Abstract
Actinobacteria are one of the most important and efficient groups of natural metabolite producers. The genus Streptomyces have been recognized as prolific producers of useful natural compounds as they produced more than half of the naturally-occurring antibiotics isolated to-date and continue as the primary source of new bioactive compounds. Lately, Streptomyces groups isolated from different environments produced the same types of compound, possibly due to frequent genetic exchanges between species. As a result, there is a dramatic increase in demand to look for new compounds which have pharmacological properties from another group of Actinobacteria, known as rare actinobacteria; which is isolated from special environments such as mangrove. Recently, mangrove ecosystem is becoming a hot spot for studies of bioactivities and the discovery of natural products. Many novel compounds discovered from the novel rare actinobacteria have been proven as potential new drugs in medical and pharmaceutical industries such as antibiotics, antimicrobials, antibacterials, anticancer, and antifungals. This review article highlights the latest studies on the discovery of natural compounds from the novel mangrove rare actinobacteria and provides insight on the impact of these findings.
Keywords: mangrove, rare actinobacteria, natural compounds, bioactivity, drug discovery
Introduction
Market demand for new drugs is extremely urgent and extensive due to common ailments and the fast spread of diseases, the development of new diseases with unknown causes, and the spread of antibiotic-resistant pathogens (Jiang et al., 2008). With the increasing advancement in science and technology, it is predicted that there would be a greater demand for new bioactive compounds synthesized by Actinobacteria from various sources including soil and marine.
Mangrove is a unique woody plant community of intertidal coasts in tropical and subtropical coastal regions. The mangrove forests are among the world's most productive ecosystems which improves coastal water, produces commercial forest products, supports coastal fisheries, and protects coastlines. Due to its properties of high salinity, strong winds, extreme tides, high temperature, anaerobic soils, and muddiness; little is known about the bacterial community living in the mangrove especially of Actinobacteria with the potential to produce bioactive metabolites.
Actinobacteria or Actinomycetes are Gram positive bacteria with high guanine (G) and cytosine (C) rations in their DNA (Goodfellow and Williams, 1983). According to Das et al. (2008), the name “Actinomycetes” was derived from two Greek words, “atkis” which means ray, and “mykes” which means fungus; and it has features of both bacteria and fungi. In the strict taxonomic sense, Actinobacteria are grouped with bacteria under the class “Schizomycetes” but limited to the order of Actinomycetales (Sivakumar et al., 2005; Gayathri and Muralikrishnan, 2013). Actinobacteria can be divided into two groups namely, Streptomyces and non-streptomyces or also known as rare actinobacteria.
Many studies have found that bioactive compounds of actinobacteria possess a wide range of biological activities. In the late Twentieth Century, some natural products produced by Actinobacteria had been used extensively in clinical settings as antibacterials, antifungals, anticancer, antitumor, and antiparasitics (Butler, 2004). It is important to discover new natural metabolites to handle the problem of antibiotic-resistant pathogens which are no longer susceptible to the antibiotics available. However, the recent search for the novel compounds always lead to the rediscovery of known compounds from the same species (Koehn and Carter, 2005).
Currently, the discoveries of new natural metabolites are focusing on non-Streptomyces or rare actinobacteria. Rare actinobacteria are always referred to as strains that are difficult to isolate and might correspond to the unmatched source of new natural metabolites (Baltz, 2006). The bioactivity studies of natural metabolites from the mangrove rare actinobacteria has become popular. Compounds discovered from the mangrove rare actinobacteria are uniquely structured and lead directly to the development of novel antibiotics that are effective against antibiotic-resistant pathogens (Lam, 2006).
Mangrove
Mangrove swamps occupy about 181,000 km2 (Jusoff, 2013) or cover approximately 75% of the world's tropical and subtropical coastlines (Yang et al., 2013). Mangroves are mostly tropical trees that grow between the high spring tide mark in stable shores, and near mean sea levels (Holguin et al., 2001). According to Tomlinson (1986), a mangrove requires five criteria to be considered a “true or strict mangrove” namely, (1) complete fidelity to the mangrove environment; (2) plays a major role in the structure of the community and has the ability to form pure stands; (3) morphological specialization for adaptation to the habitat; (4) physiological specialization for adaptation to their habitat; and (5) taxonomic isolation from terrestrial relatives. Mangroves are thus a unique environment with highly productive ecosystems (global litterfall of 100 Tg C year−1) (Jennerjahn and Ittekkot, 2002). The important physical features of the mangrove that aid their survival in the boundary zone between ocean and land are adaptations for mechanical fixation in loose soil, specialized dispersal mechanisms, breathing roots and air exchange devices, and specialized mechanisms for dealing with excess salt concentration (Spalding et al., 1997).
Microbes constitute the largest pool of metabolic pathways on earth with potential biotechnological and environmental implications. In 1988, Alongi reported that in tropical mangroves, 91% of the total microbial biomass is bacteria and fungi, another 7% is algae, and 2% is protozoa (Alongi, 1988). The microbial diversity of mangrove ecosystems provides information on their ecological role and unique biotechnological potential in the field of agriculture, industry, medicine, and pharmaceuticals (Lageiro et al., 2007). The bacterial communities in mangrove environments were firstly determined by Yan et al. (2006) and Liang et al. (2007). Both studies used molecular cloning and sequencing approaches to understand the diversity of prokaryotes in these environments (Andreote, 2012).
The mangrove ecosystem is a largely unexplored source of Actinobacteria with the potential to produce active secondary metabolites (Hong et al., 2009). Although several studies on bacterial productivity and activity within the mangrove ecosystems were conducted, little is known about their genetic and metabolic diversity.
Metagenomics of mangrove microbial communities: pyrosequencing and fosmid library
Molecular phylogenetic studies have shown that only a small number (~1%) of bacterial diversity present in environmental samples, are readily cultivable using conventional culture-based techniques (Yan et al., 2013). It is evident that more than 88% of isolates belong to four phyla known as Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes (Nikolaki and Tsiamis, 2013). The uncultivated bacteria may serve as a prolific source of new bioactive compounds as they may possess a great diversity of enzymes to be easily adapted to different environmental conditions. Unlike other strategies used for novel enzyme and microorganism identification, the metagenomic method has clear advantages as an alternative to culture dependent methods. Metagenomics is a culture-independent genomic analysis of bacteria diversity contained in a sample, reducing difficulties related to conditions for bacteria cultivation (Handelsman, 2004). Metagenomics uses different methods ranging from the generation of short sequence reads by direct use of high-throughput sequencing technologies, to the construction and sequencing of large-insert DNA libraries (Reigstad et al., 2011).
Next generation sequencing (NGS) has significantly increased sequencing output via the use of extremely parallel sequencing (Staley et al., 2013). The advancement of the next-generation DNA sequencing technologies such as high-throughput pyrosequencing and Illumina platform, increases scientific interests in understanding the microbial diversity in different environments. The study of bacterial communities from soils (Jones et al., 2009), marine water (Brown et al., 2009), wastewater (Sanapareddy et al., 2009) and human microbiomes (NIH HMP Working Group et al., 2009) have been successfully conducted using the NGS approach. High-throughput pyrosequencing is a tool for the analysis of environmental microbiome, and reveal the taxonomic diversity of a specific environment at high resolution (Zhu et al., 2011). This method enables rapid estimation of bacterial diversity and furthermore, provides information about environmental interference. The NGS approach enables the discovery of novel enzymes and molecules with potential application in the biotechnology industry (Duan and Feng, 2010). The NGS technology is known to be costly for the assessment of huge numbers of environmental samples with high sequencing depths (Zhou et al., 2011). However, the advancement in NGS technology has enabled Illumina to dominates the market with 60% of the second generation sequencer installation that successfully reducing the cost with the used of reversible terminator-based sequencing by the synthesis chemistry, small, and less expensive, Illumina MiSeq platform (Niedringhaus et al., 2011).
The application of NGS allows the study of microbial diversity without culturing the bacteria hence, avoiding the loss of unculturable bacteria on laboratory media such as fastidious bacteria. The latest metagenome study on characterization of mangrove soil collected from Rantau Abang, Malaysia via NGS was done by Chan and Ismail (2015). In this study, they showed that the Proteobacteria (43.72%) was the dominant phyla followed by Acidobacteria (17.68%), Firmicutes (13.45%), Actinobacteria (4.55%), Nitrospirae (4.22%), Planctomycetes (3.06%), Chloroflexi (2.88%), Verrucomicrobia (2.69%), Spirochaetes (1.70%), Chlamydiae (1.32%), and Bacteroidetes (1.31%). However, the unclassified bacteria found in this study were clustered as Caldithrix, Haloplasmatales, and phototrophic bacteria. A study on the profile of bacterial community in different depths of soil in Sundarbans mangrove, India was done by Basak et al. (2015). The taxanomic analysis of 2746 species showed that they belong to 33 different phyla such as Proteobacteria, Firmicutes, Chloroflexi, Bacteroidetes, Acidobacteria, Nitrospirae, and Actinobacteria. The description of mangrove microbiology has been made on the basis of pyrosequencing, based on DNA extracted directly from the sediment of four distinct mangrove areas along the coast of the Sao Paulo State in Brazil (Andreote et al., 2012). The major bacterial phyla identified from this study was Proteobacteria (47.1—56.3%) followed by Firmicutes (10.5—13.8%), Actinobacteria (5.4—12.2%), Bacteroidetes (3.8—11.8%), and Chloroflexi (1.3—5.4%); and some minor phyla such as Planctomytes, Cyanobacteria, Acidobacteria, and Archaea. Furthermore, Andreote et al. (2012) compared the mangrove metagenomes with other related studies, which indicated that the phylum Actinobacteria was mostly found in estuarine sediments. The metagenomic data from Thompson et al. (2013) showed similar predominant bacterial phyla in two different mangrove areas in Brazil, namely Rio de Janeiro and Bahia; were Proteobacteria (57.8% and 44.6%), Firmicutes (11% and 12.3%), and Actinobacteria (8.4% and 7.5%). Furthermore, the Actinobacteria, Fibrobacteres/Acidobacteria, and Firmicutes group were predominantly represented in the Bahia mangrove area. Gomes et al. (2010) performed an educated measurement of rhizosphere microbial population in comparison to bulk sediments samples. The major bacterial order discovered were Burkholderiales, Caulobacterales, and Rhizobials while the major bacterial phyla discovered were Acidobacteria, Actinobacteria, and Verrucomicrobia. The formation of metagenomic libraries using fosmid library become a method of choice for exploring environmental microbial communities that are difficult to culture and maintain.
Actinobacteria
Under the domain Bacteria, the Actinobacteria is one of the most distributed phyla among the 30 major lineage including 5 subclasses (www.bacterio.net). According to Zhi et al. (2009), under the class Actinobacteria, there are five orders known as Acidimicrobiales, Rubrobacterales, Coriobacterales, Bifidobacteriales, and Actinomycetales. However, the recent list showed that there are five new orders under Actinobacteria known as Nitriliruptorales (Sorokin et al., 2009), Solirubrobacterales (Reddy and Garcia-Pichel, 2009), Thermoleophilales (Reddy and Garcia-Pichel, 2009), Euzebyales (Kurahashi et al., 2010), and Gaiellales (Albuquerque et al., 2011), making a total of ten orders (www.bacterio.net). In nature, Actinobacteria are also known as saprophyte soil inhabitants (Naik et al., 2013), and one of the dominant colonizers in soils (McCarthy and Williams, 1992; Heuer et al., 1997; Lee et al., 2012). Actinobacteria are Gram positive bacteria with a high G+C content in their DNA, ranging from 51% in some corynebacteria, to more than 70% in Streptomyces and Frankia except for Tropheryma whipplei; an obligate pathogen which contains less than 50% G+C (Marco et al., 2007).
Actinobacteria have a variety of morphologies, from rod-coccoid (e.g., Arthrobacter spp.) or coccoid (e.g., Micrococcus spp.), to permanent or highly differentiated branch mycelium (e.g., Streptomyces spp.), or fragmenting hyphal forms (e.g., Nocardia spp.) (Atlas, 1997). They also show various physiological and metabolic properties, such as the production of extracellular enzymes and a variety of secondary metabolites such as potent antimicrobial agents (Lee et al., 2014a; Olano et al., 2014). The member of genus Streptomyces species displayed rare development characteristics with the formation of aerial mycelium with spore (Lee et al., 2014b; Ser et al., 2015), whereas certain mycobacterium exhibited a persistent non-replicating state. In common laboratory media, Actinobacteria tends to grow slowly as branching filaments and have the ability to produce motile spores (Nanjwade et al., 2010). Actinobacteria can be divided into two main groups; the Streptomyces, representing the dominant species in the group; and the rare actinobacteria.
Rare actinobacteria
In their natural habitat, Streptomyces are the dominant species of Actinobacteria. Rare actinobacteria are relatively difficult to isolate, culture, and maintain; due to the difficulties in maintaining and mimicking their natural environment. Rare actinobacteria have been considered as Actinobacteria with lower isolation rates compared to Streptomyces strains using conventional isolation methods, due to the requirement of using appropriate isolation procedures and to apply different selection conditions (Khanna et al., 2011). Until September 2010, there are approximately 220 genera of rare actinobacteria reported (Tiwari and Gupta, 2013). More evidences have shown that increasing numbers of rare actinobacteria have been discovered from different sources, thus indicating that rare actinobacteria are widely distributed in the biospheres. Even though soils and water are their major habitat, they are also isolated from different environments such as the deep ocean, desert, mangroves, plants, caves, volcanic rocks, and stones (Groth et al., 1999). In natural ecosystems, many chemical, biological, and physical elements; affect their diversity as the environmental factors such as pH, humus content, soils type, temperature, and salinity affect their distribution (Hayakawa, 2008).
The isolation of rare actinobacteria could definitely increase the chances of discovering potentially novel bioactive compounds, therefore it is vital to be equipped with better knowledge of their diversity and distribution in the environment; in order to facilitate the isolation of these strains using an efficient approach (Hong et al., 2009; Tiwari and Gupta, 2012; Lee et al., 2014a). According to Jose and Jebakumar (2013), rare actinobacteria are distributed among 24 genera: Actinomadura, Actinoplanes, Amycolatopsis, Actinokineospora, Acrocarpospora, Actinosynnema, Catenuloplanes, Cryptosporangium, Dactylosporangium, Kibdelosporangium, Kineosporia, Kutzneria, Microbiospora, Microtetraspora, Nocardia, Nonomuraea, Planomonospora, Planobispora, Pseudonocardia, Saccharomonospora, Saccharopolyspora, Saccharothrix, Streptosporangium, and Spirilliplanes.
Rare actinobacteria of mangrove origin
The mangrove ecosystem is among the world's most productive environment, and it is a huge unexplored source of Actinobacteria with a high potential to produce active secondary metabolites (Hong et al., 2009). Indeed relatively little is known about the rare actinobacteria living in the mangrove ecosystem. Recently the mangrove ecosystem has become popular for novel strains and novel bioactive compound discovery.
A few reports from different geographical locations globally have described the diversity and isolation of novel rare actinobacteria in different mangrove habitats. In 2008, Eccleston et al. reported the occurrence of Microsmonospora from the Sunshine Coast in Australia (Eccleston et al., 2008). Xie et al. (2006) and Huang et al. (2008) reported the isolation of a rifamycin-producing Micromonospora from mangrove in South China Sea. Different genera such as Brevibacterium, Dermabacter, Kytococcus, Microbacterium, Nesterenkonia, and Rothia were isolated from mangrove sediments in Brazil (Dias et al., 2009b). In China, a number of rare actinobacteria including Actinomadura, Isoptericola, Microbispora, Nocardia, Nonomuraea, and Rhodococcus; were isolated from mangrove soils and plants (Hong et al., 2009). Results from Ara et al. (2013) showed that 17 different genera of rare actinobacteria were identified from a total of 241 isolates. It is reported that the predominant genus detected was Micromonospora in both mangrove and medicinal plant rhizosphere soil samples in Dhaka, Bangladesh. This result was in line with other studies (Cross, 1981; Jiang and Xu, 1996; Hatano, 1997; Ara et al., 2002) which exhibited Micromonospora was the predominant genus in isolates from wet soil. A study done by Lee et al. (2014a) on the diversity of actinobacteria from a Malaysian mangrove forest showed that 40.2% of isolates were rare actinobacteria and the predominant genus was Mycobacterium. The study also discovered other genera of rare actinobacteria, such as Leifsonia, Streptacidiphilus, Sinomonas, and Terrabacter, which were not commonly found in mangrove environment. In this study, the understanding of rare actinobacteria diversity led to the discovery of novel genus and species strains, such as “Microbacterium mangrovi sp. nov.” (Lee et al., 2014c), “Mumia flava gen. nov., sp. nov.” (Lee et al., 2014d) and Sinomonas humi sp. nov.” (Lee et al., 2015) isolated from a mangrove forest in Tanjung Lumpur, Malaysia. The discoveries of novel rare actinobacteria from mangrove environments, from 2001 until 2015 are listed in Table 1.
Table 1.
Novel mangrove rare actinobacteria discovered between the years 2001 and 2015.
| Genus | Strain name and designation | Sources | References |
|---|---|---|---|
| Agromyces | Agromyces luteolus, 8T, Agromyces rhizospherae, 14T, Agromyces brachium, 65T | Rhizosphere of mangroves in the estuary of the Shiira River, Iriomote Island, Japan | Takeuchi and Hatano, 2001 |
| Asanoa | Asanoa iriomotensis, TT 97-02T | Soil at roots of the mangrove Bruguiera gymnorrhiza | Tamura and Sakane, 2005 |
| Polymorphospora | Polymorphospora rubra, TT 97-42T | Soil near the roots of Bruguiera gymnorrhiza and Sonneratia alba at the River Shiira, Iriomote Island, Okinawa, Japan | Tamura et al., 2006 |
| Nonomuraea | Nonomuraea Maheshkhaliensis, 16-5-14T | Mangrove forest in Maheshkhali, Cox's Bazar, Bangladesh | Ara et al., 2007 |
| Micromospora | Micromonospora pattaloongensis, TJ2-2T | Mangrove forest in Pattaloong Province, Thailand | Thawai et al., 2008 |
| Micromonospora | Micromonospora rifamycinica, AM105T | Mangrove sediment collected from the South China Sea | Huang et al., 2008 |
| Actinomadura | Actinomadura maheshkhaliensis, 13-12(50)T | Mangrove rizhosphere soils of Maheshkhali, Bangladesh | Ara et al., 2008 |
| Verrucosispora | Verrucosispora lutea, YIM 013T | Shenzhen Futian Mangrove | Liao et al., 2009 |
| Demequina | Demequina salsinemoris, KV-810T | Mangrove soil from a southern island in Japan | Matsumoto et al., 2009 |
| Sphaerisporangium | Sphaerisporangium krabiense, A-T 0308T | Tropical mangrove forest soil from Thailand | Suriyachadkun et al., 2011 |
| Isoptericola | Isoptericola chiayiensis, 06182M-1T | Mangrove soil from Chiayi Country in Taiwan | Tseng et al., 2011 |
| Micromonospora | Micromonospora rhizosphaerae, 211018T | Mangrove Excocaria agallocha rhizosphere soil | Wang et al., 2011a |
| Nonomuraea | Nonomuraea wenchangensis, 210417T | Mangrove rhizosphere soil | Wang et al., 2011b |
| Jishengella | Jishengella endophytica, 202201T | Acanthus illicifolius root from the mangrove reserve zone in Hainan, China | Xie et al., 2011 |
| Asanoa | Asanoa hainanensis, 210121T | Rhizosphere soil of the mangrove fern Acrostichum speciosum | Xu et al., 2011 |
| Microbispora | Microbispora hainanensis, 211020T | Rhizosphere mangrove soil of Exoecaria agallocha, Hainan, China | Xu et al., 2012 |
| Lysinimicrobium | Lysinimicrobium mangrovi, HI08-69T | Rhizosphere mangrove soils from Iriomote Island, Japan | Hamada et al., 2012 |
| Verrucosispora | Verrucosispora qiuiae, RtIII47T | Mangrove swamp in Sanya, Hainan Province, China | Xi et al., 2012 |
| Actinomadura | Actinomadura sediminis, YIM M 10931 T | Mangrove sediments from Dugong Creel, Little Andaman, India | He et al., 2012 |
| Agromyces | Agromyces indicus, NIO-1018T | Mangrove sediment of the Chorao Island, Goa, India | Dastager et al., 2012 |
| Micromonospora | Micromonospora haikouensis, 232617T | Composite mangrove sediment from Haikou, China | Xie et al., 2012 |
| Micromonospora | Micromonospora maritime, D10-9-5T | Mangrove soil in Samut Sakhon province, Thailand | Songsumanus et al., 2013 |
| Micromonospora | Micromonospora avicenniae, 268506T | Root of Avicennia marina collected at mangrove forest in Wengchang, Hainan province, China | Li et al., 2013a |
| Actinoallomurus | Actinoallomurus acanthiterrae, 2614A723T | Rhizosphere soil of mangrove plant Acanthus ilicifolius from Touyuan, Wenchang, Hainan province, China | Tang et al., 2013 |
| Micromonospora | Micromonospora sonneratiae, 274745T | Root of Sonneratia apetala from mangrove forest in Sanya, Hainan province, China | Li et al., 2013b |
| Micromonospora | Micromonospora wenchangensis, 2602GPT1-05T | Composite mangrove soil from Wenchang, Hainan province, China | Ren et al., 2013 |
| Microbacterium | Microbacterium mangrovi, MUSC 115T | Mangrove soil of Tanjung Lumpur river, State of Pahang, Malaysia | Lee et al., 2014c |
| Mumia | Mumia flava, MUSC 201T | Mangrove soil of Tanjung Lumpur river, State of Pahang, Malaysia | Lee et al., 2014d |
| Sinomonas | Sinomonas humi, MUSC 117T | Mangrove soil of Tanjung Lumpur river, State of Pahang, Malaysia | Lee et al., 2015 |
Natural compounds from rare actinobacteria
The discovery of the first antibiotic, penicillin, in 1929 heralded the era of antibiotics. Streptomycin was later isolated from Streptomyces griseus by Waksman in 1943, while vancomycin was discovered in 1953 as a metabolite of the rare actinobacteria strain, Amycolatopsis orientalis (Mahajan and Balachandran, 2012). The discovery of various useful antibiotics from rare actinobacteria contributes as an important part in the discovery of novel natural products (Ara et al., 2013). Researchers understand the role of microorganisms to provide rich sources of useful natural products for clinical purposes (Subramani and Aalbersberg, 2012). The demand for new drugs in the world is extremely urgent and extensive due to the fast spread of diseases, common ailments, and the fast spread of antibiotic-resistant pathogens (Jiang et al., 2008). The continuous advancement in science and technology would enable the accelerated discovery of useful bioactive compounds from prolific producers such as the Actinobacteria.
As of the year 2002, over 10,000 bioactive compounds (45% of all microbial metabolites) were produced by filamentous Actinobacteria, out of which 7600 (75%) were derived from Streptomyces, and 2500 (25%) from rare actinobacteria such as Micromonospora, Actinomadura, and Streptoverticillium (Bérdy, 2005). Actinobacteria are able to produce different types of secondary metabolites due to the function of various genes such as the non-ribosomal polyketides synthase (NRPS) and polyketide synthase (PKS) (Salomon et al., 2004; Lee et al., 2014a). In spite of the tremendous success of the past in screening for useful secondary metabolites, the chance of finding new biologically active molecules from Streptomyces has decreased (Fenical et al., 1999). As some Streptomyces strains from different environments produced the same types of compounds, possibly due to frequent genetic exchange between species (Bredholt et al., 2008; Freel et al., 2011). As a result, there is a dramatic increase in demand to look for new pharmacological important compounds.
The advancement of new selective techniques enables the isolation and screening of rare actinobacteria, which could lead to the discovery of new and useful bioactive compounds. The discovery of rare actinobacteria has enlarged the range and diversity of genetic resources available for biotechnological exploitation. It is apparent that the discovery of novel rare actinobacteria can be expected to provide new bioactive compounds (Lazzarini et al., 2000; Donadio et al., 2002; Takahashi and Ômura, 2003; Bérdy, 2005; Hong et al., 2009; Lee et al., 2014a).
The review mainly focuses on five different genera with the species found from the mangrove environment namely, Micromonospora, Jishengella, Salinispora, Saccharopolyspora, and Nocardiopsis. They are all proven to have valuable sources of potential metabolites and besides Streptomyces; they have proven to be valuable sources of potentially useful bioactive metabolites.
Micromonospora
Micromonospora is a genus of Actinobacteria from the Micromonosporaceae family. This genus was first isolated by Ørskov in 1923, is Gram positive, aerobic, and chemo-organotrophic (Ann and Maria, 2009). Currently, this genus contains 59 species and 7 subspecies (http://www.bacterio.net/). Redenbach et al. (2000) revealed that Micromonospora chalcea DSM 43026 contained a linear chromosome with a size of 7.7 Mb. Hirsch et al. (2013) reported the complete genome sequence of Micromonospora strain L5 isolated from nodules of Casuarina equisetifolia trees in Mexico. The strain Micromonospora strain L5 has a genome size of 6,907,073 bp, with 6332 predicted open reading frames (ORFs) and the GC content of 72.86%. The genome was sequenced using a combination of Illumina and 454 technologies at the Joint Genome Institute (JGI). According to Hirsch and Valdés (2009), the genome size of Micromonospora aurantica is 6.97 Mb and contains 88.64% of GC. However, both genomes of Micromonospora aurantianca ATCC 37029 and Micromonospora strain L5 have not been annotated as yet.
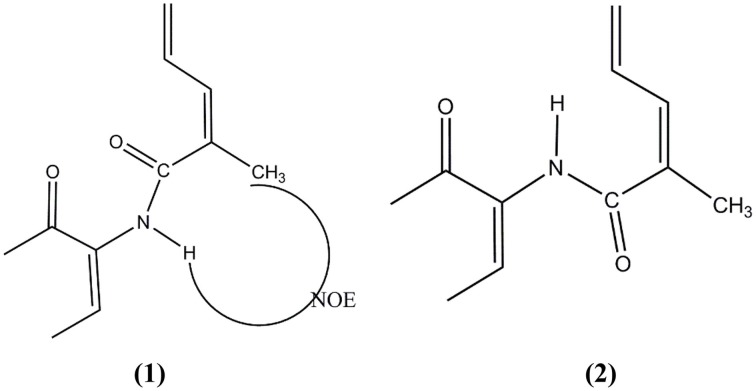
Currently, some Micromonospora from the mangrove environment has produced useful bioactive compounds for studies on drug discovery. Micromonospora rifamycinica AM105 was isolated from mangrove sediments from Hainan Island, South China Sea (Huang et al., 2009). The antimicrobial activity against methicillin-resistant Staphylococcus aereus (MRSA) using secondary metabolite extracts has led to the discovery of two antibiotic compounds (Figure 1), namely rifamycin S (1) and the geometric isomer of rifamycin S (2). There are different types of rifamycin, depending on its chemical structure such as rifamycin B, O, S, and SV. Rifamycin S has higher activity once it reduces to form rifamycin SV in aqueous solutions (Zhang, 2003). Also the antibacterial activity from a combination of rifamycin S and the geometric isomer of rifamycin S from Micromonospora rifamycinica AM105 was higher than rifamycin SV. This finding suggests that the antibacterial activity of the geometric isomer of rifamycin S was higher than rifamycin SV, and the antimicrobial activity of rifamycin SV was higher than rifamycin S.
Figure 1.
The structure of rifamycin S (1) and the geometric isomer of rifamycin S (2).
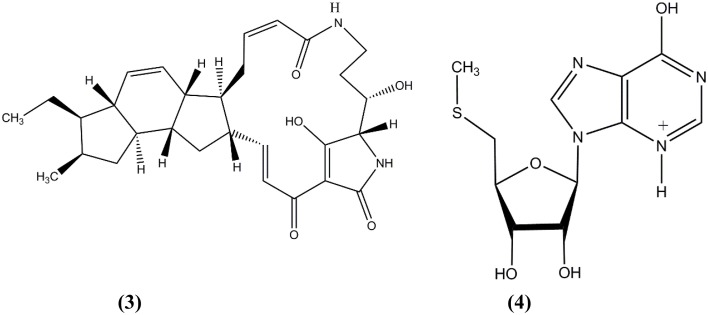
Kyeremeh et al. (2014) discovered an actinobacterial strain, Micromonospora sp. K310, from Ghanaian mangrove river sediment. The spectroscopy study of this strain led to the discovery of two new compounds (Figure 2), butremycin (3) (the (3-hydroxyl) derivative of ikarugamycin) and protonated aromatic tautomer of 5′-methylthioinosine (MTI) (4). The antibacterial activity for butremycin showed it was not potent enough to kill pathogens such as Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, and clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) strains. The antimicrobial activity of the protonated aromatic tautomer of 5′-methylthioinosine showed no antibacterial activity against these pathogens.
Figure 2.
The structure of butremycin (3) and protonated aromatic tautomer of 5′-methylthioinosine (MTI) (4).
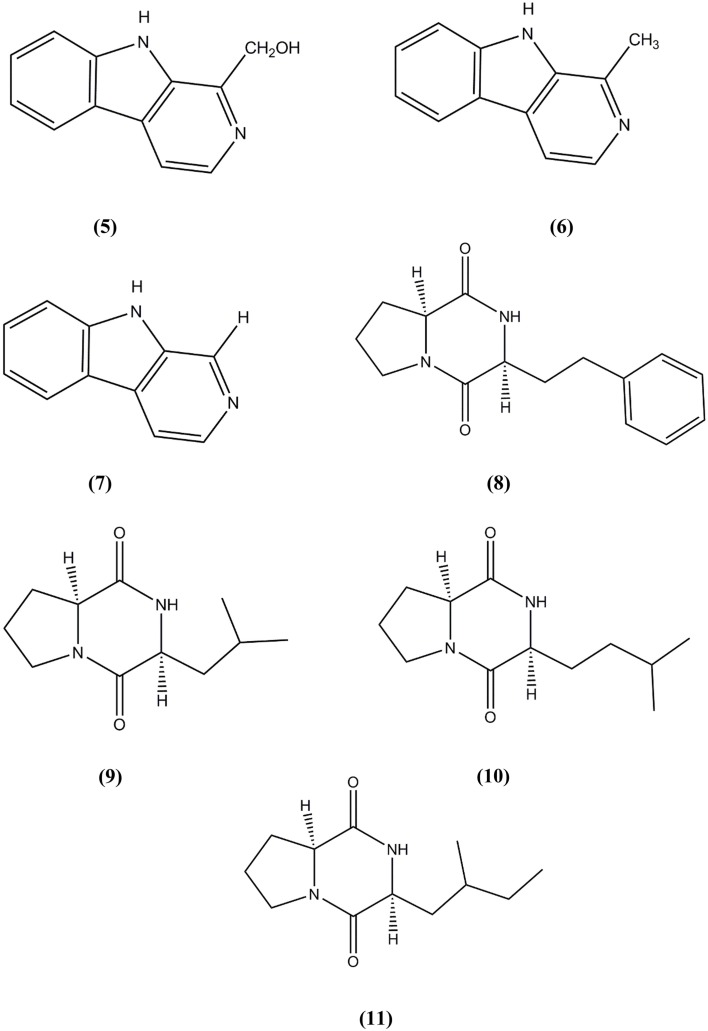
Micromonospora sp. M2DG17 was isolated from composite mangrove sediment in Haikou, China (Huang et al., 2011). The utilization of bioassay-guided fractionation enabled 5 compounds to be identified for the first time from the Micromonospora species along with two other compounds (Figure 3). The five compounds were identified as 3-hydroxymethyl-β-carboline (5), 3-methyl-β-carboline (6), β-carboline (7), Cyclo-(L-Pro-L-Phe) (8), and Cyclo-(L-Pro-L-Leu) (9) and the other two known compounds were Cyclo-(L-Pro-L-Ile) (10) and Cyclo-(L-Pro-L-Val) (11). An antitumor study using 3-methyl-β-carboline resulted in weak inhibitory on human colon carcinoma; HCT 116 cell lines (Huang et al., 2011).
Figure 3.
The structure of 3-hydroxymethyl-β-carboline (5), 3-methyl-β-carboline (6), β-carboline (7), Cyclo-(L-Pro-L-Phe) (8), and Cyclo-(L-Pro-L-Leu) (9), Cyclo-(L-Pro-L-Ile) (10), and Cyclo-(L-Pro-L-Val) (11).
Jishengella
A novel genus, Jishengella, proposed by Xie et al. (2011); is one of the rare actinobacteria within the Micromonosporaceae family. Jishengella is the genus name given by a Chinese microbiologist, Jisheng Ruan. Currently, the genus consists of one species identified as Jishengella endophytica 202201T (Xie et al., 2011).
Strain Jishengella endophytica 161111 has been able to produce a new alkaloid identified as 2-(furan-2-yl)-6-(2S,3S,4-trihydroxybutyl) pyrazine (12) (Wang et al., 2014). Extraction from the Jishengella endophytica 161111 fermentation broth resulted in the identification of three pyrazine derivatives (12, 13, 14), four pyrrololactones (15, 16, 17, 18), three β-carbolines (19, 20, 21), 1H-indole-3-carboxaldehyde (22), 2-hydroxy-1-(1H-indol-3-yl) ethanone (23), and 5-(methoxymethyl)-1H-pyrrole-2-carbaldehyde (24) (Wang et al., 2014). All compounds found in Jishengella endophytica 161111 are shown in Table 2.
Table 2.
Natural compounds discovered from Jishengella endophytica 161111.
| Compounds | References |
|---|---|
| PYRAZINE DERIVATIVES | |
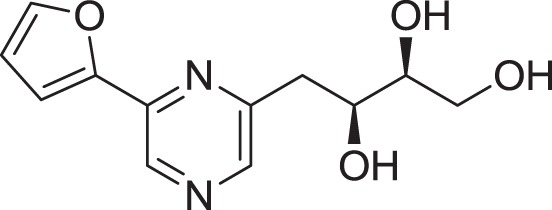 | |
| 2-(furan-2-yl)-6-(2S,3S,4-trihydroxybutyl) pyrazine (12) | Wu et al., 2007 |
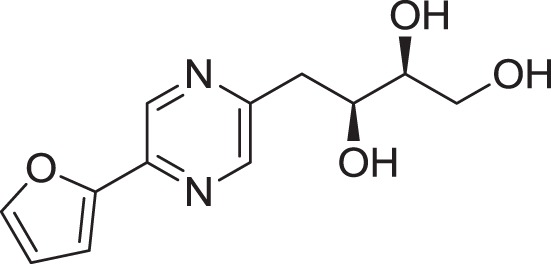 | |
| 2-(furan-2-yl)-5-(2S,3S,4-trihydroxybutyl) pyrazine (13) | Wu et al., 2007 |
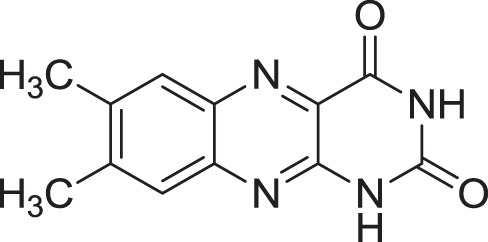 | |
| Lumichrome (14) | Ding et al., 2009 |
| PYRROLOLACTONES | |
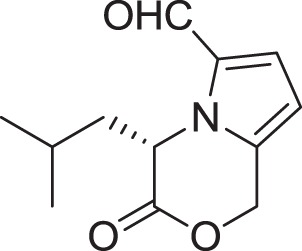 | |
| (S)-4-isobutyl-3-oxo-3,4-dihydro-1H-pyrrolo[2,1-c][1,4]oxazine-6-carbaldehyde (15) | Sannai et al., 1982 |
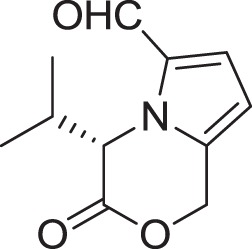 | |
| (S)-4-isopropyl-3-oxo-3,4-dihydro-1H-pyrrolo [2,1-c][1,4]oxazine-6-carbaldehyde (16) | Sannai et al., 1982 |
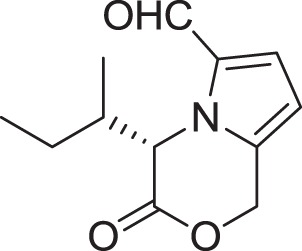 | |
| (4S)-4-(2-methylbutyl)-3-oxo-3,4-dihydro-1H-pyrrolo [2,1-c][1,4]oxazine-6-carbaldehyde (17) | Sannai et al., 1982; Tressl et al., 1995 |
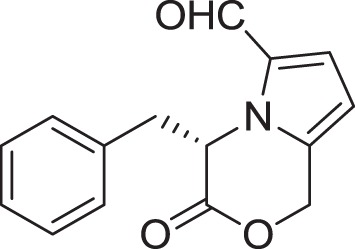 | |
| (S)-4-benzyl-3-oxo-3,4-dihydro-1H-pyrrolo[2,1-c] [1,4]oxazine-6-carbaldehyde (18) | Jeric et al., 2000 |
| β-CARBOLIN | |
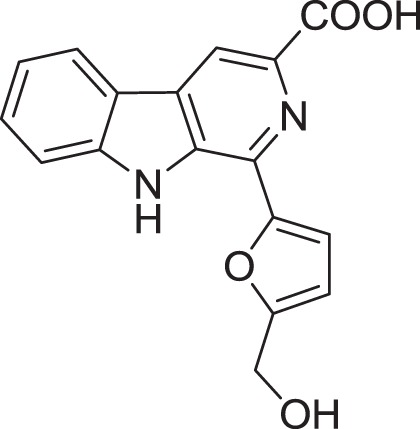 | |
| Flazin (19) | Su et al., 2002 |
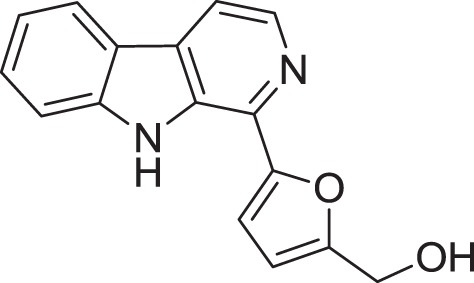 | |
| Perlolyrine (20) | Dassonneville et al., 2011 |
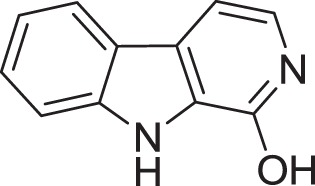 | |
| 1-hydroxy-β-carboline (21) | Jiao et al., 2010 |
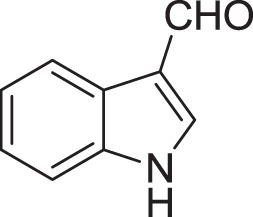 | |
| 1H-indole-3-carboxaldehyde (22) | Yang and Cordell, 1997; Ashour et al., 2007 |
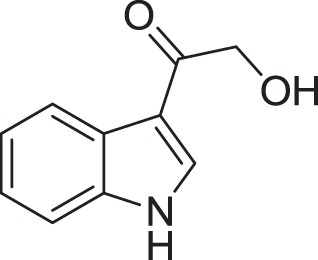 | |
| 2-hydroxy-1-(1H-indol-3-yl) ethanone (23) | Yang and Cordell, 1997; Ashour et al., 2007 |
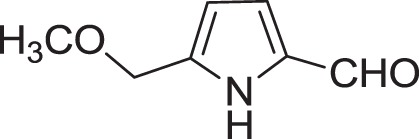 | |
| 5-(methoxymethyl)-1H-pyrrole-2-carbaldehyde (24) | Don et al., 2005 |
All compounds extracted from Jishengella endophytica 161111 were tested for antivirus effects on influenza A (H1N1) virus using the cytopathic effect (CPE) inhibition assay adapted from Grassauer et al. (2008) and Hung et al. (2009). Results showed that compounds 14, 20, 21, and 22 displayed moderate anti-H1N1 activity (Wang et al., 2014). Furthermore, all compounds were tested for cytotoxic effects as described in Mosmann (1983). The study showed that only compounds 14, 20, 21, and 22; demonstrated weak cytotoxic activity against Madin-Dady canine kidney (MDCK) normal cells.
The β-carboline alkaloids are one of the largest groups of natural and synthetic indole alkaloids that have a common tricyclic pyrido [3,4-b] indole ring structure (Allen and Holmstedt, 1980). Originally, this compound was isolated from the plant, Peganum harmala (Zygophillaceae) located in the Middle East, India, and North Africa. It was used as an effective traditional herbal drug for its abortifacient effect, and emmanagogs (Moloudizargari et al., 2013). A study done by Wang et al. (2014) showed that the simple β-carboline isolated from Jishengella endophytica 161111 was able to kill the H1N1 virus. The unsubstitued H-3 is important in anti-H1N1 activity of this kind of compounds.
Salinispora
Salinispora is a genus within the family Micromonosporaceae that was isolated from marine sediment in 2005 (Maldonado et al., 2005). Salinispora is an aerobic and Gram positive bacterium that formed extensive branched substrate hyphae with 0.25–0.5 μm in diameter. It is the first Actinobacteria that requires sea water to grow (Maldonado et al., 2005). Currently, the genus Salinispora consists of three species, namely Salinispora arenicola, Salinispora tropica, (Maldonado et al., 2005), and Salinispora pacifica (Jensen and Mafnas, 2006).
Udwary et al. (2007) reported the first complete genome of rare actinobacteria, Salinispora tropica CNB-440. This strain consists of 5,183,331 bp in size, which is approximately 3 Mbp smaller than the genome of Streptomyces (Bull and Stach, 2007). Analysis on the secondary natural product gene clusters showed that Salinispora tropica contains approximately 9.9% of the genomes dedicated to secondary metabolism, which is greater than previous Streptomyces coelicolor (≈8%) as well as other naturally-producing Actinobacteria (Udwary et al., 2007). The genome sequence of Salinispora revealed a great number of novel biosynthetic gene clusters, and based on the analysis; these findings confirms that this genus is one of the important sources of novel drug-like molecules. According to Bull and Stach (2007), 15 secondary metabolite gene clusters have been identified in Salinispora tropica.
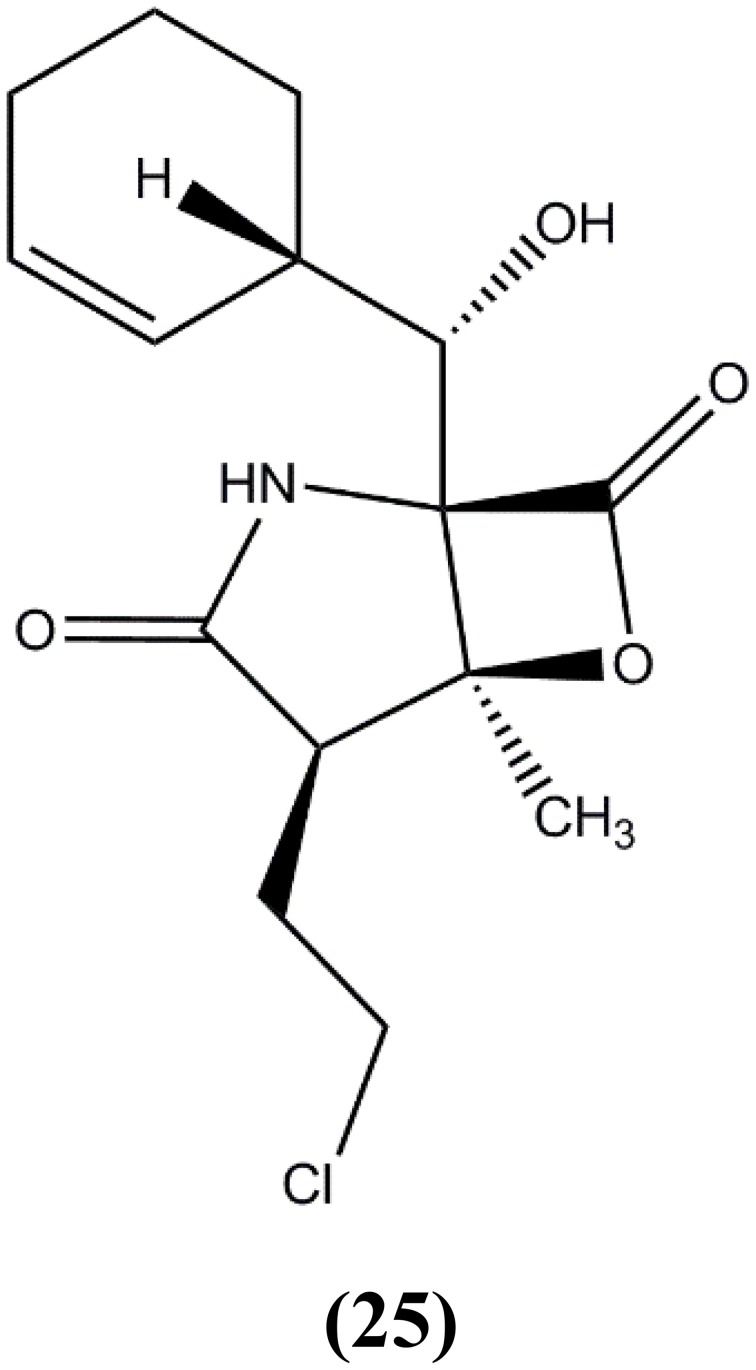
A significant feature of Salinispora tropica is its ability to produce structurally unique secondary metabolites. Feling et al. (2003) reported that the Salinispora tropica CNB-392 isolated from a mangrove area in Chub Cay, Bahamas produced a unique metabolite named salinosporamide A (25) (Figure 4) attached with γ-lactam-β-lactone bicyclic core with clasto-lactacystin-β-lactone (also called as omuralide), a metabolite discovered from terrestrial Streptomyces sp. The study showed that this compound was able to kill different degrees of cancer cells and possessed strong inhibition of the 20S proteasome. Salinosporamide A was tested for the effects on proteasome function due to its structural relationship to omuralide. Results exhibited that Salinosporamide A was 35 times more potent compared to omuralide. Currently, the Salinosporamide A is in phase I human clinical trials as a potential anticancer drug (Fenical et al., 2009).
Figure 4.
The structure of Salinosporamide A (25).
Saccharopolyspora
The genus Saccharopolyspora was discovered in 1974 by Lacey and Goodfellow from heated sugar cane bagasse (Guan et al., 2011). At the time of writing, the genus Saccharopolyspora contained 26 species and 3 subspecies isolated from various sources (http://www.bacterio.net/). The first complete genome sequence of the Saccharopolyspora strain was reported by Oliynyk et al. (2007). The genome size of Saccharopolyspora erythraea is about 8.3 Mbp predicted to encode 7264 genes. S. erythraea genome contains at least 25 gene clusters for the production of known or predicted secondary metabolites. The draft genome of Saccharopolyspora spinosa NRRL 18395 was reported by Pan et al. (2011) which consists of approximately 8.6 Mbp with a GC content of 67.94%. The latest draft genome of Saccharopolyspora rectivirgula was reported by Pettersson et al. (2014). The draft genome of this strain consists of 182 contigs totaling 3,977,051 bp with a GC content of 68.9%.
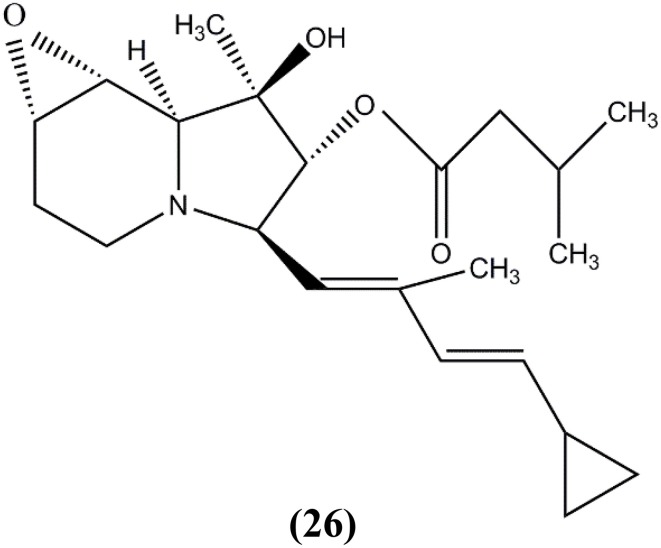
Izumikawa et al. (2012) isolated Saccharopolyspora sp. RL78 from mangrove soil in Nosoko, Ishigaki Island, Okinawa Prefecture, Japan. They discovered a new cyclizidine analog named JBIR-102 (26) (Figure 5), which exhibited cytotoxic activity against HeLa cells and human malignant pleural mesothelioma cell line, ACC-MESO-1.
Figure 5.
The structure of JBIR-102 (26).
Nocardiopsis
Nocardiopsis is a genus of the family Nocardiopsaceae discovered in 1976 by Meyer. The bacterium contains fragmenting mycelium and a cell wall containing meso-diaminopimelic acid with no diagnostically important carbohydrates (Li et al., 2012). The genus Nocardiopsis contains 45 species and 5 subspecies with validly published names (http://www.bacterio.net/). The first report on genome sequence of genus Nocardiopsis was reported by Sun et al. (2010). Members of Nocardiopsis dassonvillei have been isolated from a variety of natural habitats such as soils and marine sediments, plant, animal material, and human patients. According to Sun et al. (2010), the genome size of Nocardiopsis dassonvillei is 6,543,312 bp with a 73% GC content, and 3.3% of gene cluster is for secondary metabolite biosynthesis, transport, and catabolism. The latest complete whole genome sequence of genus Nocardiopsis was announced by Qiao et al. (2012). Nocardiopsis alba strain isolated from honeybee guts contains 5,848,211 bp with a GC content of 69.65%. Based on the Antibiotics and Secondary Metabolites Analysis Shell (antiSMASH), 18 biosynthetic gene clusters were identified and at least 8 clusters were predicted for production of known antibiotics and secondary metabolites.
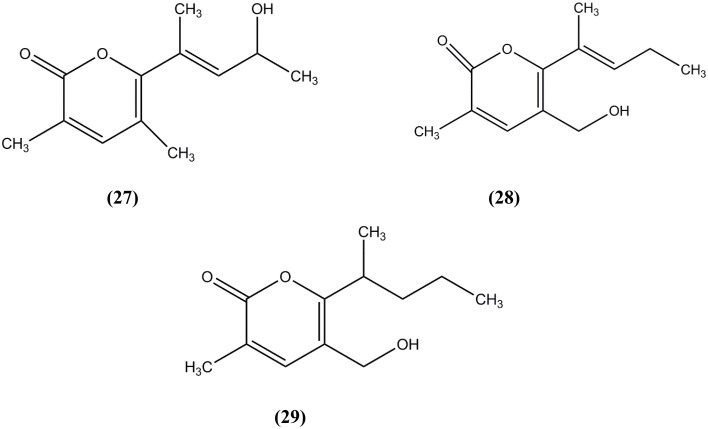
Nocardiopsis species were known to produce bioactive metabolites. Researchers discovered Nocardiopsis species were able to produce griseusin (Sun et al., 1991), apoptolidin (Kim et al., 1997), methylpendolmycin (Li et al., 2007a), thiopeptide (designated TP-1161) (Gandhimathi et al., 2009), lipopeptide biosurfactant (Ding et al., 2010), and napthhospironone A (Engelhardt et al., 2010). Three new 2-pyranone derivatives known as Nocardiatones A (27), B (28), and C (29) (Figure 6) were discovered from novel mangrove endophytic, Nocadiopsis sp. A00203 (Lin et al., 2010). This species was isolated from the leaves of Aegiceras corniculatum (Myrsinaceae). The structures of all compounds were examined using spectroscopic and mass-spectrometric analyses. The analysis of the anticancer activity of the compound was performed using HeLa cells and only Nocardiatones A showed weak cytotoxic activity against HeLa cells. No inhibition was observed, when these compounds were tested on Escherichia coli, Bacillus subtilis, Staphylococcus aureus, and yeasts.
Figure 6.
The structure of Nocardiatones A (27), B (28), and C (29).
Conclusion
The discovery of novel species and novel natural compounds from the mangrove rare actinobacteria is becoming a popular and attractive approach. Even though Strepotomyces is the genus producing various types of natural compounds for medical and pharmaceutical purposes, rare actinobacteria isolated from mangrove have a wide range of metabolites with different biological activities. Micromonospora is the largest genera among the rare actinobacteria which produces a rich source of natural products such as rifamycin, butemycin and β-carboline. Other genera of the mangrove rare actinobacteria namely, Jishengella, Salinispora, Saccharopolyspora and Nocardiopsis; exhibited various natural compounds which are potentially useful in the medical and pharmaceutical industry. Previously, one of the most common limitations of conventional metagenomics analysis is restricted taxonomy resolution. The advancement in biotechnology enables the study of strains variation and the dynamic behavior of microbial communities. The NGS technologies enabled the production of results in a rapid, low-cost and reliable manner compared to conventional approaches. These high-throughput sequencing technologies have enabled researchers to discover the microbial diversity from different environments; and also enabled the parallel sequencing of multiple bioactive strains for the identification of secondary metabolites gene clusters through comparative genomics analysis. The knowledge obtained from such studies will be vital for promoting combinatorial biosynthesis of actinobacteria related compounds.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by University of Malaya High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027 and No. A000001-50001) awarded to K-GC, and External Industry Grant (Biotek Abadi—Vote No. GBA-808138) awarded to L-HL. Authors would like to thank Dr. Sabrina Anne Jacob for improving the grammar of the manuscript.
References
- Albuquerque L., França L., Rainey F. A., Schumann P., Nobre M. F., da Costa M. S. (2011). Gaiella occulta gen.nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov. Syst. Appl. Microbiol. 34, 595–599. 10.1016/j.syapm.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Allen J. R. F., Holmstedt B. R. (1980). The simple β-carboline alkaloids. Phytochemistry 19, 1573–1582. 10.1016/S0031-9422(00)83773-5 [DOI] [Google Scholar]
- Alongi D. M. (1988). Bacterial productivity and microbial biomass in tropical mangrove sediments. Microbial. Ecol. 15, 59–79. 10.1007/BF02012952 [DOI] [PubMed] [Google Scholar]
- Andreote F. D. (2012). Metagenomics of mangrove. Encycl. Metag. 2014, 1–7. 10.1007/978-1-4614-6418-1_606-4 [DOI] [Google Scholar]
- Andreote F. D., Jiménez D. J., Chaves D., Dias A. C. F., Luvizotto D. M., Dini-Andreote F., et al. (2012). The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS ONE 7:e38600. 10.1371/journal.pone.0038600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ann M. H., Maria V. (2009). Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol. Biochem. 42, 536–542. 10.1016/j.soilbio.2009.11.023 [DOI] [Google Scholar]
- Ara I., Bakir M. A., Hozzein W. N., Kudo T. (2013). Population, morphological and chemotaxonomical characterization of diverse rare actinomycetes in the mangrove and medicinal plant rhizosphere. Afr. J. Microbiol. Res. 7, 1480–1488. 10.5897/AJMR12.452 [DOI] [Google Scholar]
- Ara I., Kudo T., Matsumoto A., Takahashi Y., Omura S. (2007). Nonomuraea maheshkhaliensis sp. nov., a novel actinomycete isolated from mangrove rhizosphere mud. J. Gen. Appl. Microbiol. 53, 159–166. 10.2323/jgam.53.159 [DOI] [PubMed] [Google Scholar]
- Ara I., Matsumoto A., Abdul Bakir M., Kudo T., Omura S., Takahashi Y. (2008). Actinomadura maheshkhaliensis sp. nov., a novel actinomycete isolated from mangrove rhizosphere soil of Maheshkhali, Bangladesh. J. Gen. Appl. Microbiol. 54, 335–342. 10.2323/jgam.54.335 [DOI] [PubMed] [Google Scholar]
- Ara I., Paramaswari S., Vikineswary S. (2002). Diversity of Micromonospora in Malaysian mangrove rhizosphere soils. Malays. J. Sci. 21, 51–59. [Google Scholar]
- Ashour M. A., Elkhayat E. S., Ebel R., Edrada R., Proksch P. (2007). Indole alkaloid from the red sea sponge Hyrtios erectus. Arkivoc 2007, 225–231. 10.3390/md11041061 [DOI] [Google Scholar]
- Atlas R. (1997). Principles of Microbiology. New York, NY: WCB McGrill-Hill. [Google Scholar]
- Baltz R. H. (2006). Combinatorial biosynthesis of novel antibiotics and other secondary metabolites in actinomycetes. SIM News 56, 148–160. [Google Scholar]
- Basak P., Pramanik A., Roy R., Chattopadhyay D., Bhattacharyya M. (2015). Cataloguing the bacterial diversity of the Sundarbans mangrove, India in the light of metagenomics. Genomics Data. 4, 90–92. 10.1016/j.gdata.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérdy J. (2005). Bioactive microbial metabolites (review). J. Antibiot. 58, 1–26. 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Bredholt H., Fjaerik E., Johnsen G., Zotchev S. B. (2008). Actinomycetes from sediments in the Trondheim Fjord, Norway: diversity and biological activity. Mar. Drugs 6, 12–24. 10.3390/md6010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. V., Philip G. K., Bunge J. A., Smith M. C., Bissett A., Lauro F. M., et al. (2009). Microbial community structure in the North Pacific Ocean. ISME J. 3, 1374–1386. 10.1038/ismej.2009.86 [DOI] [PubMed] [Google Scholar]
- Bull A. T., Stach J. E. (2007). Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 15, 491–499. 10.1016/j.tim.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Butler M. S. (2004). The role of natural products chemistry in drug discovery. J. Nat. Prod. 67, 2141–2153. 10.1021/np040106y [DOI] [PubMed] [Google Scholar]
- Chan K. G., Ismail Z. (2015). Tropical soil metagenome library reveals complex microbial assemblage. BioRxiv. (in press). 10.1101/018895 [DOI] [Google Scholar]
- Cross T. (1981). Aquatic actinomycetes: a critical survey of the occurrence, growth, and role of actinomycetes in aquatic habitats. J. Appl. Bacteriol. 50, 397–423. 10.1111/j.1365-2672.1981.tb04245.x [DOI] [PubMed] [Google Scholar]
- Das S., Lyla P., Ajmal Khan S. (2008). Distribution and generic composition of culturable marine actinobacteria from the sediments of Indian continental slope of Bay of Bengal. Chin. J. Oceanol. Limn. 26, 166–177. 10.1007/s00343-008-0166-5 [DOI] [Google Scholar]
- Dassonneville B., Witulski B., Detert H. (2011). [2+2+2] Cycloadditions of alkynylynamides – A total synthesis of perlolyrine and the first total synthesis of “isoperlolyrine”. Eur. J. Org. Chem. 2011, 2836–2844. 10.1002/ejoc.201100121 [DOI] [Google Scholar]
- Dastager S. G., Qiang Z. L., Damare S., Tang S. K., Li W. J. (2012). Agromyces indicus sp. nov., isolated from mangroves sediment in Chorao Island, Goa, India. Antonie van Leeuwenhoek 102, 345–352. 10.1007/s10482-012-9744-9 [DOI] [PubMed] [Google Scholar]
- Dias A. C. F., Andreote F. D., Dini-Andreote F., Lacava P. T., Sá A. L. B., Melo I. S., et al. (2009b). Diversity and biotechnological potential of culturable bacteria from Brazilian mangrove sediment. World J. Microbiol. Biotechnol. 25, 1305–1311. 10.1007/s11274-009-0013-7 [DOI] [Google Scholar]
- Ding Z. G., Li M. G., Zhao J. Y., Ren J., Huang R., Xie M. J., et al. (2010). Naphthospironone A: an unprecedented and highly functionalized polycyclic metabolite from an alkaline mine waste extremophile. Chemistry 16, 3902–3905. 10.1002/chem.200903198 [DOI] [PubMed] [Google Scholar]
- Ding Z. G., Zhao J. Y., Yang P. W., Li M., Huang R., Cui X. L., et al. (2009). 1H and 13C NMR assignment of eight nitrogen containing compounds from Nocardia alba sp. nov (YIM 30243T). Magn. Reson. Chem. 47, 366–70. 10.1002/mrc.2393 [DOI] [PubMed] [Google Scholar]
- Don M. J., Shen C. C., Lin Y. L., Syu W. J., Ding Y. H., Sun C. M. (2005). Nitrogen-containing compounds from Salvia miltiorrhiza. J. Nat. Prod. 68, 1066–1070. 10.1021/np0500934 [DOI] [PubMed] [Google Scholar]
- Donadio S., Monciardini P., Alduina R., Mazza P., Chiocchini C., Cavaletti L., et al. (2002). Microbial technologies for the discovery of novel bioactive metabolites. J. Biotechnol. 99,187–198. 10.1016/S0168-1656(02)00209-2 [DOI] [PubMed] [Google Scholar]
- Duan C. J., Feng J. X. (2010). Mining metagenomes for novel cellulose genes. Biotechnol. Lett. 32, 1765–1775. 10.1007/s10529-010-0356-z [DOI] [PubMed] [Google Scholar]
- Eccleston G. P., Brooks P. R., Kurtböke D. I. (2008). The occurrence of bioactive micromonosporae in aquatic habitats of the Sunshine Coast in Australia. Mar. Drugs 6, 243–261. 10.3390/md20080012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt K., Degnes K. F., Kemmler M., Bredholt H., Fjaervik E., Klinkenberg G., et al. (2010). Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 76, 4696–4976. 10.1128/AEM.00741-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feling R. H., Buchanan G. O., Mincer T. J., Kauffman C. A., Jensen P. R., Fenical W. (2003). Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl. 42, 355–357. 10.1002/anie.200390115 [DOI] [PubMed] [Google Scholar]
- Fenical W., Baden D., Burg M., de Ville de Goyet C., Grimes J. D., Katz M., et al. (1999). From Monsoons to Microbes: Understanding the Ocean's Role in Human Health. Washington, DC: National Academy Press. [PubMed] [Google Scholar]
- Fenical W., Jensen P. R., Palladino M. A., Lam K. S., Lloyd G. K., Potts B. C. (2009). Discovery and development of anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 17, 2175–2180. 10.1016/j.bmc.2008.10.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel K. C., Nam S. J., Fenical W., Jensen P. R. (2011). Evolution of secondary metabolites genes in three closely related marine actinomycetes species. Appl. Environ. Microb. 77, 7261–7270. 10.1128/AEM.05943-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhimathi R., Seghal Kiran G., Hema T. A., Selvin J., Rajeetha Raviji T., Shanmughapriya S. (2009). Production and characterization of lipopeptide biosurfactant by a sponge-associated marine actinomycetes Nocardiopsis alba MSA10. Bioprocess Biosyst. Eng. 32, 825–835. 10.1007/s00449-009-0309-x [DOI] [PubMed] [Google Scholar]
- Gayathri P., Muralikrishnan V. (2013). Isolation and characterization of endophytic actinomycetes from mangrove plant for antimicrobial activity. Int. J. Curr. Microbiol. App. Sci. 2, 78–89. [Google Scholar]
- Gomes N. C. M., Cleary D. F. R., Pinto F. N., Egas C., Almeida A., Cunha A., et al. (2010). Taking root: enduring effect of rhizosphere bacterial colonization in mangroves. PLoS ONE 5:e14065. 10.1371/journal.pone.0014065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M., Williams S. T. (1983). Ecology of actinobacteria. Annu. Rev. Microbiol. 37, 189–215. [DOI] [PubMed] [Google Scholar]
- Grassauer A., Weinmuellner R., Meier C., Pretsch A., Prieschl-Grassauer E., Unger H. (2008). Iota-carrageenan is a potent inhibitor of rhinovirus infection. Virol. J. 5, 107. 10.1186/1743-422X-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth I., Schumann P., Schuetze B., Augsten K., Kramer I., Stackebrandt E. (1999). Beutenbergia carvernae gen. nov., sp. nov., an L-lysine-containing actinomycete isolated from a cave. Int. J. Syst. Microbiol. 49, 1733–1740. [DOI] [PubMed] [Google Scholar]
- Guan T. W., Wu N., Xia Z. F., Ruan J. S., Zhang X. P., Huang Y., et al. (2011). Saccharopolyspora lacisalsi sp. nov., a novel halophilic actinomycete isolated from a salt lake in Xinjiang, China. Extremophiles 15, 373–378. 10.1007/s00792-011-0369-0 [DOI] [PubMed] [Google Scholar]
- Hamada M., Tamura T., Yamamura H., Suzuki K., Hayakawa M. (2012). Lysinimicrobium mangrovi gen. nov., sp. nov., an actinobacterium isolated from the rhizosphere of a mangrove. Int. J. Syst. Evol. Microbiol. 62, 1731–1735. 10.1099/ijs.0.035493-0 [DOI] [PubMed] [Google Scholar]
- Handelsman J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685. 10.1128/MMBR.68.4.669-685.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano K. (1997). Actinomycetes populations in mangrove rhizosphere. IFO Res. Commun. 18, 26–31. 10.5897/AJMR12.452 [DOI] [Google Scholar]
- Hayakawa M. (2008). Studies on the isolation and distribution of rare actinomycetes in soil. Actinomycetologica. 22, 12–19. 10.3209/saj.SAJ22010322773732 [DOI] [Google Scholar]
- He J., Xu Y., Sahu M. K., Tian X. P., Nie G. X., Xie Q., et al. (2012). Actinomadura sediminis sp. nov., a marine actinomycete isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 62, 1110–1116. 10.1099/ijs.0.032979-0 [DOI] [PubMed] [Google Scholar]
- Heuer H., Krsek M., Baker P., Smalla K., Wellington E. M. H. (1997). Analysis of actinobacteria communities by specific amplification genes encdofing 16S rRNA and gel electrophoresis separation in denaturing gradients. Appl. Environ. Microb. 63, 3233–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A. M., Alvarado J., Bruce D., Chertkov O., De Hoff P. L., Detter J. C., et al. (2013). Complete genome sequence of Micromonospora strain L5, a potential plant-growth-regulating actinomycete, originally isolated from Casuarina equisetifolia root nodules. Genome Announc. 1:e00759-13. 10.1128/genomeA.00759-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A. M., Valdés M. (2009). Micromospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol. Biochem. 42, 536–542. 10.1016/j.soilbio.2009.11.023 [DOI] [Google Scholar]
- Holguin G., Vazquez P., Bashan Y. (2001). The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol. Fertil. Soils. 33, 265–278. 10.1007/s003740000319 [DOI] [Google Scholar]
- Hong K., Gao A. H., Xie Q. Y., Gao H., Zhuang L., Lin H. P., et al. (2009). Actinobacteria for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44. 10.3390/md7010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. Q., Lv J. S., Hu Y. H., Fang Z., Zhang K. S., Bao S. X. (2008). Micromonospora rifamycinica sp. nov., a novel actinomycete from mangrove sediment. Int. J. Syst. Evol. Microbiol. 58, 17–20. 10.1099/ijs.0.64484-0 [DOI] [PubMed] [Google Scholar]
- Huang H., Wu X., Yi S., Zhou Z., Zhu J., Fang Z., et al. (2009). Rifamycin S and its geometric isomer produced by a newly found actinomycetes, Microsmonospora rifamycinica. Antonie van Leeuwenhoek. 95, 143–148. 10.1007/s10482-008-9297-0 [DOI] [PubMed] [Google Scholar]
- Huang Z. Y., Tang J. S., Gao H., Li Y. J., Hong K., Li J., et al. (2011). Studies on the cytotoxic constituents from marine actinomycete Micromonospora sp. M2DG17. Chin. Mar. Drugs 30, 29–33. 10.3390/md12052590 [DOI] [Google Scholar]
- Hung H. C., Tseng C. P., Yang J. M., Ju Y. W., Tseng S. N., Chen Y. F., et al. (2009). Aurintricarboxylic acid inhibits influenza virus neuraminidase. Antiviral. Res. 81, 123–131. 10.1016/j.antiviral.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M., Hosoya T., Takagi M., Shin-ya K. (2012). A new cyclizidine analog-JBIR-102-from Saccharopolyspora sp. RL78 isolated from mangrove soil. J. Antibiot. (Tokyo). 65, 41–43. 10.1038/ja.2011.99 [DOI] [PubMed] [Google Scholar]
- Jennerjahn T. C., Ittekkot V. (2002). Relevance of mangroves for the production and deposition of organic matter along tropical continental margins. Naturwissenschaften 89, 23–30. 10.1007/s00114-001-0283-x [DOI] [PubMed] [Google Scholar]
- Jensen P. R., Mafnas C. (2006). Biogeography of the marine actinomycete Salinispora. Environ. Microbiol. 8, 1881–1888. 10.1111/j.1462-2920.2006.01093.x [DOI] [PubMed] [Google Scholar]
- Jeric I., Simicic L., Stipetic M., Horvat S. (2000). Synthesis and reactivity of the monosaccharide esters of amino acids as models of teichoic acid fragment. Glycocon. J. 17, 273–282. 10.1023/A:1007157018792 [DOI] [PubMed] [Google Scholar]
- Jiang C. L., Xu L. H. (1996). Diversity of aquatic actinomycetes in lakes of the middle plateau, Yunnan, China. Appl. Environ. Microbiol. 62, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Xu P., Lou K., Xu L. H., Liu Z. H. (2008). Problems and countermeasure on development of pharmaceuticals from actinobacteria resource. Microbiology 35, 272–274. 10.4236/aim.2013.31001 [DOI] [Google Scholar]
- Jiao W. H., Gao H., Li C. Y., Zhou G. X., Kitanaka S., Ohmura A., et al. (2010). Beta-carboline alkaloids from the stems of Picrasma quassioides. Magn. Reson. Chem. 48, 490–495. 10.1002/mrc.2602 [DOI] [PubMed] [Google Scholar]
- Jones R. T., Robeson M. S., Lauber C. L., Hamady M., Knight R., Fierer N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453. 10.1038/ismej.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose P. A., Jebakumar S. R. D. (2013). Non-streptomycete actinomycetes nourish the current microbial antibiotic drug discovery. Front. Microbiol. 4:240. 10.3389/fmicb.2013.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusoff K. (2013). Malaysian mangrove forests and their significance to the coastal marine environment. Pol. J. Environ. Stud. 22, 979–1005. [Google Scholar]
- Khanna M., Solanki R., Lal R. (2011). Selective isolation of rare actinomycetes producing novel antimicrobial compounds. Int. J. Adv. Biotechnol. Res. 2, 357–375. 10.1016/j.micres.2012.06.005 [DOI] [Google Scholar]
- Kim J. W., Adachi H., Shin-ya K., Hayakawa Y., Seto H. (1997). Apoptolidin, a new apoptosis inducer in transformed cells from Nocardiopsis sp. J. Antibiot. (Tokyo). 50, 628–630. 10.716/antibiotics.50.628 [DOI] [PubMed] [Google Scholar]
- Koehn F. E., Carter G. T. (2005), The evolving role of natural products in drug discovery. Nat. Rev. Drug. Discov. 4, 206–220. 10.3390/md8030438 [DOI] [PubMed] [Google Scholar]
- Kurahashi M., Fukunaga Y., Sakiyama Y., Harayama S., Yokota A. (2010). Euzebya tangerine gen. nov., sp. nov., a deeply branching marine actinobacterium isolated from the sea Holothuria edulis, and proposal of Euzebyaceae ifam. Nov., Euzebyales ord. nov. and Nitriliruptoridae subclassis nov. Int. J. Syst. Evol. Microbiol. 60, 2314–2319. 10.1099/ijs.0.016543-0 [DOI] [PubMed] [Google Scholar]
- Kyeremeh K., Acquah K. S., Sazak A., Houssen W., Tabudravu J., Deng H., et al. (2014). Butremycin, the 3-hydroxyl derivative of ikarugamycin and a protonated aromatic tautomer of 5′-methylthioinosine from a Ghanaian Micromonospora sp. K310. Mar. Drugs 12, 999–1012. 10.3390/md12020999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageiro M. M., Moura M. J., Reis A., Ferreira M. J. C. (2007). Microbial proteases application in leather industry. J. Biotechnol. 131, S239–S240. 10.1016/j.jbiotec.2007.07.717 [DOI] [Google Scholar]
- Lam K. S. (2006). Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 9, 245–251. 10.1016/j.mib.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Lazzarini A., Cavaletti L., Toppo G., Marinelli F. (2000). Rare genera of actinomycetes as potential sources of new antibiotics. Antonie van Leeuwenhoek 78, 399–405. 10.1023/A:1010287600557 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Azman A.-S., Zainal N., Eng S.-K., Ab Mutalib N.-S., Yin W.-F., et al. (2014c). Microbacterium mangrovi sp. nov., an amylolytic actinobacterium isolated from Tanjung Lumpur mangrove forest. Int. J. Syst. Evol. Microbiol. 64, 3513–3519. 10.1099/ijs.0.062414-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Azman A.-S., Zainal N., Yin W.-F., Ab Mutalib N.-S., Chan K.-G. (2015). Sinomonas humi sp. nov., an amylolytic actinobacterium isolated from mangrove forest soil. Int. J. Syst. Evol. Microbiol. 65, 996–1002. 10.1099/ijs.0.000053 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Cheah Y.-K., Mohd Sidik S., Ab Mutalib N.-S., Tang Y.-L., Lin H.-P., et al. (2012). Molecular characterization of Antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotechnol. 28, 2125–2137. 10.1007/s11274-012-1018-1 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Zainal N., Azman A.-S., Ab Mutalib N.-S., Hong K., Chan K.-G. (2014d). Mumia flava gen. nov., sp. nov., an actinobacterium of the family Nocardioidaceae. Int. J. Syst. Evol. Microbiol. 64, 1461–1467. 10.1099/ijs.0.058701-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Zainal N., Azman A.-S., Eng S.-K., Ab Mutalib N.-S., Yin W.-F., et al. (2014b). Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 64, 3297–3306. 10.1099/ijs.0.065045-0 [DOI] [PubMed] [Google Scholar]
- Lee L.-H., Zainal N., Azman A.-S., Eng S.-K., Goh B.-H., Yin W.-F., et al. (2014a). Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Scientific World J. 2014:698178. 10.1155/2014/698178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang J., Zhu W. Y., He J., Tian X. P., Xie Q., et al. (2012). Nocardiopsis coralliicola sp. nov., isolated from the gorgonian coral, Menella praelonga. Int. J. Syst. Evol. Microbiol. 62, 1653–1658. 10.1099/ijs0.035402-0 [DOI] [PubMed] [Google Scholar]
- Li L., Mao Y. J., Xie Q. Y., Deng Z., Hong K. (2013a). Micromonospora avicenniae sp. nov., isolated from a root of Avicennia marina. Antonie van Leeuwenhoek 103, 1089–1096. 10.1007/s10482-013-9888 [DOI] [PubMed] [Google Scholar]
- Li L., Tang Y. L., Wei B., Xie Q. Y., Deng Z., Hong K. (2013b). Micromonospora sonneratiae sp. nov., isolated from a root of Sonneratia apetala. Int. J. Syst. Evol. Microbiol. 63, 2383–2388. 10.1099/ijs.0.043570-0 [DOI] [PubMed] [Google Scholar]
- Li Y. Q., Li M. G., Li W., Zhao J. Y., Ding Z. G., Cui X. L., et al. (2007a). Griseusin D, a new pyranonaphthoquinone derivative from a alkaphilic Nocardiopsis sp. J. Antibiot. (Tokyo). 60, 757–761. 10.1038/ja.2007.100 [DOI] [PubMed] [Google Scholar]
- Liang J. B., Chen Y. Q., Lan C. Y., Tam N. F. Y., Zan Q. J., Huang L. N. (2007). Recovery of novel bacterial diversity from mangrove sediment. Mar. Biol. 150, 739–747. 10.1007/s00227-006-0377-2 [DOI] [Google Scholar]
- Liao Z. L., Tang S. K., Guo L., Zhang Y. Q., Tian X. P., Jiang C. L., et al. (2009). Verrucosispora lutea sp. nov., isolated from a mangrove sediment sample. Int. J. Syst. Evol. Microbiol. 59, 2269–2273. 10.1099/ijs.0.008813-0 [DOI] [PubMed] [Google Scholar]
- Lin C., Lu C., Shen Y. (2010). Three new 2-pyranone derivatives from mangrove endophytic actinomycete strain Nocardiopsis sp. A00203. Rec. Nat. Prod. 4, 176–179. [Google Scholar]
- Mahajan G. B., Balachandran L. (2012). Antibacterial agents from actinomycetes- a review. Front. Biosci. 4:e373. 10.2741/E373 [DOI] [PubMed] [Google Scholar]
- Maldonado L. A., Fenical W., Jensen P. R., Kauffman C. A., Mincer T. J., Ward A. C., et al. (2005). Salinispora arenicola gen. nov., sp. nov. and Salinispora tropicasp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 55, 1759–1766. 10.1099/ijs.0.63625-0 [DOI] [PubMed] [Google Scholar]
- Marco V., Carlos C., Andreas T., Govind C., Gerald F. F., Keith F. C., et al. (2007). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. 10.1128/MMBR.00005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Nakai K., Morisaki K., Omura S., Takahashi Y. (2009). Demequina salsinemoris sp. nov., isolated on agar media supplemented with ascorbic acid or rutin. Int. J. Syst. Evol. Microbiol. 60, 1206–1209. 10.1099/ijs.0.012617-0 [DOI] [PubMed] [Google Scholar]
- McCarthy A. J., Williams S. T. (1992). Actinobacteria as agents of biodegradation in the environment—a review. Gene 115, 189–192. 10.1016/0378-1119(92)90558-7 [DOI] [PubMed] [Google Scholar]
- Moloudizargari M., Mikaili P., Aghajanshakeri S., Asghari M. H., Shayegh J. (2013). Pharmacological and therapeutic effects of Peganum harmala and its main alkaloids. Pharmacogn. Rev. 7, 199–212. 10.4103/0973-7847.120524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- Naik G., Shukla S., Mishra S. K. (2013). Isolation and characterization of actinomyctes isolates for production of antimicrobial compounds. J. Microbiol. Biotech. Res. 3, 33–36. 10.1016/S2221-1691(13)60092-1 [DOI] [Google Scholar]
- Nanjwade B. K., Chandrashekhara S., Goudanavar P. S., Shamarez A. M., Manvi F. V. (2010). Production of antibiotics from soil-isolated actinomycetes and evaluation of their antimicrobial activities. Trop. J. Pharm. Res. 9, 373–377. 10.4314/tjpr.v9i4.58933 [DOI] [Google Scholar]
- Niedringhaus T. P., Milanova D., Kerby M. B., Synder M. P., Barron A. E. (2011). Landscape of Next-Generation sequencing technologies. Anal. Chem. 83, 4327–4342. 10.1021/ac2010857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH HMP Working Group Peterson, J., Garges S., Giovanni M., Mclnnes P., Wang L., et al. (2009). The NIH human microbiome project. Genome Res. 19, 2317–2323. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaki S., Tsiamis G. (2013). Microbial diversity in the era of omic technologies. BioMed. Res. Int. 2013:958719. 10.1155/2013/958719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano C., García I., Gonzáles A., Rodriguez M., Rozas D., Rubio J., et al. (2014). Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotech. 7, 242–256. 10.1111/1751-7915.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliynyk M., Samborskyy M., Lester J. B., Mironenko T., Scott N., Dickens S., et al. (2007). Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL 23338. Nat. Biotech. 25, 447–453. 10.1038/nbt1297 [DOI] [PubMed] [Google Scholar]
- Pan Y., Yang X., Zhang R., Hu Y., Zhou Y., Wang J., et al. (2011). Genome sequencing of the spinosyns-producing bacterium Saccharopolyspora spinosa NRRL 18395. J. Bacteriol. 193, 3150–3151. 10.1128/JB.00344-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson B. M. F., Behra P. R. K., Manduva S., Das S., Dasgupta S., Bhattacharya A., et al. (2014). Draft genome sequence of Saccharopolyspora rectivirgula. Genome Announc. 2:e01117-13. 10.1128/genomeA.01117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Chen L., Li Y., Wang J., Zhang W., Chen S. (2012). Whole-genome sequence of Nocardiopsis alba strain ATCC BAA-2165, associated with honeybees. J. Bacteriol. 194, 6358–6359. 10.1128/JB.01522-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G. S. N., Garcia-Pichel F. (2009). Description of Patulibacter americanus sp. nov., isolated from biological soil crusts, emended description of the genus Patulibacter Takahashi et al. 2006 and proposal of Solirubrobacterales ord. nov. and Thermoleophilales ord. nov. Int. J. Syst. Evol. Microbiol. 59, 87–94. 10.1099/ijs.0.64185-0 [DOI] [PubMed] [Google Scholar]
- Redenbach M., Scheel J., Schmidt U. (2000). Chromosome topology and genome size of selected actinomycetes species. Antonie van Leeuwenhoek 78, 227–235. 10.1023/A:1010289326752 [DOI] [PubMed] [Google Scholar]
- Reigstad L. J., Bartossek R., Schleper C. (2011). Preparation of high-molecular weight DNA and metagenomic libraries from soils and hot springs. Methods Enzymol. 496, 319–344. 10.1016/B978-0-12-386489-5.00013-0 [DOI] [PubMed] [Google Scholar]
- Ren J., Li L., Wei B., Tang Y. L., Deng Z. X., Sun M., et al. (2013). Micromonospora wenchangensis sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 63, 2389–2395. 10.1099/ijs.0.045476-0 [DOI] [PubMed] [Google Scholar]
- Salomon C. E., Magarvey N. A., Sherman D. H. (2004). Merging the potential of microbial genetics with biological and chemical diversity: an even brighter future for marine natural product drug discovery. Nat. Prod. Rep. 21, 105–121. 10.1039/B301384G [DOI] [PubMed] [Google Scholar]
- Sanapareddy N., Hamp T. J., Gonzales L. C., Hilger H. A., Fodor A. A., Clinton S. M. (2009). Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl. Environ. Microbiol. 75, 1688–1696. 10.1128/AEM.01210-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannai Y., Fujimori T., Kato K. (1982). Studies on flavor components of roasted chicory root. Agric. Biol. Chem. 46, 429–433. 10.1080/00021369.1982.10865066 [DOI] [Google Scholar]
- Ser H.-L., Zainal N., Palanisamy U. D., Goh B.-H., Yin W.-F., Chan K.-G., et al. (2015). Streptomyces gilvigriseus sp. nov., a novel actinobacterium isolated from mangrove forest soil. Antonie van Leeuwenhoek 107, 1369–1378. 10.1007/s10482-015-0431-5 [DOI] [PubMed] [Google Scholar]
- Sivakumar K., Sahu M. K., Kathiresan K. (2005). Isolation and characterization of streptomycetes producing antibiotics from mangrove environment. Asian J. Microbiol. Biotechnol. Environ. Sci. 7, 457–464. [Google Scholar]
- Songsumanus A., Tanasupawat S., Igarashi Y., Kudo T. (2013). Micromonospora maritima sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 63, 554–559. 10.1099/ijs.0.039180-0 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., van Pelt S., Tourova T. P., Evtushenko L. I. (2009). Nitriliruptor alkaliphilus gen.nov., sp. nov., a deep-lineage haloalkaliphlic actinobacterium from soda lakes capable of growth on aliphatic nitriles, and proposal of Nitriliruptoraceae fam. nov. and Nitriliruptorales ord. nov. Int. J. Syst. Evol. Microbiol. 59, 248–253. 10.1099/ijs.0.002204-0 [DOI] [PubMed] [Google Scholar]
- Spalding M., Blasco F., Field C. (1997). World Mangrove Atlas. Japan: The International Society for Mangrove Ecosystem. [Google Scholar]
- Staley C., Unno T., Gould T. J., Jarvis B., Phillips J., Cotner J. B., et al. (2013). Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J. Appl. Microbiol. 115, 1147–1158. 10.1111/jam.12323 [DOI] [PubMed] [Google Scholar]
- Su B. N., Chang L. C., Park E. J., Cuendet M., Santarsiero B. D., Mesecar A. D., et al. (2002). Bioactive constituents of the seeds of Brucea javanica. Planta. Med. 68, 730–733. 10.1055/s-2002-33798 [DOI] [PubMed] [Google Scholar]
- Subramani R., Aalbersberg W. (2012). Marine actinomycetes: an ongoing source of novel bioactive metabolite. Microbiol. Res. 167, 571–580. 10.1016/j.micres.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Sun H., Lapidus A., Nolan M., Lucas S., Glavina Del Rio T., Tice H., et al. (2010). Complete genome sequence of Nocardiopsis dassonvillei type strain (IMRU 509T). Stand. Genomic. Sci. 3, 325–336. 10.4056/sigs.1363462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. H., White C. B., Dedinas J., Cooper R., Sedlock D. M., et al. (1991). Methylpendolmycin, an indolactam from a Nocardiopsis sp. J. Nat. Prod. 54, 1440–1443. 10.1021/np50077a040 [DOI] [PubMed] [Google Scholar]
- Suriyachadkun C., Chunhametha S., Ngaemthao W., Tamura T., Kirtikara K., Sanglier J. J., et al. (2011). Sphaerisporangium krabiense sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 61, 2890–2894. 10.1099/ijs.0.027151-0 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Ômura S. (2003). Isolation of new actinobacteria strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol. 49, 141–154. 10.2323/jgam.49.141 [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Hatano K. (2001). Agromyces luteolus sp. nov., Agromyces rhizospherae sp. nov. and Agromyces bracchium sp. nov., from the mangrove rhizosphere. Int. J. Syst. Evol. Microbiol. 51, 1529–1537. 10.1099/00207713-51-4-1529 [DOI] [PubMed] [Google Scholar]
- Tamura T., Hatano K., Suzuki K. (2006). A new genus of the family Micromonosporaceae, Polymorphospora gen. nov., with description of Polymorphospora rubra sp. nov. Int. J. Syst. Evol. Microbiol. 56, 1959–1964. 10.1099/ijs.0.64046-0 [DOI] [PubMed] [Google Scholar]
- Tamura T., Sakane T. (2005). Asanoa iriomotensis sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 55, 725–727. 10.1099/ijs.0.02982-0 [DOI] [PubMed] [Google Scholar]
- Tang Y. L., Lin H. P., Xie Q. Y., Li L., Peng F., Deng Z., et al. (2013). Actinoallomurus acanthiterrae sp. nov., an actinomycete isolated from rhizosphere soil of the mangrove plant Acanthus ilicifolius. Int. J. Syst. Evol. Microbiol. 63, 1874–1879. 10.1099/ijs.0.043380-0 [DOI] [PubMed] [Google Scholar]
- Thawai C., Tanasupawat S., Kudo T. (2008). Micromonospora pattaloongensis sp. nov., isolated from a Thai mangrove forest. Int. J. Syst. Evol. Microbiol. 58, 1516–1521. 10.1099/ijs.0.65410-0 [DOI] [PubMed] [Google Scholar]
- Thompson C. E., Beys-da-Silva W. O., Santi L., Berger M., Vainstein M. H., Guima rães J. A., et al. (2013). A potential source for cellulolytic enzyme discovery and environmental aspects revealed through metagenomics of Brazilian mangroves. AMB Express 3:65. 10.1186/2191-0855-3-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari K., Gupta R. K. (2012). Rare actinomycetes: a potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 32, 108–132. 10.3109/07388551.2011.562482 [DOI] [PubMed] [Google Scholar]
- Tiwari K., Gupta R. K. (2013). Diversity and isolation of rare actinomycetes: an overview. Crit. Rev. Biotechnol. 39, 256–294. 10.3109/1040841X.2012.709819 [DOI] [PubMed] [Google Scholar]
- Tomlinson P. B. (1986). The Botany of Mangroves. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Tressl R., Nittka C., Kersten E. (1995). Formation of isoleucine-specific maillard products from [l-13C]-d-glucose and [l-13C]-D-fructose. J. Agric. Food Chem. 43, 1163–1169. 10.1021/jf00053a009 [DOI] [Google Scholar]
- Tseng M., Liao H. C., Chiang W. P., Yuan G. F. (2011). Isoptericola chiayiensis sp. nov., isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 61, 1667–1670. 10.1099/ijs.0.022491-0 [DOI] [PubMed] [Google Scholar]
- Udwary D. W., Zeigler L., Asolkar R. N., Singan V., Lapidus A., Fenical W., et al. (2007). Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U.S.A. 104, 10376–10381. 10.1073/pnas.0700962104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xu X. X., Qu Z., Wang H. L., Lin H. P., Xie Q. Y., et al. (2011a). Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int. J. Syst. Evol. Microbiol. 61, 320–324. 10.1099/ijs.0.020461-0 [DOI] [PubMed] [Google Scholar]
- Wang F., Xu X. X., Qu Z., Wang C., Lin H. P., Xie Q. Y., et al. (2011b). Nonomuraea wenchangensis sp. nov., isolated from mangrove rhizosphere soil. Int. J. Syst. Evol. Microbiol. 61, 1304–1308. 10.1099/ijs.0.025742-0 [DOI] [PubMed] [Google Scholar]
- Wang P., Kong F., Wei J., Wang Y., Wang W., Hong K., et al. (2014). Alkaloids from the Mangrove-Derived Actinomycete Jishengella endophytica 161111. Mar. Drugs 12, 477–490. 10.3390/md12010477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. A., Zhao Y. M., Yu N. J. (2007). A novel analgesic pyrazine derivative from the leaves of Croton tiglium L. J. Asian. Nat. Prod. Res. 9, 437–441. 10.1080/10286020500384781 [DOI] [PubMed] [Google Scholar]
- Xi L., Zhang L., Ruan J., Huang Y. (2012). Description of Verrucosispora qiuiae sp. nov., isolated from mangrove swamp sediment, and emended description of the genus Verrucosispora. Int. J. Syst. Evol. Microbiol. 62, 1564–1569. 10.1099/ijs.0.033787-0 [DOI] [PubMed] [Google Scholar]
- Xie Q. Y., Qu Z., Lin H. P., Li L., Hong K. (2012). Micromonospora haikounsis sp. nov., isolated from mangrove soil. Antonie van Leeuwenhoek 101, 649–655. 10.1007/s10482-011-9682-y [DOI] [PubMed] [Google Scholar]
- Xie Q. Y., Wang C., Wang R., Qu Z., Lin H. P., Goodfellow M., et al. (2011). Jishengella endophytica gen.nov., sp. nov., a new member of the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 61, 1153–1159. 10.1099/ijs.0.025288-0 [DOI] [PubMed] [Google Scholar]
- Xie X. C., Mei W. L., Zhao Y. X., Hong K., Dai H. F. (2006). A new degraded sesquiterpene from marine actinomycete Streptomyces sp. 0616208. Chin. Chem. Lett. 17, 1463–1465. [Google Scholar]
- Xu X. X., Qu Z., Wang H., Lin H. P., Wang C., Xie Q. Y., et al. (2011). Asanoa hainanensis sp. nov., isolated from rhizosphere soil of Acrostichum speciosum in a mangrove, and emended description of the genus Asanoa. Int. J. Syst. Evol. Microbiol. 61, 2384–2388. 10.1099/ijs.0.025825-0 [DOI] [PubMed] [Google Scholar]
- Xu X. X., Wang H. L., Lin H. P., Wang C., Qu Z., Xie Q. Y., et al. (2012). Microbispora hainanensis sp. nov., isolated from rhizosphere soil of Excoecaria agallocha in a mangrove. Int. J. Syst. Evol. Microbiol. 62, 2430–2434. 10.1099/ijs.0.037267-0 [DOI] [PubMed] [Google Scholar]
- Yan B., Hong K., Yu Z. N. (2006). Achaeal communities in mangrove soil characterized by 16S rRNA gene clones. J. Microbiol. 44, 566–571. [PubMed] [Google Scholar]
- Yan X., Geng A., Zhang J., Wei Y., Zhang L., Qian C., et al. (2013). Discovery of (hemi-) cellulase genes in a metagenomic library from a biogas digester using 454 pyrosequencing. Appl. Microbiol. Biotechnol. 97, 8173–8182. 10.1007/s00253-013-4927-5 [DOI] [PubMed] [Google Scholar]
- Yang J., Gao J., Cheung A., Liu B., Schwendenmann L. (2013). Vegetation and sediment characteristics in expanding mangrove forest in New Zealand. Eastuar. Coast. Shelf. S. 134, 11–18. 10.1016/j.ecss.2013.09.017 [DOI] [Google Scholar]
- Yang S. W., Cordell G. A. (1997). Metabolism studies of indole derivatives using a staurosporine producer, Streptomyces staurosporeus. J. Nat. Prod. 60, 44–48. 10.1021/np960566u [DOI] [PubMed] [Google Scholar]
- Zhang Z. P. (2003). Microbial Pharmacology. Beijing: Chemical Industry Press. [Google Scholar]
- Zhi X. Y., Li W. J., Stackebrandt E. (2009). An update of the structure and 16S rRNA gene sequence-based definitions of higher ranks of the class Actinobacteria with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 59, 589–608. 10.1099/ijs.0.65780-0 [DOI] [PubMed] [Google Scholar]
- Zhou H. W., Li D. F., Tam N. F. Y., Jiang X. T., Zhang H., Sheng H. F., et al. (2011). BIPES, a cost-effective high-throughput methods for assessing microbial diversity. ISME J. 5, 741–749. 10.1038/ismej.2010.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Wu Q., Dai J., Zhang S., Wei F. (2011). Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108, 17714–17719. 10.1073/pnas.1017956108 [DOI] [PMC free article] [PubMed] [Google Scholar]