deciphered the different phenomena associated with virulence, environmental stresses and cell morphology in 16 MΔotpR and its parental strain 16 M. This study provided the important information for understanding the detailed OtpR-regulated interaction networks and Brucella pathogenesis.
Additional Information
How to cite this article: Liu, W. et al. RNA-seq reveals the critical role of OtpR in regulating Brucella melitensis metabolism and virulence under acidic stress. Sci. Rep. 5, 10864; doi: 10.1038/srep10864 (2015).
Supplementary Material
Acknowledgments
This study was in partial sponsored by National Basic Research Program of China, Program 973 (2010CB530202); the National Natural Science Foundation of China (30871882). The China Scholarship Council supported W.L.’s tenure at the University of Michigan for her bioinformatics training and data analysis. This project was also supported by a University of Michigan Medical School bridging fund to Y.H.
Footnotes
Author Contributions W.X.L. and Q.M.W. conceived and designed the experiments. W.X.L., H.D. and J.L. performed the experiments. W.X.L., Q.X.O., Y.J.L., X.L.W. and H.D. analyzed the data. Q.M.W. and Y.Q.H. contributed reagents/materials/analysis tools. Z.S.X. assisted in the usage of analysis tools and writing Perl scripts for data processing. W.X.L., Y.Q.H. and Q.M.W. participated in data interpretation. W.X.L. wrote the paper. Y.Q.H. and Q.M.W. edited the paper. All authors read and approved the final manuscript.
References
- DelVecchio V. G. et al. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci U S A 99, 443–448. (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschiroli M. L., Foulongne V. & O’Callaghan D. Brucellosis: a worldwide zoonosis. Curr Opin Microbiol 4, 58–64. (2001). [DOI] [PubMed] [Google Scholar]
- Phillips R. W. & Roop R. M. 2nd Brucella abortus HtrA functions as an authentic stress response protease but is not required for wild-type virulence in BALB/c mice. Infect Immun 69, 5911–5913 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte F., Liautard J. P. & Kohler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun 67, 4041–4047. (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren J., Li N., Liu W. & Wu Q. Disruption of the BMEI0066 gene attenuates the virulence of Brucella melitensis and decreases its stress tolerance. Int J Biol Sci 5, 570–577 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrica Mdel C., Fernandez I., Sieira R., Paris G. & Goldbaum F. A. The two-component systems PrrBA and NtrYX co-ordinately regulate the adaptation of Brucella abortus to an oxygen-limited environment. Mol Microbiol 88, 222–233 (2013). [DOI] [PubMed] [Google Scholar]
- Dozot M. et al. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol 8, 1791–1802 (2006). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. OtpR regulated the growth, cell morphology of B. melitensis and tolerance to beta-lactam agents. Vet Microbiol 159, 90–98 (2012). [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39, D225–229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop R. M. 2nd, Gaines J. M., Anderson E. S., Caswell C. C. & Martin D. W. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198, 221–238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atluri V. L., Xavier M. N., de Jong M. F., den Hartigh A. B. & Tsolis R. M. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol 65, 523–541 (2011). [DOI] [PubMed] [Google Scholar]
- Kulakov Y. K., Guigue-Talet P. G., Ramuz M. R. & O’Callaghan D. Response of Brucella suis 1330 and B. canis RM6/66 to growth at acid pH and induction of an adaptive acid tolerance response. Res Microbiol 148, 145–151 (1997). [DOI] [PubMed] [Google Scholar]
- Arenas G. N., Staskevich A. S., Aballay A. & Mayorga L. S. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect Immun 68, 4255–4263 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschiroli M. L. et al. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A 99, 1544–1549 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira-Gomes A. P., Cloeckaert A. & Zygmunt M. S. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis. Infect Immun 68, 2954–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpuche Aranda C. M., Swanson J. A., Loomis W. P. & Miller S. I. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A 89, 10079–10083 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arocena G. M., Zorreguieta A. & Sieira R. Expression of VjbR under nutrient limitation conditions is regulated at the post-transcriptional level by specific acidic pH values and urocanic acid. PloS one 7, e35394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M. et al. Impact of Hfq on global gene expression and intracellular survival in Brucella melitensis. PloS one 8, e71933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A. C., Melo-Barbosa H. P., Miyoshi A., Silva A. & Azevedo V. Application of RNA-seq to reveal the transcript profile in bacteria. Genet Mol Res 10, 1707–1718 (2011). [DOI] [PubMed] [Google Scholar]
- Kimbrel J. A., Di Y., Cumbie J. S. & Chang J. H. RNA-Seq for Plant Pathogenic Bacteria. Genes (Basel) 2, 689–705 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J. & Thomson N. R. Studying bacterial transcriptomes using RNA-seq. Curr Opin Microbiol 13, 619–624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Liu W., Peng X., Jing Z. & Wu Q. The effects of MucR on expression of type IV secretion system, quorum sensing system and stress responses in Brucella melitensis. Vet Microbiol 166, 535–542 (2013). [DOI] [PubMed] [Google Scholar]
- Rodriguez M. C. et al. Evaluation of the effects of erythritol on gene expression in Brucella abortus. PloS one 7, e50876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. et al. Deep-sequencing analysis of the mouse transcriptome response to infection with Brucella melitensis strains of differing virulence. PloS one 6, e28485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. et al. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC genomics 12, 451 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Liao J. M., Zeng S. X., Zhou X. & Lu H. Global effect of inauhzin on human p53-responsive transcriptome. PloS one 7, e52172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., X. Z., He Y. Towards a semantic web application: Ontology-driven ortholog clustering analysis. International Conference on Biomedical Ontologies (ICBO), University at Buffalo, NY, July 26-30, 2011. Full length paper. Page 33-40 (2011).
- Robey M. & Cianciotto N. P. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect Immun 70, 5659–5669 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M. A., Plikaytis B. B. & Shinnick T. M. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 184, 4025–4032 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchhardt J. et al. Normalization strategies for cDNA microarrays. Nucleic Acids Res 28, E47 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Git A. et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 16, 991–1006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev Genet 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. C. & Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol 42, 395–413 (2001). [DOI] [PubMed] [Google Scholar]
- van den Ent F. et al. Structural and mutational analysis of the cell division protein FtsQ. Mol Microbiol 68, 110–123 (2008). [DOI] [PubMed] [Google Scholar]
- Holtzendorff J. et al. Diel expression of cell cycle-related genes in synchronized cultures of Prochlorococcus sp. strain PCC 9511. J Bacteriol 183, 915–920 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Niesel D. W., Peterson J. W. & Klimpel G. R. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect Immun 66, 5196–5201 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H. et al. The role of a Brucella abortus lipoprotein in intracellular replication and pathogenicity in experimentally infected mice. Microb Pathog 54, 34–39 (2013). [DOI] [PubMed] [Google Scholar]
- Kim J. Y. et al. Immunoproteomics of Brucella abortus RB51 as candidate antigens in serological diagnosis of brucellosis. Vet Immunol Immunopathol (2014). [DOI] [PubMed] [Google Scholar]
- Abu Kwaik Y. & Engleberg N. C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress conditions. Mol Microbiol 13, 243–251 (1994). [DOI] [PubMed] [Google Scholar]
- Lin J., Adams L. G. & Ficht T. A. Immunological response to the Brucella abortus GroEL homolog. Infect Immun 64, 4396–4400 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol Rev 12, 165–178 (1993). [Google Scholar]
- Schafer G., Engelhard M. & Muller V. Bioenergetics of the Archaea. Microbiol Mol Biol Rev : MMBR 63, 570–620 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haine V., Dozot M., Dornand J., Letesson J. J. & De Bolle X. NnrA is required for full virulence and regulates several Brucella melitensis denitrification genes. J Bacteriol 188, 1615–1619 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese I. et al. The Ton system, an ABC transporter, and a universally conserved GTPase are involved in iron utilization by Brucella melitensis 16M. Infect Immun 72, 5783–5790 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M. F., Sun Y. H., den Hartigh A. B., van Dijl J. M. & Tsolis R. M. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol 70, 1378–1396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue R. M. et al. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol 7, 1151–1161 (2005). [DOI] [PubMed] [Google Scholar]
- Rambow-Larsen A. A., Rajashekara G., Petersen E. & Splitter G. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J Bacteriol 190, 3274–3282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow-Larsen A. A., Petersen E. M., Gourley C. R. & Splitter G.A. Brucella regulators: self-control in a hostile environment. Trends Microbiol 17, 371–377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier E., Pappas G. & Gorvel J. P. Virulence factors in brucellosis: implications for aetiopathogenesis and treatment. Expert Rev Mol Med 9, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- Xiang Z., Tian Y. & He Y. PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol 8, R150 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Analyses of Brucella Pathogenesis, Host Immunity, and Vaccine Targets using Systems Biology and Bioinformatics. Front. Cell. Infect. Microbiol 2, 2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker J. M., Prasol M. S., Perchuk B. S., Biondi E. G. & Laub M. T. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3, e334 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E. D. Microbial pathogens with impaired ability to acquire host iron. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine 13, 85–89 (2000). [DOI] [PubMed] [Google Scholar]
- Arocena G. M., Sieira R., Comerci D. J. & Ugalde R. A. Identification of the quorum-sensing target DNA sequence and N-Acyl homoserine lactone responsiveness of the Brucella abortus virB promoter. J Bacteriol 192, 3434–3440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieira R., Arocena G. M., Bukata L., Comerci D. J. & Ugalde R. A. Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters. J Bacteriol 192, 217–224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieira R., Comerci D. J., Pietrasanta L. I. & Ugalde R. A. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol Microbiol 54, 808–822 (2004). [DOI] [PubMed] [Google Scholar]
- Leonard S. et al. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J Bacteriol 189, 131–141 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzureau S. et al. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J Bacteriol 189, 6035–6047 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.