Abstract
Biosensors based on the principle of Förster (or fluorescence) resonance energy transfer (FRET) have been developed to visualize spatio-temporal dynamics of signalling molecules in living cells. Many of them adopt a backbone of intramolecular FRET biosensor with a cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) as donor and acceptor, respectively. However, there remains the difficulty of establishing cells stably expressing FRET biosensors with a YFP and CFP pair by lentiviral or retroviral gene transfer, due to the high incidence of recombination between YFP and CFP genes. To address this, we examined the effects of codon-diversification of YFP on the recombination of FRET biosensors introduced by lentivirus or retrovirus. The YFP gene that was fully codon-optimized to E.coli evaded the recombination in lentiviral or retroviral gene transfer, but the partially codon-diversified YFP did not. Further, the length of spacer between YFP and CFP genes clearly affected recombination efficiency, suggesting that the intramolecular template switching occurred in the reverse-transcription process. The simple mathematical model reproduced the experimental data sufficiently, yielding a recombination rate of 0.002–0.005 per base. Together, these results show that the codon-diversified YFP is a useful tool for expressing FRET biosensors by lentiviral or retroviral gene transfer.
Biosensors based on the principle of Förster (or fluorescence) resonance energy transfer (FRET) have shed new light on the spatiotemporal dynamics of signalling molecules in a living cell. The FRET biosensors are largely grouped into intermolecular and intramolecular FRET biosensors. A number of intramolecular FRET biosensors, which comprise both the donor and the acceptor fluorophores within a single protein, have been developed to visualize signalling molecules such as Ca2+1, phospholipids2,3, small GTPases4, protein kinases5 and so on6,7. It is widely accepted that the intramolecular FRET biosensors enjoy higher sensitivity and easier loading to cells and mice as compared with intermolecular FRET biosensors, which consist of a pair of donor and acceptor fluorophores8,9.
A critical drawback of the intramolecular FRET biosensors is that conventional gene-delivery techniques including the transfection of linearized DNAs and viral vectors of Retroviridae often fail to generate stable cell lines expressing FRET biosensors10. In many cases, the generated cell lines express only the donor or acceptor fluorescent protein. This phenomenon may be due to recombination between the donor and acceptor fluorescent proteins. We recently found that piggyBac11 or Tol212 transposon-mediated gene transfer can be used to establish cell lines and transgenic mice expressing FRET biosensors13,14. Live cell FRET imaging of up to several days has revealed the fluctuation of signalling molecules such as ERK and Rac110,13,15. The transposon-mediated gene transfer technique, however, is applied only to cells into which plasmids are efficiently transfected by lipofection or electroporation. In contrast, retroviral vectors can infect a wide range of cells in vitro and in vivo.
It is known that recombination occurs between homologous RNA sequences in the process of retrovirus-mediated gene transfer16,17. Among several models of such recombination, the most widely accepted is the template switching model. In this model, RNA-dependent DNA polymerase, i.e., reverse transcriptase, jumps from one template RNA to the other of the two single-stranded genomic RNAs in each retrovirus. The high sequence similarity is considered to guide the template switching. Currently, most intramolecular FRET biosensors adopt a cyan fluorescent protein (CFP) and a yellow fluorescent protein (YFP) as the donor and acceptor fluorescent proteins, respectively. Because both CFP and YFP are derived from GFP, they share high nucleotide sequence homology, which presumably causes the recombination. Consistent with this idea, tandem GFP genes or tandem-dimer Tomato (tdTomato) genes have shown clear recombination during the process of lentiviral or retroviral gene transfer18,19. Further evidence is provided by the results that FRET biosensors carrying a YFP and a coral-derived teal fluorescent protein (TFP) are readily expressed by retrovirus-mediated gene transfer without any recombination20,21,22. The nucleotide sequence homology between codon-humanized TFP (hTFP) and codon-humanized YFP (hYFP) (61%) is markedly lower than the homology between codon-humanized CFP (hCFP) and hYFP (96%) (Supplementary Fig. S1). Although TFP has some advantages over CFP as a FRET donor23, the substitution of CFP to TFP decreases FRET gain more than in most FRET biosensors containing YFP as the acceptor, which could be due to the weak or absent dimerization of YFP and TFP13.
In consideration of these facts, a few research groups have successfully utilized a pair of codon-diversified YFP mutant and CFP to establish stable cell lines expressing FRET biosensors by retroviral transduction22,24. However, there have been no reports analyzing the effect of codon diversification on the efficiency of recombination in FRET biosensors transduced by retrovirus systematically. Therefore, we examined recombination in FRET biosensors with codon-diversified YFP mutants delivered by two retroviral vectors, a Murine leukemia virus (MuLV)-derived pCX4 retroviral vector25 and a human immunodeficiency virus (HIV)-derived pCSII lentiviral vector26. In addition, based on the experimental data, we evaluated the recombination rate in lentiviral or retroviral gene transfer by mathematical modelling and statistical analysis.
Results
Construction of codon-diversified YFP genes
We used a FRET biosensor for protein kinase A, AKAR3EV13, to examine the contribution of nucleotide sequence homology in recombination between CFP and YFP (Fig. 1A). AKAR3EV comprised a YFP-derived YPet27, and a CFP-derived nTurquoise-GL28 as the acceptor and donor, respectively. These fluorescent proteins sandwich the phosphate-binding domain of FHA1, EV linker, and a substrate peptide of PKA (Fig. 1B). The nuclear export signal (NES) was included at the C-terminus of the biosensor. Both the YPet and the nTurquoise-GL genes have been codon-optimized for humans. The homology between the humanized YPet, called hYPet hereafter, and nTurquoise-GL was 96%. As the codon-diversified YFP, we chose a YPet gene optimized for E. coli, called eYPet hereafter. The nucleotide sequence homology between eYPet and nTurquoise-GL was 68% (Supplementary Fig. S1). We constructed six YPet chimeras between hYPet and eYPet: h75-e25YPet, h50-e50YPet, h25-e75YPet, e75-h25YPet, e50-h50YPet, and e25-h75YPet (Fig. 1B, and see Methods). The order of h and e and their numbers indicated the order and the percentage ratio of hYPet to eYPet, respectively. For instance, h75-e25YPet was composed of the first 75% of the hYPet gene DNA sequence, followed by the last 25% of the eYPet gene DNA sequence. h100YPet and e100YPet are identical to the authentic hYPet and eYPet, respectively. These genes for FRET biosensors were inserted into either the MuLV-derived retroviral vector or HIV-derived lentiviral vector, which were transfected into 293T cells to generate retroviral or lentiviral vectors.
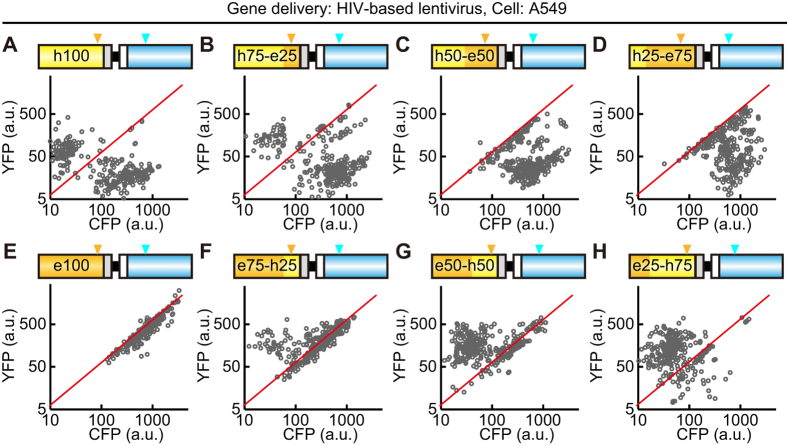
Figure 1. Recombination of FRET biosensors during lentiviral or retroviral infection.

(A) Schematic representation of the recombination between YFP and CFP genes in FRET biosensors in the process of lentiviral or retroviral gene transfer. Two copackaged genomic RNAs encoding FRET biosensors are included in a virus particle. After infection, cells express only YFP or CFP. (B) FRET biosensors with different YFP variants. A PKA FRET biosensor, AKAR3EV, is composed of YPet (YFP), a FHA1 domain, linker, PKA substrate, nTurquoise-GL (CFP), and a nuclear export sequence (NES). In this study, YPet is replaced with h100YPet, H75-e25YPet, h50-e50YPet, h25-e75YPet, e100YPet, e75-h25YPet, e50-h50YPet, and e25-h75YPet.
A template-switching model for the gene recombination during lentivirus - or retrovirus-mediated gene transfer
Recombination of retroviral genomes could be caused by template switching/jumping during minus-strand DNA synthesis, or reverse transcription, on the RNA genome and plus-strand DNA replication. Figure 2 shows the case of template switching/jumping during reverse transcription. This model provided a proper interpretation of the results with the series of lentiviral or retroviral vectors: Assuming that e100YPet is not recombined with nTurquoise-GL and that the hues of YFP and CFP are determined primarily by the T203Y substitution and the Y66W substitution, respectively29, we can expect the outcomes of recombination between the YPet chimeras and nTurquoise-GL illustrated in Fig. 2: First, h25-e75YPet is recombined with nTurquoise-GL in the first-quarter segment. In this case, the recombined fluorescent protein gene will carry the nucleotides for Y66W, but not for T203Y, and therefore encodes CFP (Fig. 2A). Second, e75-h25YPet is recombined with nTurquoise-GL in the fourth-quarter segment. In this case, the resulting fluorescent protein will miss the nucleotides for Y66W and may or may not contain the nucleotides for T203Y, and therefore encodes either GFP or YFP (Fig. 2B). The recombination between YPet and nTurquoise-GL generate 4 types of chimeric GFP (Supplementary Fig. S2A and S2B). It is expected that chimeric GFP with H148G mutation fluoresced less than the wild-type GFP and YFP, because of the higher pKa value30. In fact, we confirmed that the chimeric GFP emitted less fluorescence than original YPet in our experimental condition (Supplementary Fig. S2B). Therefore, the decrease of fluorescence was caused by substation the nTurquoise-GL sequence for YPet sequence.
Figure 2. Working model for the recombination in FRET biosensors.

(A) The recombination of FRET biosensors with h25-e75YPet generates CFP, which includes the critical amino acid substitution of Y66W from GFP. (B) The recombination of FRET biosensors with e75-h25YPet generates GFP or YFP, which includes the critical amino acid substitution of T203Y from GFP.
Recombination between YFP and CFP genes expressed by the lentiviral vector
We infected cells of the human lung adenocarcinoma cell line A549 with HIV-derived viruses carrying one of the eight FRET biosensor genes at a multiplicity of infection (MOI) of approximately 0.7. Approximately one week after infection without drug selection, the cells were imaged with epi-fluorescence microscopy. The fluorescence intensities of YFP and CFP in each cell were quantified and are shown in scatter plots.
Most cells infected with the h100YPet-carrying lentivirus demonstrated clear evidence of recombination between the YFP and CFP genes, i.e., cells expressed either YFP or CFP, but not both (Fig. 3A). In cells infected with the h75-e25YPet-carrying virus, the fraction of cells emitting YFP fluorescence alone was smaller than that in h100YPet-carrying virus-infected cells, and cells emitting both CFP and YFP fluorescences were detectable (Fig. 3B). In cells infected with the h50-e50YPet-carrying virus or h25-e75YPet-carrying virus, all cells emitted either CFP fluorescence alone or both CFP and YFP fluorescences (Fig. 3C,D). As expected, the e100YPet-carrying lentivirus did not show any sign of recombination and emitted equal amounts of CFP and YFP fluorescence (Fig. 3E). In cells infected with the e75-h25YPet-carrying virus, a small fraction of cells emitted YFP fluorescence alone (Fig. 3F). The fraction of cells that emitted YFP fluorescence alone was increased in the e50-h50YPet-carrying virus and e25-h75YPet-carrying virus, concomitant with the decrease in cells emitting both CFP and YFP fluorescences (Fig. 3G,H). Of note, the fluorescence intensity of chimeric fluorescent proteins was less than the prototype as described; the average intensities of YFP channel was highest in the prototype e100YPet-carrying virus (Fig. 3E–H). Overall, very similar results were obtained when HeLa cells were infected at an MOI of less than 1.0 (Supplementary Fig. S3A–S3H). These results are in line with the model shown in Fig. 2, supporting the idea that the recombination is caused by template switching/jumping during reverse transcription.
Figure 3. Recombination between the YFP and CFP genes by lentiviral gene transfer.
(A–H) A549 cells were infected with lentivirus encoding 8 different FRET biosensors as shown in Fig. 1B. At least 4 days after infection, the cells were imaged with an epi-fluorescence microscope. The average fluorescence intensities of CFP and YFP are represented as a log-log plot. Each dot corresponds to an A549 cell. Three hundred cells were analyzed from two independent experiments. Red lines are the fitted line with the e100YPet data. Orange and cyan arrowheads indicate the T203Y and Y66W positions, respectively.
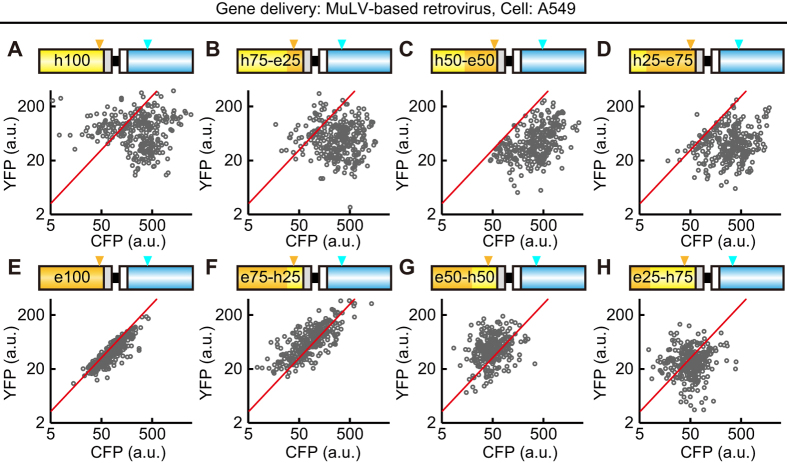
Recombination between YFP and CFP genes expressed by the retroviral vectors
We then used a MuLV-derived retroviral vector to express the FRET biosensors stably, and analyzed the results as described above (Fig. 4). The MOI was approximately 0.5. Cells infected with the h100YPet-carrying retrovirus did not show clear cell populations, although significant fractions of the cells emitted CFP fluorescence much more strongly than they did YFP fluorescence (CFP-dominant cells), or emitted YFP fluorescence much more strongly than they did CFP fluorescence (YFP-dominant cells) (Fig. 4A). In cells infected with the h75-e25YPet-carrying virus, the CFP-dominant cells were increased with a concomitant decrease in YFP-dominant cells (Fig. 4B). In cells infected with the h50-e50YPet-carrying virus or h25-e75YPet-carrying virus, the CFP-dominant cells were further increased with the disappearance of YFP-dominant cells (Fig. 4C,D). Again, the e100YPet -carrying retrovirus did not show any sign of recombination and emitted equal amounts of CFP and YFP fluorescence (Fig. 4E). The YFP-dominant cells appeared in cells infected with the e75-h25YPet-carrying virus (Fig. 4F) and were further increased in those infected with the e50-h50YPet-carrying virus (Fig. 4G). Cells infected with the e25-h75YPet-carrying virus did not exhibit any clear subpopulations (Fig. 4G,H).
Figure 4. Recombination between the YFP and CFP genes by retroviral gene transfer.
(A–H) A549 cells were infected with retrovirus encoding 8 different FRET biosensors as shown in Fig. 1B. At least 4 days after infection, the cells were imaged with an epi-fluorescence microscope. The average fluorescence intensities of CFP and YFP are represented as a log-log plot. Each dot corresponds to an A549 cell. Three hundred cells were analyzed from two independent experiments. Red lines are the fitted line with the e100YPet data. Orange and cyan arrowheads indicate the T203Y and Y66W positions, respectively.
When using HeLa cells as a host, we obtained similar but slightly different results (Supplementary Fig. S3I–S3P). For example, the e100YPet-carrying retrovirus also showed some sign of recombination; i.e., the appearance of CFP-dominant cells. These observations suggested that the recombination was affected to some extent by viral vectors, and/or host cells. Finally, we confirmed that the recombination was not observed in cells transfected with the expression plasmids used for the preparation of lentivirus and retrovirus (Supplementary Fig. S4).
The effect of the spacer between YFP and CFP on the recombination
We next examined whether the length of the spacer between YFP and CFP affected the recombination. For this purpose, we developed an h100YPet-carrying lentiviral vector, in which h100YPet and nTurquoise-GL were linked with a GGSGG linker (15 bases) (Fig. 5A). Cells infected with the h100YPet-GGSGG-nTurquoise-GL-carrying lentivirus exhibited markedly less, but not negligible, recombination (Fig. 5B) than did those infected with the h100YPet–carrying lentivirus, in which the length between the two fluorescent genes was 812 bases (Fig. 5C). This result may suggest that the recombination took place in a single copy of the lentiviral gene, rather than between the 2 copies of the lentiviral gene in a single virus (see Discussion).
Figure 5. Effect of a short spacer between YFP and CFP on recombination.
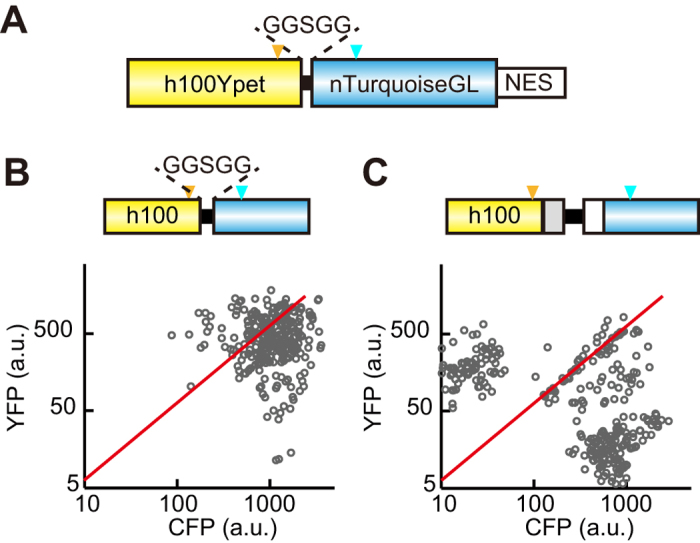
(A) Schematic representation of a FRET biosensor with YPet, GGSGG linker, nTurquoise-GL, and NES. (B,C) HeLa cells were infected with lentivirus encoding FRET biosensor with a short spacer, 15 bases (B) and full spacer, 812 bases (C). At least 4 days after infection, 300 cells were imaged with an epi-fluorescence microscope, and represented as in Fig. 3. Note that panel C is the same graph as in Supplementary Fig. 2A. Orange and cyan arrowheads indicate the T203Y and Y66W positions, respectively.
The validation of the recombination
To validate the recombination between YPet and nTurquoise-GL genes, we sequenced the viral DNAs integrated into the genomes. We sorted A549 cells infected with h100YPet-carrying lentivirus depending on the fluorescence of YFP, followed by genomic DNA extraction, PCR amplification of the recombined fluorescent protein genes, and sequencing. As expected, recombination was found between YPet and nTurquoise-GL genes (Supplementary Fig. S5A).The frequency of recombination was apparently correlated with the length of homology regions in between the differences of YPet and nTurquoise-GL genes (Supplementary Fig. S5B and S5C). However, the recombination in 448–609 base region occurred more frequent considerably than in the other regions. These results suggested that recombination took place almost randomly in between the identical nucleotide sequences, and the secondary structure of RNA viral genomes affected the recombination frequency to some extent.
Mathematical modelling of recombination
Finally, we attempted to quantify the recombination rate by mathematical model and statistical analysis. The following two assumptions were made in the computer simulation: First, based on the results in Supplementary Figure S5, we assumed that the CFP (nTurquoise-GL) gene is recombined randomly with the identical nucleotide of hYFP, but not eYFP. Second, one copy of the retroviral gene is integrated into the genome of the host cells under each condition. The mathematical model included only one parameter to be fitted—the recombination rate, r (/base), which represented the probability of recombination per base. The parameter r was obtained by the maximal likelihood estimation method (for more details, see Methods and Supplementary Fig. S6).
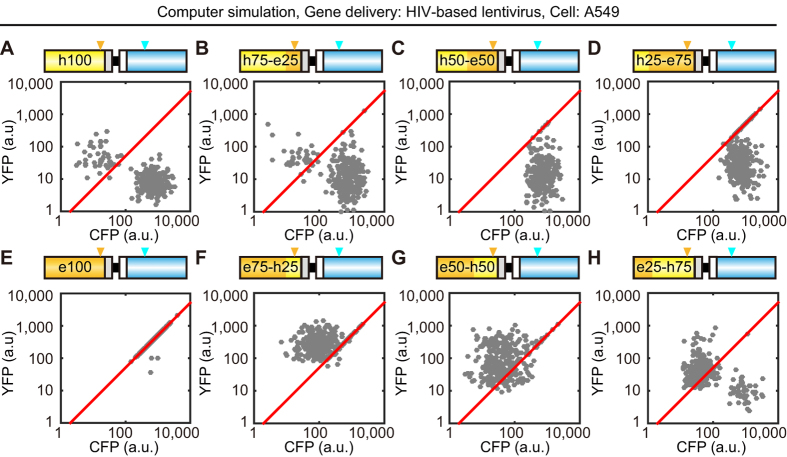
The simple mathematical model qualitatively reproduced the four experimental data sets of A549 cells and HeLa cells infected with lentiviral or retroviral vectors (Fig. 6 and Supplementary Fig. S7). These data validated our hypothesis that the recombination was governed by a random process within the homology region. Interestingly, the recombination rate of lentivirus did not differ from that of retrovirus; both were 0.002 ~ 0.005 /base (Table 1 and Supplementary Fig. S8). Further, the lentivirus with shorter spacer between YFP and CFP genes indeed showed smaller recombination rate than that with longer spacer (Table 1).
Figure 6. Computer simulation of recombination between the YFP and CFP genes.
The recombination of YFP and CFP genes in A549 cells infected with the indicated lentivirus was simulated by computer with a recombination rate of 0.0043 (/base), which showed maximal likelihood estimation. To reproduce the experimental data, 9 parameters were extracted from the experimental data set in Fig. 3 (see Supplementary Fig. S6 and the Methods for details). Red lines were fitted with the e100YPet data. Orange and cyan arrowheads indicate the T203Y and Y66W positions, respectively.
Table 1. Recombination rates calculated by the mathematical model.
| Recombination rate (/base) |
||
|---|---|---|
| Lentivirus | Retrovirus | |
| A549 cells | 0.0031 | 0.0024 |
| HeLa cells | 0.0044, 0.0021* | 0.0021 |
*Recombination rate of a FRET biosensor with YPet, GGSGG linker, nTurquoise-GL, and NES in Fig. 5B.
Discussion
Here, we demonstrated that a FRET biosensor comprised of e100YPet and nTurquoise-GL could be applied to lentivirus- or retrovirus-mediated gene transfer, and such transfer had the lowest possibility of recombination when performed in HeLa cells and A549 cells. Previous studies have proven the impact of codon usage on the gene expression31, and therefore we expected that the substitution of h100YPet partly or totally by e100YPet would decrease the expression of the FRET biosensor. Unexpectedly, we did not detect any difference in the fluorescence intensity between the FRET biosensors codon-optimized for humans and bacteria (Figs 3,4, and Supplementary Figs S3, S4). Because the YFP transcript is relatively short (714 bases), the effect of codon usage bias may not substantially affect the expression level of the FRET biosensor. Furthermore, considering the fact that the difference in cell lines did not change the results of recombination in the series of FRET biosensors, we could conclude that e100YPet enabled us to establish stable cell lines expressing highly sensitive FRET biosensors with the YPet and CFP pair13 by lentivirus- or retrovirus-mediated gene transfer.
The finding that the recombination rate was reduced when we used a short spacer between h100YPet and nTurquoise-GL (Fig. 5) may suggest that the recombination between the YFP and CFP genes occurs by template switching within a single RNA genome but not between the two RNA genomes in a single retroviral particle. Because the template switching within the RNA template is expected to suffer from steric hindrance, the shorter spacer between h100YPet and nTurquoise-GL may dampen the recombination efficiency. There is additional evidence that supports the template switching within a single RNA genome. If the template switching between the two RNA genomes occurred, then the h25-e75YPet-carrying virus should generate not only cells expressing CFP alone, but also cells expressing the biosensor with two copies of YFP and a single copy of CFP. However, we could not detect such cells by quantitative imaging (Figs 3D and 4D). Hu et al. reported that deletion of repeated regions occurs by an intramolecular template switch within the same RNA in the process of reverse transcription32. The rate of intramolecular recombination is much more efficient than that of intermolecular recombination, and the value of the intramolecular recombination rate has been roughly estimated as 30–41% per 110 bases, and thus 0.27–0.37% per base32,33. In agreement with these facts, these values were almost identical to our data (Table 1), strongly supporting the notion that the intramolecular recombination took place in FRET biosensors transduced by lentivirus or retrovirus. Even with the aforementioned evidence, we still could not completely neglect the template switching between the two RNA genomes. A potential merit of template switching between the two RNA genomes is to short-cut the reverse transcription in a viral particle. Because Hu et al. examined the recombination in a 110-bp direct repeat, there is little advantage to the minus-strand DNA synthesis. This idea might also explain why shortening of the spacer between YPet and nTurquoise-GL decreased the rate of recombination.
We observed the recombination even in the FRET biosensor with h25-e75YPet or e75-h25YPet, which shares high nucleotide sequence homology with nTurquoise-GL only in the quarter of the YPet genome (Figs 3,4 and Supplementary Figs S3, S4). This is also consistent with the model of template switching, because the reverse transcriptase jumps to the other RNA template through the binding of minus-strand DNA to the complementary RNA18. Although we do not know the length of homologous DNA sequence required, a length of at least 178 bases, which corresponds to 25% of the YFP gene, is sufficient for the recombination mediated by lentiviral or retroviral gene transfer. Salamongo et al. have reported that template jumping occurred between the short identical sequences (21 bases) at both the N-terminal and C-terminal sequences in GFP and Tomato19, suggesting that codon diversification, but not domain swapping, is necessary to avoid recombination.
In conclusion, here we have shown that, in addition to transposon-mediated gene transfer, lentivirus- or retrovirus-mediated gene delivery is also available for establishing a stable cell line expressing FRET biosensors with the YFP and CFP pair. This technique will provide new insight into not only the effect of molecular-targeted drugs on the signalling molecules13, but also intravital imaging in living animals.
Methods
FRET biosensor construction
The FRET biosensors developed in this study were originated from pAKAR3EV, a PKA biosensor13. To generate e100YPet, h75-e25YPet, h50-e50YPet, h25-e75YPet, e75-h25YPet, e50-h50YPet, and e25-h75YPet, the YPet-based Rac1 biosensor Raichu-Rac1, which was a kind gift from Dr. Wang of University of California at San Diego34 and contained e100YPet, was used as a PCR template. The hYPet gene in the original pAKAR3EV was replaced with these YPet variants. These FRET biosensor genes were inserted into pCX4bsr25 or pCSIIbsr, which was derived from pCSII-EF1 (a kind gift from Dr. Miyoshi, RIKEN) with IRES-bsr (blasticidin S-resistant gene).
Cell culture
HeLa cells were purchased from the Human Science Research Resources Bank (Sennanshi, Japan). HEK-293T cells were obtained from Invitrogen as Lenti-X 293 cells (Carlsbad, CA). A549 cells were obtained from the American Tissue Culture Collection. HeLa, HEK-293T, and A549 cells were maintained in DMEM (Wako, Osaka, Japan) supplemented with 10% FBS. For imaging, these cells were plated on 35-mm glass base dishes (Asahi Techno Glass, Tokyo, Japan). One hour before observation, HeLa and A549 cells were maintained with phenol red-free Medium 199 (Invitrogen) containing 0.1% bovine serum albumin and 20 mM HEPES.
Lentivirus or retrovirus-mediated gene transfer
For lentiviral production, HEK-293T cells were cotransfected with the pCSIIbsr vector, psPAX2 (Addgene plasmid 12260), and pCMV-VSV-G-RSV-Rev by using Polyethyleneimine “Max” MW 40,000 (Polyscience Inc., Warrington, PA). For retrovirus production, pCX4bsr, pGP, and pCMV-VSV-G-RSV-Rev were introduced into HEK-293T cells. Virus-containing media were collected at 48 hours after transfection, filtered, and used to infect target cells with 8 μg/mL polybrene. At least 4 days after infection, the infected cells without drug selection were imaged with a fluorescence microscope.
Spectroscopy by fluorescence microscope and analysis
CFP and YFP images of HeLa cells and A549 cells were obtained by using an inverted microscope (IX81-ZDC; Olympus, Tokyo, Japan) equipped with a cooled CCD camera (Cool SNAP-K4; Roper Scientific), an illumination system (Spectra-X light engine; Lumencore, OR), an IX2-ZDC2 laser-based autofocusing system (Olympus), a MAC5000 controller for filter wheels and XY stage (Ludl Electronic Products, Hawthorne, NY), an incubator chamber system (Tokai Hit, Shizuoka, Japan) and a GM-4000 CO2 supplier (Tokai-Hit, Fujinomiya, Japan). The following filters were used for the dual emission imaging studies: FF01–438/24–25 (Semrock, Rochester, NY, USA), and FF01–475/28–25 (Semrock) excitation filter for CFP and YFP/GFP, a U-MREF glass reflector (Olympus) as a dichroic mirror, an FF01–483/32–25 emission filter (Semrock) for CFP and an FF01–542/27–25 emission filter (Semrock) for YFP. The microscope was controlled by MetaMorph software (Universal Imaging, West Chester, PA). The average fluorescence intensities of CFP and YFP in each cell were measured by manually delineating a region of interest at the cytoplasm with MetaMorph software.
FACS and purification of genomic DNA
A549 cells infected with h100YPet-carrying lentivirus were sorted with a FACSAria™ III (BD Biosciences). YFP/CFP fluorescences were detected using a 445 nm laser, a 530/30 nm emission filter for YFP and a 480/20 nm emission filter for CFP. The genomic DNAs of these bulk cells were extracted by QuickExtract™ DNA Extraction Solution (Epicentre, Madison, WI, USA) in accord with manufacture’s protocol. The primers for amplification were listed as follows; the forward primer 5′-TCTTCCATTTCAGGTGTCGTGAACACGC-3′, and the reverse primer 5′-GCGGCCGCCCAGCTCGTCCATGCCGAGAGT-3′.
Mathematical modeling of gene recombination and statistical analysis
A mathematical model of recombination between the YFP and CFP genes was built as follows: First, the CFP (nTurquoise-GL) gene was recombined randomly with the identical nucleotide of hYFP, but not eYFP, according to the recombination rate, r (/base). If the reverse-transcript of CFP is recombined with hYFP between the first nucleotide and nt 199, the recombinant gene product emits CFP fluorescence, because the Y66W mutation is responsible for the cyan fluorescence. If CFP is recombined with hYFP after nt 198, the chimeric GFP or YFP gene product emits yellow fluorescence. Depending on the recombination site, fluorescence intensity of chimeric GFP gene reduces based on the experimental results in Supplementary Figure S2B. To recapitulate the experimental data set, fluorescence intensities and deviations for intercellular heterogeneity were obtained from the experimental data, and applied to simulation (Supplementary Fig. S6). The parameter r was obtained by the maximal likelihood estimation method. Given r, the log likelihood value was calculated as follows
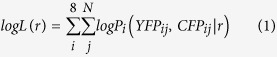 |
where Pi is a probability density obtained by simulation with the given r value, and YFPij and CFPij are YFP and CFP fluorescence intensities at the j-th cell in experimental data of the i-th condition among eight different YFP genes. To obtain P, the recombination events were repeated in at least 30,000 cells under each condition. In computer simulation, the value of r was varied in the range of 0.0001–0.01 to obtain the r value showing maximal log likelihood value.
Additional Information
How to cite this article: Komatsubara, A. T. et al. Quantitative analysis of recombination between YFP and CFP genes of FRET biosensors introduced by lentiviral or retroviral gene transfer. Sci. Rep. 5, 13283; doi: 10.1038/srep13283 (2015).
Supplementary Material
Acknowledgments
We thank the members of the Matsuda Laboratory for their helpful discussions. K.A. and M.M. were supported by the Platform for Dynamic Approaches to Living System from the Ministry of Education, Culture, Sports, and Science, Japan. K.A. was supported by JSPS KAKENHI Grant Number 25136706.
Footnotes
Author Contributions K.A. designed research; A.T.K. performed experiments; K.A. analysed mathematical model and simulation; A.T.K., M.M. and K.A. wrote the paper.
References
- Miyawaki A. et al. Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin. 388, 882–887 (1997). [DOI] [PubMed]
- Sato M., Ueda Y., Takagi T. & Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 5, 1016–22 (2003). [DOI] [PubMed] [Google Scholar]
- Nishioka T. et al. Rapid turnover rate of phosphoinositides at the front of migrating MDCK cells. Mol. Biol. Cell 19, 4213–23 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N. et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411, 1065–8 (2001). [DOI] [PubMed] [Google Scholar]
- Aoki K. et al. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell 52, 529–40 (2013). [DOI] [PubMed] [Google Scholar]
- Miyawaki A. Visualization of the Spatial and Temporal Dynamics of Intracellular Signaling. Dev. Cell 4, 295–305 (2003). [DOI] [PubMed] [Google Scholar]
- Aoki K., Kiyokawa E., Nakamura T. & Matsuda M. Visualization of growth signal transduction cascades in living cells with genetically encoded probes based on Förster resonance energy transfer. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 2143–51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa E., Aoki K., Nakamura T. & Matsuda M. Spatiotemporal Regulation of Small GTPases as Revealed by Probes Based on the Principle of Förster Resonance Energy Transfer (FRET): Implications for Signaling and Pharmacology. Ann. Rev. Pharmacol. Toxicol. 51, 337–58 (2011) [DOI] [PubMed] [Google Scholar]
- Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev. Cell 4, 295–305 (2003). [DOI] [PubMed] [Google Scholar]
- Aoki K., Komatsu N., Hirata E., Kamioka Y. & Matsuda M. Stable expression of FRET biosensors: a new light in cancer research. Cancer Sci. 103, 614–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–83 (2005). [DOI] [PubMed] [Google Scholar]
- Kawakami K. & Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics 166, 895–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N. et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka Y. et al. Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct. Funct. 37, 65–73 (2012). [DOI] [PubMed] [Google Scholar]
- Yukinaga H. et al. Fluctuation of Rac1 activity is associated with the phenotypic and transcriptional heterogeneity of glioma cells. J. Cell Sci. 127, 1805–15 (2014). [DOI] [PubMed] [Google Scholar]
- Lai M. M. RNA recombination in animal and plant viruses. Microbiol. Rev. 56, 61–79 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E. & Holmes E. C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 9, 617–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. & Zhang J. Intramolecular recombinations of Moloney murine leukemia virus occur during minus-strand DNA synthesis. J. Virol. 76, 9614–23 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamango D. J., Evans D. A., Baluyot M. F., Furlong J. N. & Johnson M. C. Recombination can lead to spurious results in retroviral transduction with dually fluorescent reporter genes. J. Virol. 87, 13900–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga-Udagawa R. et al. The scaffold protein Shoc2/SUR-8 accelerates the interaction of Ras and Raf. J. Biol. Chem. 285, 7818–26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka Y., Yasuda S., Fujita Y., Aoki K. & Matsuda M. Multiple decisive phosphorylation sites for the negative feedback regulation of SOS1 via ERK. J. Biol. Chem. 285, 33540–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mues M. et al. Real-time in vivo analysis of T cell activation in the central nervous system using a genetically encoded calcium indicator. Nat. Med. 19, 778–83 (2013). [DOI] [PubMed] [Google Scholar]
- Ai H., Henderson J. N., Remington S. J. & Campbell R. E. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 400, 531–40 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak A. L. et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–94 (2004). [DOI] [PubMed] [Google Scholar]
- Akagi T., Sasai K. & Hanafusa H. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. USA. 100, 13567–72 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Blömer U., Takahashi M., Gage F. H. & Verma I. M. Development of a self-inactivating lentivirus vector. J. Virol. 72, 8150–7 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. W. & Daugherty P. S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23, 355–60 (2005). [DOI] [PubMed] [Google Scholar]
- Goedhart J. et al. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods 7, 137–9 (2010). [DOI] [PubMed] [Google Scholar]
- Sawano A. & Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28, E78 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Llopis J., Deveraux Q. L., Tsien R. Y. & Reed J. C. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat. Cell Biol. 2, 318–25 (2000). [DOI] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S. & Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–53 (2004). [DOI] [PubMed] [Google Scholar]
- Hu W. S., Bowman E. H., Delviks K. A. & Pathak V. K. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J. Virol. 71, 6028–36 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K. & Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. USA. 87, 6019–23 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M., Sun J., Chien S. & Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. USA. 105, 14353–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.