Supplemental Digital Content is available in the text.
Keywords: genetics; genetic association studies; magnetic resonance imaging; polymorphism, single nucleotide; stroke, lacunar
Background and Purpose—
Lacunar strokes comprise ≈20% of all strokes. Despite this frequency, their pathogenesis is poorly understood. Previous genome-wide association studies in lacunar stroke have been disappointing, which may be because of phenotypic heterogeneity. Pathological and radiological studies suggest that there may be different pathologies underlying lacunar strokes. This has led to the suggestion of 2 subtypes: isolated lacunar infarcts and multiple lacunar infarcts and leukoaraiosis.
Methods—
We performed genome-wide analyses in a magnetic resonance imaging–verified cohort of 1012 younger onset lacunar stroke cases and 964 controls. Using these data, we first estimated the heritability of lacunar stroke and its 2 hypothesized subtypes, and secondly, we determined whether this is enriched for regulatory regions in the genome, as defined by data from Encyclopedia of DNA Elements (ENCODE) and other sources. Finally, we determine the evidence for a polygenic contribution from rare variation to lacunar stroke and its subtypes.
Results—
Our results indicate a substantial heritable component to magnetic resonance imaging–verified lacunar stroke (20%–25%) and its 2 subtypes (isolated lacunar infarct, 15%–18%; multiple lacunar infarcts/leukoaraiosis, 23%–28%). This heritable component is significantly enriched for sites affecting expression of genes. In addition, we show that the risk of the 2 subtypes of lacunar stroke in isolation, but not in combination, is associated with rare variation in the genome.
Conclusions—
Lacunar stroke, when defined on magnetic resonance imaging, is a highly heritable complex disease. Much of this heritability arises from regions of the genome affecting gene regulation. Rare variation affects 2 subtypes of lacunar in isolation, suggesting that they may have distinct genetic susceptibility factors.
Lacunar infarcts resulting from occlusion of the small penetrating arteries of the brain comprise ≈20% of all ischemic strokes, a proportion similar to those resulting from cardioembolism or large artery atherosclerosis.1 Despite this, comparatively little is known about their underlying pathogenesis. Epidemiological studies have established hypertension and diabetes mellitus as important risk factors,2–4 but pathological studies have been hampered by methodological inconsistencies and inadequate classification of the disease, as well as limited pathological tissue because of the low early mortality rate.5
One approach to identifying the underlying causes of complex diseases, such as lacunar stroke, is through genetic studies. In the recent past, genome-wide association studies (GWAS) have transformed our understanding of complex diseases and have begun to identify the common genetic component to ischemic and hemorrhagic strokes.6,7 Despite these advances, no genetic variants have yet been identified that specifically confer risk of lacunar stroke in white populations. Indeed, although family history data suggest that genetic predisposition may be particularly important for lacunar stroke,8 estimates of the heritability of lacunar stroke from GWAS have been low compared with those of the other subtypes.9,10 Multiple factors might explain this disparity.
One factor which might be important is disease heterogeneity. Pathological studies have shown different vascular lesions in patients presenting with lacunar stroke, with 2 main pathologies reported, namely focal microatheroma and a diffuse small-vessel arteriopathy.11 The former has been associated with larger single lacunar infarcts and the latter with multiple smaller lacunes and leukoaraiosis.11,12 These 2 subtypes have also been shown to have differing risk factor profiles.13 These sources point to the existence of pathophysiological subtypes of lacunar stroke,12 each of which might be presumed to have distinct genetic susceptibility factors. Another factor might be inadequate disease classification. Lacunar infarcts are small and frequently not seen on computed tomography. Despite this, in GWAS, to date, lacunar stroke has been often been diagnosed based on computed tomography, with a clinical lacunar syndrome and no infarcts visible on computed tomography being used as criteria for lacunar stroke. It has been shown that this can lead to a marked overdiagnosis of lacunar stroke.14,15 Diagnostic accuracy is much improved with magnetic resonance imaging (MRI).
In this study, we use a large, highly phenotyped cohort of MRI-confirmed lacunar stroke cases and controls to investigate the genetic architecture of lacunar stroke and its subtypes. We investigate whether MRI-confirmed lacunar stroke and its subtypes are heritable and whether this variation in enriched for sites in the genome affecting the expression and regulation of genes. Finally, we investigate whether the risk of the 2 subtypes of lacunar stroke is conferred through rare genetic variation.
Materials and Methods
Study Population
A total of 1029 white patients with lacunar stroke, aged ≤70 years, were recruited from 72 specialist’s stroke centers throughout the United Kingdom (online-only Data Supplement), between 2002 and 2012, as part of the Young Lacunar Stroke DNA Resource. This study was approved by the Multi-Center Research Ethics Committee for Scotland (04/MRE00/36), and informed consent was obtained from all participants. Lacunar stroke was defined as a clinical lacunar syndrome,16 with an anatomically compatible lesion on MRI (subcortical infarct, ≤15 mm in diameter). Time from event to MRI was variable; the median time to MRI was 5 days. All patients underwent full stroke investigation, including brain MRI, imaging of the carotid arteries, and ECG. Echocardiography was performed when appropriate. All MRIs and clinical histories were reviewed centrally by 1 physician (H.S.M.). Exclusion criteria were stenosis >50% in the extra- or intracranial cerebral vessels, or previous carotid endarterectomy; cardioembolic source of stroke, defined according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) criteria17 as high or moderate probability; cortical infarct on MRI; subcortical infarct >15 mm in diameter, as these can be caused by embolic mechanisms (striatocapsular infarcts); and any other specific cause of stroke (eg, lupus anticoagulant, cerebral vasculitis, dissection, and monogenic cause of stroke). All cases were screened for NOTCH3 cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy and Fabry disease mutations, and positive cases were excluded. An additional 82 white patients of all ages with lacunar stroke were recruited from St George’s Hospital, London. The same investigations and exclusions were made as in the DNA-lacunar study.
Unrelated white controls, free of clinical cerebrovascular disease, were obtained by random sampling, stratified for age and sex, from general practice lists from the same geographical location as the patients. All patients and controls underwent a standardized clinical assessment and completed a standardized study questionnaire. MRI was not performed in controls.
The data set was genotyped on the Illumina HumanExomeCore array, which contains both exome content (≈250 000 single-nucleotide polymorphisms [SNPs]) and common tag SNPs (≈250 000 SNPs) found on conventional GWAS arrays, and imputed to 1000-genome phase 1. Full details are provided in the online-only Data Supplement.
Risk Factors
Hypertension was defined as elevation of systolic blood pressure of >140 mm Hg or diastolic blood pressure of >90 mm Hg persisting >7 days after stroke onset or before stroke treatment with antihypertensive drugs.18 Diabetes mellitus was defined as a previous diagnosis of type I or type II diabetes mellitus or at least 2 random glucose readings of >11.1 mmol/L or fasting blood glucose readings of >7.0 mmol/L after the acute phase of stroke.19 Hypercholesterolemia was defined as serum cholesterol of >5.2 mmol/L or prestroke treatment with a cholesterol-lowering agent.20 A positive smoking history was recorded in those who had smoked at any time in their lives.
Subtyping of Lacunar Stroke
Leukoaraiosis was graded on MRI using the semiquantitative Fazekas scale, which has been shown to reflect pathological severity of small-vessel disease in a postmortem validation study.21 On the basis of the leukoaraiosis grade, patients were subtyped into 2 groups: (1) isolated lacunar infarct (ILI): single lacunar infarct with absent or mild leukoaraiosis (equivalent to Fazekas periventricular score of <2); (2) multiple lacunar infarcts (MLI) or lacunar infarct with moderate or severe confluent leukoaraiosis (equivalent to Fazekas grade of >2) according to a previously validated method.8,22 Twenty MRI scans were randomly selected on a second occasion by the same rater, and there was perfect agreement in assignment of subtype (κ=1).
Heritability Estimates
To assess the heritability of lacunar stroke, we first set to missing all imputed genotypes with a probability of <0.9 and discarded all SNPs that met that criteria in <90% of individuals. We then calculated the genetic relationships between all individuals across all 8 122 203 remaining SNPs using the GCTA package.23 After removing distantly related individuals (>0.125), we used genetic restricted maximum likelihood (GREML) methods to estimate the proportion of phenotypic variance on the liability scale explained by the genetic relationships between individuals based on common SNPs (here termed heritability), as implemented in the GCTA package.23 We performed the analysis for 3 phenotypes: first for all MRI-defined lacunar stroke cases versus controls, then for cases with ILI versus controls, and for cases with MLI or extensive leukoaraiosis versus controls. We included the first principal component as a covariate in the model in all analyses. We calculated the heritability for prevalence of stroke (K) of 1% and 3%, assuming that lacunar strokes comprise 20% of all cases.24
Heritability Estimates, Partitioned on Functional Status
Recently, the Encyclopedia of DNA Elements (ENCODE) project has generated a huge wealth of data describing functional sites in the human genome.25 The project aimed to identify all functional sites in the genome through a series of experiments in many tissue types, including chromatin immunoprecipitation sequencing, DNase I hypersensitive sites sequencing, and formaldehyde-assisted isolation of regulatory elements sequencing. Each technique uses a distinct approach to identify the location of regulatory regions in the genome. This is of particular interest to genetic studies because previous analyses have shown that GWAS associations are enriched for such functional sites.26–28 In addition, genotype-tissue expression studies provide a complementary approach to identifying SNP variants that affect expression of genes. In such studies, mRNA expression levels of genes are compared with SNP genotypes, thereby determining SNPs that affect expression. Such SNPs are often termed expression quantitative trait loci (eQTLs).
In this experiment, we investigated whether the heritability of lacunar stroke was enriched for regulatory sites in the autosome. To do this, we used information from the RegulomeDB database,29 which catalogues data from the ENCODE project and others, determining the evidence that each SNP in the genome affects the regulation of genes. The database separates SNPs into categories based on the available evidence. The group with the strongest evidence, which we term eQTLs, includes SNPs that have been shown to affect the levels of an mRNA molecule in any tissue and overlap any transcriptional factor–binding site, transcription factor motif, DNase footprint, or DNase peak from ENCODE or other experiments. We first partitioned our data on this group, including all such SNPs, as well as tagging SNPs with r2>0.98, based on linkage disequilibrium from European samples from the 1000-genome data set.30 A total of 24 722 SNPs were included.
Secondly, we partitioned our data on regulatory regions from RegulomeDB.29 This group includes all SNPs that overlap a transcription factor–binding site and DNase peak, as well as either having a matched transcription factor motif or a matched DNase footprint. Therefore, these SNPs represent regions where regulatory factors are thought to bind to the genome. As before, we included all such SNPs, as well as tagging SNPs with r2>0.98. A total of 938 693 SNPs were included.
We then used the GCTA package to calculate the heritability, partitioned on (1) eQTLs and (2) regulatory regions for lacunar stroke and its 2 subtypes (ILI and MLI/leukoaraiosis). We compared our estimates with the proportion of overall heritability that would be expected for the number of SNPs analyzed.
Polygenic Contribution From Rare Variation
If we define protective variants as those where the major allele is associated with disease and risk variants as those where the minor allele is associated with disease, then under the null, an equal proportion of genetic associations should be from either protective or risk variants. An increase in the ratio of risk to protective variants at low allele frequencies can indicate a polygenic contribution from low-frequency variants to disease risk for 2 reasons31; (1) risk SNPs are under negative selection or (2) cases are sampled from the extreme end of the disease liability distribution, meaning that the increase in minor allele counts of a risk variant in the case group has a comparatively stronger effect on power. Therefore, we tested whether genome-wide associations from the lacunar stroke cohort were enriched for rare risk variants compared with protective variants, calculating the ratio of risk to protective variants for allele frequency windows for SNPs below a given P-value threshold (P<0.05).
We first performed association analysis on lacunar stroke case/control status using SNPTEST version 2.4, including the first 2 ancestry informative principal components, as derived using EIGENSTRAT, as covariates.32,33 We excluded all poorly imputed SNPs (SNPTEST info measure <0.5) and low-frequency variants (minor allele frequency<0.01). We calculated the ratio of risk to protective variants for all SNPs with P<0.05 at allele frequency bands. We next generated 1000 simulations for a GWAS data set of the same number of cases and controls as our datasets and calculated the ratio of risk to protective variants at P<0.05 for each simulation. We tabulated the number of simulations in which the risk to protective risk ratio was greater than that observed in our data and divided by the number of simulations to generate an empirical P value.
We first analyzed all lacunar stroke cases versus controls and then performed analyses for the MLI/leukoaraiosis and ILI subtypes versus controls. We evaluated the enrichment for risk allele frequencies between 1% to 5% and 5% to 10% separately. In addition, we evaluated the enrichment for risk allele frequencies between 30% and 50% as a negative control. All analyses were performed using the pisa java package.31
Results
Study Characteristics
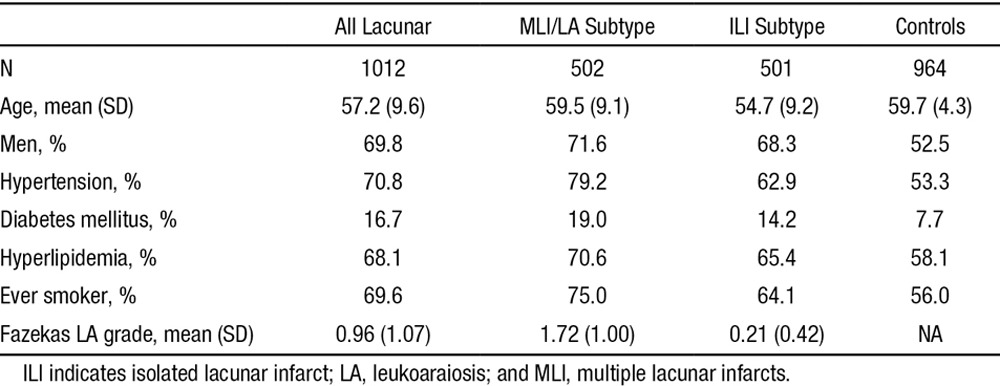
After quality control steps, a total of 1012 cases and 964 controls remained for analysis. Characteristics of this cohort are given in Table 1.
Table 1.
Cohort Characteristics
Heritability Estimates
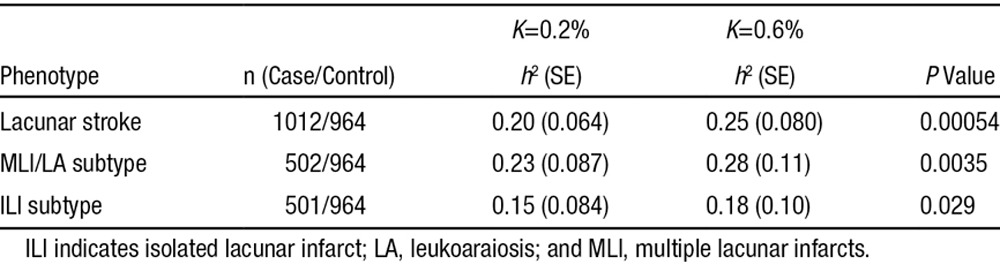
We estimated the proportion of variance of lacunar stroke status explained by the relatedness between individuals using GREML methods,34,35 as implemented in the GCTA package.23 All analyses showed that lacunar stroke and its subtypes were significantly heritable (Table 2). We determined the heritability of lacunar stroke to be 0.20 (0.064) assuming prevalence of 0.2% and 0.25 (0.080) assuming prevalence of 0.6%. For the subtypes of lacunar stroke, heritability estimates were higher for the MLI/leukoaraiosis subtype and slightly lower for the ILI subtype, although this difference was not significant (P>0.05). We performed sensitivity analyses to determine the influence of the genetic relatedness threshold and number of principal components included in the model. Both parameters had minimal influence on the results (Tables I and II in the online-only Data Supplement).
Table 2.
Heritability Estimates, Based on Assumption of Prevalence of Lacunar Stroke (K)
Heritability Estimates, Partitioned on Functional Status
We next estimated the heritability of lacunar stroke and its subtypes explained by eQTLs and regulatory regions. An excess of heritability was explained by either eQTLs or regulatory regions when compared with what would be expected by chance. Partitioning on eQTLs, we found that 2.3% of the heritability of lacunar stroke was explained by these SNPs (Figure). This value is equivalent to 11.5% of the total heritability, meaning that although eQTLs only make up 3.0% of all SNPs, they explain a considerable proportion of the heritability. Similar results were obtained for the subtypes.
Figure.
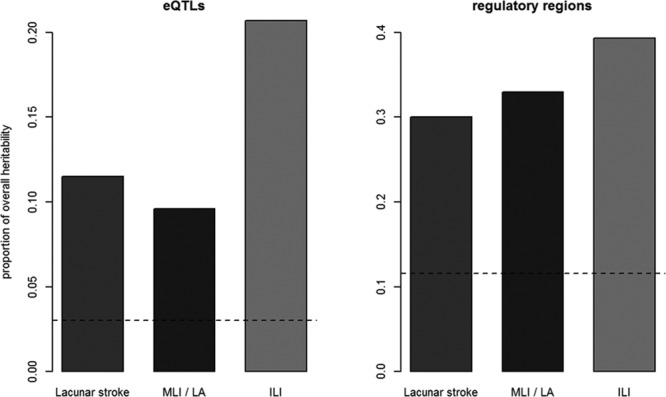
Proportion of overall heritability explained by expression quantitative trait loci (eQTLs) and regulatory regions for lacunar stroke and its subtypes, with horizontal line indicating the expected proportion of heritability for the number of single-nucleotide polymorphisms included in analysis. ILI indicates isolated lacunar infarct; LA, leukoaraiosis; and MLI, multiple lacunar infarcts.
Similarly, when partitioning on regulatory regions, 6.0% of the heritability lacunar stroke status was explained by the SNPs, equating to 30.0% of the heritability from only 11.6% of the SNPs. Similar results were again obtained for the subtypes.
Polygenic Contribution From Rare Variation
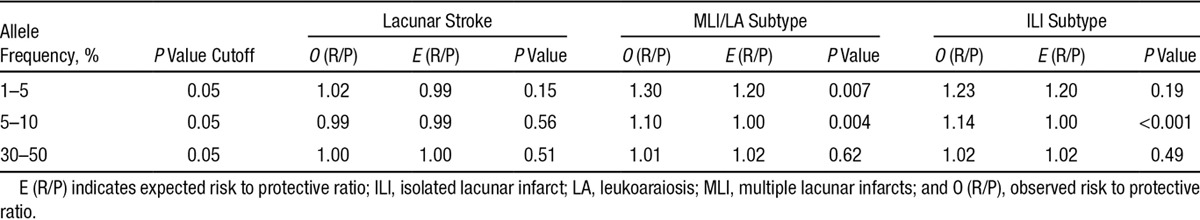
We used the pisa package to calculate the excess of risk to protective variants with P<0.05 at low allele frequencies (1%–5% and 5%–10%, separately).31 Our simulations indicate an excess of risk to protective variants at P<0.05 for frequencies between 5% and 10% in the 2 subtypes of lacunar stroke but not in all lacunar stroke itself (ILI subtype, P<0.001; MLI/leukoaraiosis subtype, P=0.004; lacunar, P=0.22; Table 3). We also found a significant association with the MLI/leukoaraiosis subtype at frequencies between 1% and 5% but not with the ILI subtype or all lacunar strokes (ILI, P=0.19; MLI/leukoaraiosis, P=0.007; lacunar, P=0.15). The results indicate that there is a polygenic contribution from rare variants to the 2 subtypes of lacunar stroke, but this cannot be detected when all lacunar strokes are considered together. This suggests that distinct pathophysiological mechanisms lead to the 2 diseases.
Table 3.
Evidence for Contribution of Rare Variants to Disease Risk
Discussion
Using a genome-wide approach from a large younger onset lacunar stroke population, our results show that lacunar stroke, when verified by MRI, is highly heritable. Our estimates are greater than those previously reported from GWAS data for lacunar stroke,9,10 suggesting that detailed phenotyping of cases, including MRI, is important for identification of genetic associations with the disease. In addition, we show that 2 subtypes of lacunar stroke, ILI and MLI/leukoaraiosis, are also highly heritable. Estimates of heritability for the MLI/leukoaraiosis subtype were higher, which may indicate a stronger genetic component, although more evidence is needed to determine this. The estimates of heritability for each of the analyses are comparable with those for Alzheimer disease (24%),36 schizophrenia (23%),37 and multiple sclerosis (30%),36 in which large-scale GWAS have been highly successful.
We also show that a significant proportion of the heritability of lacunar stroke, and each of its subtypes, is from SNPs affecting the regulation of genes. This was true for both eQTLs, where a SNP has been shown to directly affect the expression of a given gene, and for regulatory regions, where experiments from ENCODE indicate a region where regulatory factors bind to the genome. This is an important finding because it indicates a mechanism by which genetic variation leads to increased risk of lacunar stroke. Our findings mirror those from other complex diseases26,28 and support the notion that much of the genetic variation in liability to lacunar stroke is through subtle differences in gene expression and regulation.
Finally, our results indicate that rare genetic variants contribute to risk of lacunar stroke subtypes, ILI and MLI/leukoaraiosis, but do not have a significant effect on lacunar stroke as a whole. The strongest evidence was for the MLI/leukoaraiosis subtype, where significant enrichment of risk variants was observed for SNPs with a minor allele frequency of <10%. A significant result was only observed in the ILI subtype for SNPs with minor allele frequency between 5% and 10%. This might be because of power, as there is a greater number of SNPs in this frequency band. This result is important for several reasons. Firstly, it suggests that there exist genetic associations that are specific to these subtypes of lacunar stroke and, therefore, that some of the genetic contribution is not shared between the 2 diseases. This has important consequences for future studies. First of all, it shows that for genetic studies of lacunar stroke, detailed phenotyping is important. By splitting analyses into the 2 subtypes of lacunar stroke, novel associations might be identified. Secondly, as the focus of genetic studies turns from common to rare variants, our results show that the greatest benefits will be reaped from detailed phenotyping of lacunar stroke populations.
Our study has several strengths. All data in this multicenter study were prospectively collected using uniform data collection proformas. MRI was used in all cases to confirm lacunar stroke and was centrally reviewed by a single rater. Twenty randomly selected scans showed perfect agreement for determination of lacunar stroke subtype, indicating high reliability. Similarly, our study has limitations. The sample size used was relatively small for genetic studies, meaning that confidence intervals around estimates of heritability were moderately large. Similarly, we were underpowered to directly estimate the genetic correlation between the 2 subtypes based on all SNPs using GREML approaches (18% power to detect genetic correlation=0.5, using GCTA-GREML power calculator).38 An important extension of this work would be to perform such an analysis in a larger population with sufficient power to determine the degree to which the 2 subphenotypes share pathogenesis. In addition, our assessment of the proportion of heritability explained by eQTLs is limited by the information currently available. As eQTL studies grow in size, more SNPs will be identified that affect mRNA expression, and this will likely affect our results. Another important point to consider is that we were unable to obtain MRIs for the controls. It is possible that a proportion of these might have had silent subcortical infarcts, which may have a small effect of the results. Finally, the GREML approach used to estimate heritability has limitations. The estimates of heritability are derived from common SNPs meaning that the contributions from rare and structural variations are underestimated because of incomplete tagging of causal variation. Similarly, a proportion of the heritability might be because of susceptibility to risk factors for lacunar stroke, such as hypertension. An extension to this work would be to estimate the proportion of the observed heritability that acts through susceptibility to such risk factors.
In summary, we show that lacunar stroke, when diagnosed using MRI and detailed phenotyping, is highly heritable and that much of this heritability can be partitioned on regions of the genome affecting the regulation of genes. Our results suggest that rare variation affects 2 subtypes of lacunar in isolation, but not with lacunar stroke as a whole, suggesting that these 2 subtypes might have distinct genetic susceptibility factors.
Acknowledgments
A full list of the centers from which patients were recruited for DNA-lacunar study is given in the online-only Data Supplement.
Sources of Funding
H.S. Markus was supported by a National Institute for Health Research (NIHR) Senior Investigator award. H.S. Markus and S. Bevan were supported by the NIHR Cambridge University Hospitals Comprehensive Biomedical Research Centre. Collection of the UK Young Lacunar Stroke Resource was primarily supported by the Wellcome Trust with additional support from the Stroke Association. Genotyping and M. Traylor were supported by a project grant from the Stroke Association (TSA 2013/01).
Disclosures
None.
Supplementary Material
Footnotes
The online-only Data Supplement is available with this article at http://stroke.ahajournals.org/lookup/suppl/doi:10.1161/STROKEAHA.115.009485/-/DC1.
References
- 1.Sudlow CL, Warlow CP. Comparable studies of the incidence of stroke and its pathological types: results from an international collaboration. International Stroke Incidence Collaboration. Stroke. 1997;28:491–499. doi: 10.1161/01.str.28.3.491. [DOI] [PubMed] [Google Scholar]
- 2.Kase CS, Wolf PA, Chodosh EH, Zacker HB, Kelly-Hayes M, Kannel WB, et al. Prevalence of silent stroke in patients presenting with initial stroke: the Framingham Study. Stroke. 1989;20:850–852. doi: 10.1161/01.str.20.7.850. [DOI] [PubMed] [Google Scholar]
- 3.Ohira T, Shahar E, Chambless LE, Rosamond WD, Mosley TH, Jr, Folsom AR. Risk factors for ischemic stroke subtypes: the Atherosclerosis Risk in Communities study. Stroke. 2006;37:2493–2498. doi: 10.1161/01.STR.0000239694.19359.88. doi: 10.1161/01.STR.0000239694.19359.88. [DOI] [PubMed] [Google Scholar]
- 4.Jackson CA, Hutchison A, Dennis MS, Wardlaw JM, Lindgren A, Norrving B, et al. Differing risk factor profiles of ischemic stroke subtypes: evidence for a distinct lacunar arteriopathy? Stroke. 2010;41:624–629. doi: 10.1161/STROKEAHA.109.558809. doi: 10.1161/STROKEAHA.109.558809. [DOI] [PubMed] [Google Scholar]
- 5.Bailey EL, Smith C, Sudlow CL, Wardlaw JM. Pathology of lacunar ischemic stroke in humans–a systematic review. Brain Pathol. 2012;22:583–591. doi: 10.1111/j.1750-3639.2012.00575.x. doi: 10.1111/j.1750-3639.2012.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Australian Stroke Genetics Collaborative; Wellcome Trust Case Control Consortium 2 (WTCCC2); International Stroke Genetics Consortium. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, et al. International Stroke Genetics Consortium. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–521. doi: 10.1016/j.ajhg.2014.02.012. doi: 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerrard-Dunne P, Cloud G, Hassan A, Markus HS. Evaluating the genetic component of ischemic stroke subtypes: a family history study. Stroke. 2003;34:1364–1369. doi: 10.1161/01.STR.0000069723.17984.FD. doi: 10.1161/01.STR.0000069723.17984.FD. [DOI] [PubMed] [Google Scholar]
- 9.Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. doi: 10.1161/STROKEAHA.112.665760. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 10.Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, Sturm JW, et al. Australian Stroke Genetics Collaborative; International Stroke Genetics Consortium; Wellcome Trust Case Control Consortium 2. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44:1147–1151. doi: 10.1038/ng.2397. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher CM. Lacunar strokes and infarcts: a review. Neurology. 1982;32:871–876. doi: 10.1212/wnl.32.8.871. [DOI] [PubMed] [Google Scholar]
- 12.Boiten J, Lodder J, Kessels F. Two clinically distinct lacunar infarct entities? A hypothesis. Stroke. 1993;24:652–656. doi: 10.1161/01.str.24.5.652. [DOI] [PubMed] [Google Scholar]
- 13.Khan U, Porteous L, Hassan A, Markus HS. Risk factor profile of cerebral small vessel disease and its subtypes. J Neurol Neurosurg Psychiatry. 2007;78:702–706. doi: 10.1136/jnnp.2006.103549. doi: 10.1136/jnnp.2006.103549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajajee V, Kidwell C, Starkman S, Ovbiagele B, Alger J, Villablanca P, et al. UCLA MRI Acute Stroke Investigators. Diagnosis of lacunar infarcts within 6 hours of onset by clinical and CT criteria versus MRI. J Neuroimaging. 2008;18:66–72. doi: 10.1111/j.1552-6569.2007.00150.x. doi: 10.1111/j.1552-6569.2007.00150.x. [DOI] [PubMed] [Google Scholar]
- 15.Markus HS, Khan U, Birns J, Evans A, Kalra L, Rudd AG, et al. Differences in stroke subtypes between black and white patients with stroke: the South London Ethnicity and Stroke Study. Circulation. 2007;116:2157–2164. doi: 10.1161/CIRCULATIONAHA.107.699785. doi: 10.1161/CIRCULATIONAHA.107.699785. [DOI] [PubMed] [Google Scholar]
- 16.Bamford J, Sandercock P, Dennis M, Burn J, Warlow C. Classification and natural history of clinically identifiable subtypes of cerebral infarction. Lancet. 1991;337:1521–1526. doi: 10.1016/0140-6736(91)93206-o. [DOI] [PubMed] [Google Scholar]
- 17.Adams HP, Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 18.Whitworth JA World Health Organization, International Society of Hypertension Writing Group. 2003 World Health Organization (WHO)/International Society of Hypertension (ISH) statement on management of hypertension. J Hypertens. 2003;21:1983–1992. doi: 10.1097/00004872-200311000-00002. doi: 10.1097/01.hjh.0000084751.37215.d2. [DOI] [PubMed] [Google Scholar]
- 19.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 20.Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive summary of the third report of the national cholesterol education program (ncep) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel iii). JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 21.Fazekas F, Kleinert R, Offenbacher H, Schmidt R, Kleinert G, Payer F, et al. Pathologic correlates of incidental MRI white matter signal hyperintensities. Neurology. 1993;43:1683–1689. doi: 10.1212/wnl.43.9.1683. [DOI] [PubMed] [Google Scholar]
- 22.Hassan A, Hunt BJ, O’Sullivan M, Parmar K, Bamford JM, Briley D, et al. Markers of endothelial dysfunction in lacunar infarction and ischaemic leukoaraiosis. Brain. 2003;126(pt 2):424–432. doi: 10.1093/brain/awg040. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Chapman AM, Plested M, Jackson D, Purroy F. The Incidence, Prevalence, and Mortality of Stroke in France, Germany, Italy, Spain, the UK, and the US: a literature review. Stroke Res Treat. 2012;2012:436125. doi: 10.1155/2012/436125. doi: 10.1155/2012/436125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, et al. Tobacco and Genetics Consortium; Bipolar Disorder Psychiatric Genomics Consortium; Schizophrenia Psychiatric Genomics Consortium. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan Y, Lim ET, Sandholm N, Wang SR, McKnight AJ, Ripke S, et al. DIAGRAM Consortium; GENIE Consortium; GIANT Consortium; IIBDGC Consortium; PGC Consortium. An excess of risk-increasing low-frequency variants can be a signal of polygenic inheritance in complex diseases. Am J Hum Genet. 2014;94:437–452. doi: 10.1016/j.ajhg.2014.02.006. doi: 10.1016/j.ajhg.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 33.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SH, Harold D, Nyholt DR, Goddard ME, Zondervan KT, Williams J, et al. ANZGene Consortium; International Endogene Consortium; Genetic and Environmental Risk for Alzheimer’s disease Consortium. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ); International Schizophrenia Consortium (ISC); Molecular Genetics of Schizophrenia Collaboration (MGS) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visscher PM, Hemani G, Vinkhuyzen AA, Chen GB, Lee SH, Wray NR, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10:e1004269. doi: 10.1371/journal.pgen.1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]


