Abstract
Objective
Accurate laboratory testing is a critical component of dengue surveillance and control. The objective of this programme was to assess dengue diagnostic proficiency among national-level public health laboratories in the World Health Organization (WHO) Western Pacific Region.
Methods
Nineteen national-level public health laboratories performed routine dengue diagnostic assays on a proficiency testing panel consisting of two modules: one containing commercial serum samples spiked with cultured dengue viruses for the detection of nucleic acid and non-structural protein 1 (NS1) (Module A) and one containing human serum samples for the detection of anti-dengue virus antibodies (Module B). A review of logistics arrangements was also conducted.
Results
All 16 laboratories testing Module A performed reverse transcriptase polymerase chain reaction (RT–PCR) for both RNA and serotype detection. Of these, 15 had correct results for RNA detection and all 16 correctly serotyped the viruses. All nine laboratories performing NS1 antigen detection obtained the correct results. Sixteen of the 18 laboratories using IgM assays in Module B obtained the correct results as did the 13 laboratories that performed IgG assays. Detection of ongoing/recent dengue virus infection by both molecular (RT–PCR) and serological methods (IgM) was available in 15/19 participating laboratories.
Discussion
This first round of external quality assessment of dengue diagnostics was successfully conducted in national-level public health laboratories in the WHO Western Pacific Region, revealing good proficiency in both molecular and serological testing. Further comprehensive diagnostic testing for dengue virus and other priority pathogens in the Region will be assessed during future rounds.
Introduction
Dengue is a mosquito-borne viral infection associated with significant morbidity and mortality caused by any of four closely related virus serotypes (DENV-1,-2,-3 and −4), all of which circulate in the World Health Organization (WHO) Western Pacific Region.1,2 Dengue presentation is broad and non-specific, which may confound clinical diagnosis. The majority (~75%) of infections in humans are asymptomatic, but a small proportion of symptomatic patients develops severe dengue characterized by rapid progression into shock, severe bleeding and/or multiorgan impairment, which leads to death if unattended or mismanaged.2,3
In the Western Pacific Region, dengue outbreaks occur yearly in multiple countries, driven by a complex interplay of virus, vector and host biology, climatic and socioeconomic factors as well as international travel and trade.1,4–7 Different case definitions are used for dengue surveillance throughout the Region; some countries (e.g. Singapore, Australia) include only laboratory-confirmed cases, while others include all clinical diagnoses with only a subset (e.g. paediatric patients) being laboratory-confirmed. In 2013, outbreaks resulted in 44 098 dengue cases in the Lao People's Democratic Republic, 39 222 cases in Malaysia, 10 548 cases in New Caledonia and 22 170 cases in Singapore.8,9 Analysis of the outbreaks in Singapore, Malaysia and later Fiji (> 20 000 cases as of 22 April 2014) revealed DENV serotype switches from the previous year.10,11 Secondary heterotypic infection is believed to foreshadow larger numbers of dengue and severe dengue cases.12 Surveillance detection of a switch in the prevalent serotype within a population is thus cause for concern.
Accurate laboratory testing is a critical component of dengue surveillance and control. During the acute phase of infection, detection is targeted to DENV RNA and/or the virus non-structural protein 1 (NS1), while anti-DENV antibodies IgM and/or high titre IgG are the diagnostic targets in the convalescent phase. Several commercial diagnostic tests for dengue are available that detect DENV RNA or determine serotype using reverse transcription polymerase chain reaction (RT–PCR), or detect NS1, or IgG and IgM antibodies against the virus. A common mechanism used by laboratories to maintain accuracy and quality of diagnosis is external quality assessment (EQA) or proficiency testing, whereby an external agency distributes blinded samples to a laboratory for analysis and then verifies and reports the results. EQA can be used to compare laboratory performance, reveal potential problems associated with diagnostic kits or procedures, indicate areas in a laboratory requiring improvement and identify training needs.13
The WHO Regional Office for the Western Pacific recently launched an EQA for dengue diagnostics testing in 2013, under the Asia Pacific Strategy for Emerging Diseases (APSED) 2010.14 This EQA is based largely on the WHO EQA for influenza15 and uses proficiency testing to assess national-level public health laboratory performance in detecting DENV nucleic acid, NS1 antigen and anti-DENV antibodies using molecular and serological assays. It is proposed that it will be an annual exercise, free of charge or at low cost to the laboratories and with the gradual inclusion of other pathogens. As well as ensuring the accurate diagnosis of dengue, the EQA programme also links participating laboratories with international reference laboratories that can assist in more specialized diagnostics or analytical functions as required.
The objective of this manuscript is to summarize the first round of EQA of dengue diagnostics undertaken in the WHO Western Pacific Region in 2013.
Methods
Participating laboratories
Nineteen national-level public health laboratories from 18 countries and areas (two in Viet Nam) in the WHO Western Pacific Region where dengue is endemic or where imported cases have been detected were invited to participate in the EQA; all 19 agreed (Fig. 1). An EQA panel was dispatched to these laboratories between May and July 2013.
Fig. 1.
National-level public health laboratories that participated in EQA of dengue diagnostics, WHO Western Pacific Region, 2013
SAR, Special Administrative Region.
Preparation of EQA panel
The WHO Collaborating Centre for Reference and Research of Arbovirus and their Associated Vectors, located at the Environmental Health Institute of the National Environment Agency in Singapore, was selected as the EQA provider as it had the necessary technical expertise, access to samples and the required resources.
The panel for the 2013 EQA of dengue diagnostics consisted of two modules (A and B) containing serum with inactivated DENV (Module A) and serum samples from a dengue patient (Module B) (Table 1). All samples were heat-inactivated and contained no detectable HIV, hepatitis B surface antigen or hepatitis C virus antibody.
Table 1. Characteristics of modules used in EQA of dengue diagnostics, WHO Western Pacific Region, 2013.
| Module | Sample ID | Contents | Serotype | Antibodies |
|---|---|---|---|---|
| Viral RNA/NS1 antigen (Module A) | A2013-V01 | Inactivated dengue virus in serum | DENV-2 | – |
| A2013-V02 | Serum alone | Not applicable | – | |
| A2013-V03 | Inactivated dengue virus in serum | DENV-1 | – | |
| Antibody (Module B) | B2013-S01* | Convalescent serum | – | IgM, IgG |
| B2013-S02* | Convalescent serum | – | IgM, IgG | |
| B2013-S03 | Negative human serum | – | Negative control |
* B2013-S01 and B2013-S02 were the same sample collected from a recently recovered dengue patient used to assess the reproducibility of laboratory results.
ID, identification; NS1, non-structural protein 1.For Module A, samples A2013-V01 and A2013-V03 contained at least 106 RNA copies/mL of in vitro-cultured DENV of different serotypes – DENV-1 genotype III and DENV-2 cosmopolitan clade II, deposited in Genbank with accession numbers KP685233 and KP685236, respectively. These were diluted in pathogen-free human serum (SeraCare Life Sciences, Milford, Massachusetts, USA). The presence of NS1 antigen in the samples was confirmed using commercial dengue NS1 assays, and virus non-infectivity after heat-inactivation was verified through three passages of an in-house cell-based viral infectivity assay. Sample A2013-V02 (serum only) was confirmed DENV-negative by real-time RT–PCR16 and commercial dengue NS1 assays, and negative for anti-dengue antibodies using commercial enzyme-linked immunosorbent assay (ELISA) and the plaque-reduction neutralization technique (PRNT).17
For Module B, samples B2013-S01 and B2013-S02 were split serum samples from a convalescent dengue patient included to assess reproducibility of testing by the participating laboratories. These samples were confirmed by PRNT to contain neutralizing antibodies against DENV 1–4 (> 1:1000) and confirmed DENV-negative as described above. They were additionally verified using several commercial dengue antibody-based detection assays (Alere, Waltham, Massachusetts, USA; Standard Diagnostics Inc., Yongin-si, Gyeonggi-do, Republic of Korea; Focus Diagnostics Inc., Cypress, California, USA; and Bio-Rad Laboratories Inc., Hercules, California, USA). SeraCare human serum was used as the negative sample B2013-S03.
All EQA samples were confirmed externally by an independent International Organization for Standardization (ISO) 15189-accredited laboratory before dispatch to participating laboratories.
Participating laboratories could request to receive either one or both of the modules shipped on dry ice from the EQA provider by courier. The laboratories were requested to inform the EQA provider when they received the panels and to report whether the samples arrived frozen. Participants were provided with a unique identifier, an instruction and results submission form, a good laboratory practices survey and quality of shipment and feedback forms. Participants were requested to test samples in triplicate independent runs (to assess reproducibility) by the routine methods used in their laboratories and to submit background technical information on methods, kits and reagents used. Test results were required within 30 days.
Analysis of results
In Module A, two points each were awarded for the correct detection of DENV by RT–PCR or NS1 assay and accurate serotyping of DENV. In Module B, two points each were awarded for the correct detection of anti-DENV IgG and IgM antibodies. Using in-date reagents scored an additional three points. Awardable points were based solely on the assays performed on each sample. The final score was the proportion of points earned out of the possible awardable points. Accuracy for each assay (e.g. serotyping) was defined as the proportion of laboratories scoring 100% for that assay.
Quantitative data (RT–PCR cycle threshold values and ELISA values) submitted were used for reference and for assessing reproducibility of laboratory results. For ELISA assays, the percentage coefficient of variation (CV) was calculated from values recorded in triplicate runs to evaluate the reproducibility of results. A limit of ≤ 15% CV was used,18 mirroring manufacturers’ guidelines on inter-/intra-sample variation specified in the product inserts accompanying commercial ELISA kits. Large variations were flagged for attention in assessment reports sent to each laboratory.
Results
Laboratory proficiency in dengue diagnostics
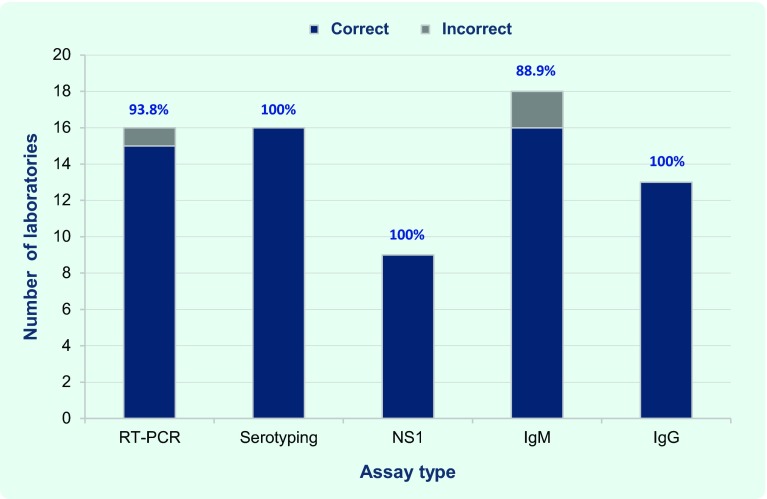
The most common assay performed was the anti-DENV IgM ELISA; 16 of the 18 laboratories that conducted this test detected IgM in all of the samples (two laboratories detected IgM in only one of the split samples), achieving an overall accuracy of 88.9% for this assay (Fig. 2).
Fig. 2.
Proportion of participating laboratories by test conducted and results, EQA of dengue diagnostics, WHO Western Pacific Region, 2013
NS1, non-structural protein 1; RT–PCR, reverse transcription polymerase chain reaction.
Sixteen laboratories used RT–PCR for nucleic acid detection. As one laboratory reported equivocal results for the negative sample in Module A, the overall accuracy for RT–PCR was 93.8%. These 16 laboratories also conducted virus serotyping with 100% accuracy (Fig. 2). One laboratory conducted RT–PCR only, while another used an expired reagent for a viral RNA assay, although this had no effect on accuracy. The capacity to detect ongoing or recent dengue infection was demonstrated by the 15 laboratories that conducted both RT–PCR and IgM anti-DENV ELISA.
Testing for anti-DENV IgG and NS1 antigen were the next most common assays, employed by 13 and nine laboratories, respectively, with 100% accuracy for both tests (Fig. 2). The seven laboratories that conducted all five assays (RT–PCR, serotyping, NS1, IgM and IgG assays) achieved 100% accuracy in each of them.
Module A: Viral RNA and NS1 antigen
Of those laboratories using RT–PCR in Module A, the majority (11/16) used the QIAmp Viral RNA Mini Kit (Qiagen, Valencia, California, USA) for extraction and purification of DENV RNA and commercial kits to perform RT–PCR. More than half (56.3%) used real-time RT–PCR, while the remainder used conventional RT–PCR methods (Table 2). The laboratory reporting equivocal results for the negative sample in Module A used the real-time methodology. Most laboratories (87.5%) used in-house positive controls for viral detection and serotyping. DENV genome regions targeted for virus detection and serotyping varied, with non-structural protein 5 and capsid being the most commonly used. To detect NS1 antigen, seven of nine laboratories employed the Platelia Dengue NS1 Ag kit (Bio-Rad Laboratories Inc.), one used the SD Dengue NS1 Ag ELISA (Standard Diagnostics Inc.) and the other an in-house ELISA.
Table 2. Number and proportion of participating laboratories by assay type used, EQA of dengue diagnostics, WHO Western Pacific Region, 2013.
| Assay type | Number/total | % |
|---|---|---|
| Viral nucleic acid detection (RT–PCR) | 16/19 | 84.2 |
| Real-time RT–PCR methodology | 9/16 | 56.3 |
| Conventional RT–PCR methodology | 7/16 | 43.8 |
| Commercial RT–PCR kits | 11/16 | 68.8 |
| In-house RT–PCR controls | 14/16 | 87.5 |
| Viral antigen detection (NS1) | 9/19 | 47.4 |
| Commercial NS1 ELISA kits | 8/9 | 88.9 |
| Antibody detection | 18/19 | 94.7 |
| IgM antibody detection | 18/18 | 100.0 |
| ELISA methodology | 18/18 | 100.0 |
| Commercial IgM ELISA kits | 15/18 | 83.3 |
| IgG antibody detection | 13/18 | 72.2 |
| ELISA methodology | 11/13 | 84.6 |
| Commercial IgG ELISA kits | 11/11 | 100.0 |
ELISA, enzyme-linked immunosorbent assay; NS1, non-structural protein 1; and RT–PCR, reverse transcriptase polymerase chain reaction.
Of seven laboratories detecting NS1 in triplicate runs using commercial ELISA kits, three demonstrated lower reproducibility (up to 30% CV) between runs. Though the final interpretation of results was not affected, large deviations in CV warrant greater adherence to work processes.
Module B: Serology
All 18 laboratories that requested Module B chose the ELISA methodology to detect anti-DENV IgM (Table 2). Half used the Panbio Dengue IgM Capture ELISA (Alere Inc.) with two also using a rapid diagnostic test (Panbio Dengue Duo Cassette, Alere Inc.). The two laboratories detecting anti-DENV IgM in only one of the split samples used an in-house IgM MAC-ELISA protocol and a commercial Dengue IgM ELISA kit (Euroimmun, Luebeck, Germany), respectively. These were not used by any other laboratories. ELISA was also the methodology of choice for anti-DENV IgG detection, with 11 out of 13 (84.6%) of laboratories using commercial indirect ELISA and/or high titre IgG capture ELISA kits. The remaining two laboratories performed in-house DENV haemagglutination inhibition assays.
Participating laboratories demonstrated reproducible IgG assay results (≤ 15% CV on average) in Module B; however, a ≥ 30% CV between sample runs for IgM assays was observed in a third of the participating laboratories. This included the two laboratories with incorrect results for the split samples.
Logistics
Most (17/19) laboratories returned results within the month allotted; the average time between receipt of samples and completed results was 27.8 days. One laboratory was five days late and another requested a 13-day extension while waiting for the delivery of reagents. There were no major logistics issues with shipping the panels to participating countries; all deliveries arrived on time and with cold chain intact. Flight rescheduling was announced ahead of time and deliveries were targeted to ensure a laboratory member was available and that national holidays or weekends were avoided.
Obtaining import permits from respective governments or agencies added a significant amount of time to the preparatory work before sending the panels. Eleven laboratories had to request permits, which took a median of 1.5 months to obtain (ranging from one week to 2.5 months). One laboratory had a standing import permit. The EQA time frame was also delayed as some participating laboratories had to be recruited through official ministry/department of health channels rather than directly; the longest recruitment took 1.5 months.
Discussion
This study reports on an EQA programme established for dengue diagnostics for national-level public health laboratories in the Western Pacific Region. It provided the first indication of the proficiency of the participating laboratories in diagnosing dengue samples and demonstrated the range of assays used by participants to diagnose dengue. It also facilitated communication between national laboratories and the WHO Collaborating Centre for Reference and Research of Arbovirus and their Associated Vectors, which will be useful for future public health emergencies.
The appropriate dengue diagnostic tools must be employed at the correct time to ensure the most effective diagnostic capability.19 It is therefore important for national/reference laboratories to be equipped with the tools to detect both DENV RNA/NS1 antigen and anti-dengue antibodies. It is encouraging that 15 of the 19 participating laboratories employed assays to detect both DENV RNA and anti-DENV IgM as part of their routine diagnostic algorithm for ongoing/recent dengue infection. Of the remaining four laboratories, one performed RT–PCR but not antibody testing, and three performed antibody testing but not RT–PCR. The diagnostic capacity of these laboratories could be quickly strengthened through the use of commercial ELISA assays for the detection of NS1 antigen or anti-DENV antibodies.
Anti-DENV IgM assays were performed by all 18 laboratories that tested Module B. Using commercial ELISAs for anti-DENV IgM detection was the most common approach and the majority of laboratories delivered accurate and reproducible results on almost all samples. Discrepancies reported in anti-DENV IgM assay results may be attributed to operational issues (such as unfamiliarity with ELISA, insufficient adherence to work processes, inadequate reagent handling skills and pipetting techniques). In-house ELISAs were used by three laboratories, one of which reported incorrect results. While in-house assays may appear to be economical, the maintenance of test validity, reagent quality and appropriately trained staff must remain a priority. As anticipated, dengue rapid diagnostic tests were rarely employed at the national laboratory level.
Thirteen of the 18 laboratories participating in Module B also performed assays for the detection of anti-DENV IgG. Two types of commercial anti-DENV IgG ELISAs were employed; four laboratories used high titre IgG ELISAs suitable for detecting ongoing/recent infections, six used low titre IgG ELISAs for the detection of a prior dengue infection (such as in seroprevalence studies) and one laboratory used both. High titre IgG ELISAs, when used on acute-phase sera, can differentiate between primary and secondary dengue infections in endemic areas; however, low titre ELISAs have no diagnostic value unless they are used in conjunction with an IgM ELISA. The presence of IgM alone is highly indicative of an ongoing/recent infection, whereas detection of IgG at low titre can occur indefinitely after dengue infection. As national laboratories are more likely to test samples from ongoing/recent infections, this may explain the more prevalent use of IgM kits. Several of the laboratories that did not test for anti-DENV IgG reported this was because they did not have IgG kits available or did not routinely test for IgG.
Reproducibility is also an important component of EQA. High variability (≥ 15% CV) between experimental runs was observed in several laboratories participating in Module B, particularly in two laboratories incorrectly diagnosing the split samples in Module B. This highlights the importance of using validated assays and adhering to standard operating procedures to ensure accurate and reproducible test results, as well as the continual training of laboratory technicians. The interpretation of this calculation is limited due to the small number of samples used (the two samples repeated in triplicate gives only six data points per laboratory); however, the results have provided an indication of variability and potential operational issues, which was the aim of this initial exercise. Participation in audits, such as EQA, is useful for laboratories to ascertain areas requiring improvement.
The high accuracy of participating laboratories to diagnose dengue using serological and molecular tests were similar to that observed by the European Network for Diagnostics of “Imported” Viral Diseases (ENIVD) for their initial four-sample panel for serology but not for their EQA panel of 20 samples where 79% of participating laboratories required improvement in correctly detecting anti-DENV antibodies.20 Likewise, the recent ENIVD EQA for molecular detection of DENV found that 80.4% of laboratories needed improvement in identifying dengue and non-dengue samples and serotypes.21 In contrast to our EQA, participating laboratories were in countries where dengue is not endemic, and samples were composed of different dilutions of DENV or patient serum and included other arboviruses or anti-sera against them as controls. Panels in upcoming rounds of the EQA in the Western Pacific Region will be composed of more dengue serotypes and titre ranges, as well as other arboviruses of priority to the Region.
Despite encountering no major logistical issues and EQA being executed mostly as intended, valuable administrative lessons were learnt. The delays in acquiring import permits and recruiting laboratories through government channels were unexpected. More time to accommodate these steps will therefore be allotted in the future.
This first round of EQA of dengue diagnostics had some limitations. The modules comprised three samples each, limiting the variety of samples that could be included such as blinded samples to assess reproducibility. Module size also prevented the inclusion of other arboviruses or anti-sera against them and the inclusion of multiple titrations of virus for determining assay sensitivity. However, the aim of this first round of EQA was to attain an initial overview of dengue diagnostic testing in the Region. The findings presented here need to be further substantiated during upcoming rounds of EQA with more comprehensive panels.
This first round of EQA in the Western Pacific Region showed that using the existing influenza EQA programme facilitated EQA for another priority pathogen. Despite the small number of samples tested, this exercise showed that laboratory diagnosis of dengue in the Western Pacific Region is good and provided lessons for subsequent iterations. Therefore this ongoing EQA programme for dengue, which will be expanded to include other priority pathogens, should strengthen the regional public health laboratory system for detecting emerging infectious diseases, in line with APSED (2010).
List of participating laboratories
PathWest Laboratory Medicine, QEII Medical Centre (Australia), Ministry of Health, Department of Laboratory Services (Brunei Darussalam), Institut Pasteur du Cambodge (Cambodia), Chinese Center for Disease Control and Prevention, National Institute for Viral Diseases Control and Prevention (China), Fiji Centre for Communicable Disease Control (Fiji), Institut Louis Malardé (French Polynesia), Public Health Laboratory Centre, Virology Division (Hong Kong Special Administrative Region SAR, China), National Institute of Infectious Diseases, Virology 1st (Japan), Korea National Institute of Health, Division of Arboviruses (Republic of Korea), National Center for Laboratory and Epidemiology (the Lao People's Democratic Republic), Health Bureau, Public Health Laboratory (Macau SAR, China), Department of Medical Microbiology, University of Malaya (Malaysia), National Center for Zoonotic Diseases, Ministry of Health (Mongolia), Institut Pasteur de Nouvelle-Calédonie, Laboratoire de Biologie Médicale (New Caledonia), Institute of Environmental Science and Research Ltd, Clinical Virology (New Zealand), Papua New Guinea Institute of Medical Research, Environmental & Emerging Diseases Unit (Papua New Guinea), Research Institute for Tropical Medicine, Department of Virology (the Philippines), National Institute of Hygiene and Epidemiology, Virology Department, and Pasteur Institute in Ho Chi Minh, Laboratory of Arboviruses (Viet Nam).
Acknowledgements
The authors are grateful to the national-level public health laboratories that participated in the EQA and to Professor Leo Yee Sin of the Tan Tock Seng Hospital, Singapore, for assistance in obtaining dengue antibody-positive serum samples. We would also like to thank Sandy Walker for critical reading of the manuscript.
Conflicts of interest
None declared.
Funding
This programme was in large part financially supported by the U. S. Agency for International Development Emerging Pandemic Threats (EPT) programme’s IDENTIFY project.
References
- 1.Arima Y, et al. Epidemiologic update on the dengue situation in the Western Pacific Region, 2012. Western Pacific Surveillance and Response Journal. 2015;6(2) doi: 10.5365/wpsar.2014.5.4.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dengue: guidelines for diagnosis, treatment, prevention and control. Geneva: World Health Organization; 2009. http://www.who.int/tdr/publications/documents/dengue-diagnosis.pdf accessed 17 June 2015. [PubMed] [Google Scholar]
- 3.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KS, et al. Dengue virus surveillance in Singapore reveals high viral diversity through multiple introductions and in situ evolution. Infection, Genetics and Evolution. 2012;12:77–85. doi: 10.1016/j.meegid.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Ritchie SA. Dengue vector bionomics: why Aedes aegypti is such a good vector. In: Gubler DJ, Ooi EE, Vasudevan S, Farrar J, editors. Dengue and dengue hemorrhagic fever. Oxfordshire: CABI; 2014. pp. 455–80. [DOI] [Google Scholar]
- 6.Banu S, et al. Dengue transmission in the Asia-Pacific region: impact of climate change and socio-environmental factors. Tropical Medicine & International Health. 2011;16:598–607. doi: 10.1111/j.1365-3156.2011.02734.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilder-Smith A, Gubler DJ. Geographic expansion of dengue: the impact of international travel. The Medical Clinics of North America. 2008;92:1377–90, x. doi: 10.1016/j.mcna.2008.07.002. [x.] [DOI] [PubMed] [Google Scholar]
- 8.Dengue Situation Update– 25 December 2013. Manila: World Health Organization Regional Office for the Western Pacific; 2013. http://www.wpro.who.int/entity/emerging_diseases/Dengue.Biweekly.24Dec2013.pdf accessed 17 June 2015. [Google Scholar]
- 9.Communicable Disease Surveillance in Singapore 2013. Singapore: Ministry of Health; 2014. https://www.moh.gov.sg/content/moh_web/home/Publications/Reports/2014/communicable-diseases-surveillance-in-singapore-2013.html accessed 25 June 2015. [Google Scholar]
- 10.Governments of Malaysia and Singapore. Joint Media release: UNITEDengue cross-border data sharing provides countries with timely risk alerts. Singapore: National Environment Agency; 2014. http://www.moh.gov.my/index.php/database_stores/attach_download/337/573 accessed 17 June 2015. [Google Scholar]
- 11.Dengue Situation Update - 22 April 2014. Manila: World Health Organization Regional Office for the Western Pacific; 2014. http://www.wpro.who.int/emerging_diseases/Dengue.Biweekly.22Apr2014.pdf accessed 17 June 2015. [Google Scholar]
- 12.Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Archives of Virology. 2013;158:1445–59. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 13.Laboratory quality management system. Geneva: World Health Organization; 2011. http://www.who.int/ihr/training/laboratory_quality/10_b_eqa_contents.pdf accessed 17 June 2015. [Google Scholar]
- 14.Asia Pacific Strategy for Emerging Diseases. (2010). Manila: World Health Organization Regional Office for the Western Pacific; 2011. http://www.wpro.who.int/emerging_diseases/documents/ASPED_2010/en/ accessed 17 June 2015. [Google Scholar]
- 15.WHO External Quality Assessment Project for the detection of influenza virus type A by PCR. Manila: World Health Organization Regional Office for the Western Pacific; 2012. http://www.who.int/influenza/gisrs_laboratory/external_quality_assessment_project/en/ accessed 17 June, 2015. [Google Scholar]
- 16.Lai YL, et al. Cost-effective real-time reverse transcriptase PCR (RT-PCR) to screen for Dengue virus followed by rapid single-tube multiplex RT-PCR for serotyping of the virus. Journal of Clinical Microbiology. 2007;45:935–41. doi: 10.1128/JCM.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morens DM, et al. Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: comparison of the BHK suspension test with standard plaque reduction neutralization. Journal of Clinical Microbiology. 1985;22:250–4. doi: 10.1128/jcm.22.2.250-254.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Food and Drug Administration. Guidance for Industry: bioanalytical method validation. Maryland: United States Department of Health and Human Services; 2001. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070107.pdf accessed 17 June 2015. [Google Scholar]
- 19.Peeling RW, et al. Evaluation of diagnostic tests: dengue. Nature Reviews Microbiology. 2010;8(Suppl):S30–8. doi: 10.1038/nrmicro2459. [DOI] [PubMed] [Google Scholar]
- 20.Donoso Mantke O, et al. Quality control assessment for the serological diagnosis of dengue virus infections. Journal of Clinical Virology. 2004;29:105–12. doi: 10.1016/S1386-6532(03)00110-0. [DOI] [PubMed] [Google Scholar]
- 21.Domingo C, et al. 2nd International external quality control assessment for the molecular diagnosis of dengue infections. PLoS Neglected Tropical Diseases. 2010;4:e833. doi: 10.1371/journal.pntd.0000833. [DOI] [PMC free article] [PubMed] [Google Scholar]