We show a lack of ascertainment bias, a greater clustering of cases, and a greater risk of secondary infection in blood-relatives for human H5N1 infections compared to H7N9, which suggests possibly a greater potential pandemic risk for H7N9 than H5N1.
Keywords: influenza A(H7N9), influenza A(H5N1), clinical epidemiology, cluster
Abstract
Background. The pandemic potential of avian influenza viruses A(H5N1) and A(H7N9) remains an unresolved but critically important question.
Methods. We compared the characteristics of sporadic and clustered cases of human H5N1 and H7N9 infection, estimated the relative risk of infection in blood-related contacts, and the reproduction number (R).
Results. We assembled and analyzed data on 720 H5N1 cases and 460 H7N9 cases up to 2 November 2014. The severity and average age of sporadic/index cases of H7N9 was greater than secondary cases (71% requiring intensive care unit admission vs 33%, P = .007; median age 59 years vs 31, P < .001). We observed no significant differences in the age and severity between sporadic/index and secondary H5N1 cases. The upper limit of the 95% confidence interval (CI) for R was 0.12 for H5N1 and 0.27 for H7N9. A higher proportion of H5N1 infections occurred in clusters (20%) compared to H7N9 (8%). The relative risk of infection in blood-related contacts of cases compared to unrelated contacts was 8.96 for H5N1 (95% CI, 1.30, 61.86) and 0.80 for H7N9 (95% CI, .32, 1.97).
Conclusions. The results are consistent with an ascertainment bias towards severe and older cases for sporadic H7N9 but not for H5N1. The lack of evidence for ascertainment bias in sporadic H5N1 cases, the more pronounced clustering of cases, and the higher risk of infection in blood-related contacts, support the hypothesis that susceptibility to H5N1 may be limited and familial. This analysis suggests the potential pandemic risk may be greater for H7N9 than H5N1.
We know from the history of influenza pandemics that novel influenza A viruses that cross the species barrier from animals to humans represent one of the greatest threats to global public health. Avian influenza viruses H5N1 and H7N9 have caused a large number of human infections over an extended period of time and with high reported case fatality [1–3]. As such these viruses are of major concern as potential pandemic threats, yet the pandemic potential of these viruses remains an unresolved question.
The Influenza Risk Assessment Tool of the US Centers for Disease Control and Prevention (CDC) identifies 10 criteria with which to evaluate the pandemic risk of novel influenza viruses [4]. Among these 10 criteria, key epidemiological criteria include the severity of disease, the susceptibility of the population, and the transmissibility of the virus between humans. Disease severity, especially infection or symptomatic case fatality risks (CFRs), remains uncertain for both H5N1 and H7N9 viruses since the number of subclinical or mild cases remains a subject of debate [5–7].
The transmissibility between humans of H5N1 and H7N9 is perhaps the most important determinant of pandemic risk and has become worrisome given some cases occurred in clusters of 2 or more epidemiologically linked cases [8–11]. Clusters may arise from a number of factors acting singly or in combination, including the play of chance, a common exposure, person-to-person transmission, or familial susceptibility [12]. The extent of observed clustering is also affected by the process of case ascertainment. Understanding which of these explanations is more likely is important because it can provide insights into not only the transmissibility of the viruses but also the susceptibility of the human population, and of the completeness of surveillance data, thereby informing assessments of the potential threat to public health.
We conducted a comparative analysis of the characteristics of the complete series of laboratory-confirmed sporadic and clustered cases of human H5N1 and H7N9 infection worldwide up to 2 November 2014. Our objective was to inform assessments of the relative severity of infection, the transmissibility of the viruses, and to inform understanding of the susceptibility of the population. A joint analysis was conducted to provide insights beyond an analysis of one virus alone, because some factors, such as chance and surveillance biases, may be acting similarly for both viruses.
METHODS
Data Sources
In summary, data on all Chinese H7N9 cases and H5N1 cases as of 2 November 2014 were provided by Chinese CDC. Data on human H5N1 cases in Vietnam and Azerbaijan as of 2 November 2014 were provided by the Vietnam National Institute of Hygiene and Epidemiology and the Azerbaijan Ministry of Health, respectively. Information on all other cases were obtained from various publically available sources including the World Health Organization updates, local health authority's news releases, ProMed posts, and published literature. Details of data sources, case definitions, and exposure definitions are available in the Supplementary Materials. A cluster was defined as a group of 1 or more confirmed cases of H5N1 or H7N9 virus infection and additional confirmed or probable cases associated with a specific setting, such as a household, hospital, other residential institution, military barracks, recreational camp, or a neighborhood, and with the onset of cases occurring within 2 weeks of each other [9].
Statistical Methods
Fisher exact test or the Mann–Whitney U test were used, as appropriate, to compare the characteristics of clustered and sporadic cases. We used the data from the clustered cases with complete information on household contacts to estimate the relative risk (RR) of infection of blood-relatives vs unrelated contacts of the index case under the assumption that the probability of detecting infection is the same for related and unrelated close contacts. Definitions of blood-relatives and unrelated contacts are available in the Supplementary Materials. The RR of infection in related vs unrelated contacts was calculated as:
To explore the conditions that might lead to the observed level of clustering, we estimated the probability of infection given exposure, under the assumption of equal susceptibility of every individual and equal probability of detecting sporadic and clustered cases. For a given probability of infection given exposure π, under the assumption of no genetic effect, the number of infection in a household with size m follows Bin(m,π). Hence we can simulate the expected proportion of infections occurring in household clusters based on the household structure data of China. We used the method of Cauchemez et al to estimate the reproduction number R (the average number of people infected by a single human case) for H5N1 and H7N9, using the proportion of all detected sporadic and index cases with any reported exposure to the animal reservoir (G) to provide an upper estimate of R (R = 1-G), and also the proportion of index cases in a cluster with any reported exposure to the animal reservoir (F) to provide a lower estimate of R (R = 1-F) [13]. We assessed R for different scenarios of the case detection rate (0.01% to 10%) and case-to-case variation in infectiousness.
Ethical Approval
The National Health and Family Planning Commission of China, the Ministry of Health of Vietnam, and the Ministry of Health of Azerbaijan determined that the collection of data from human cases of avian influenza infection was part of the public health investigation of an outbreak and was exempt from institutional review board assessment. All other data were obtained from publicly available data sources. All data were supplied and analyzed in an anonymous format, without access to personal identifying information.
RESULTS
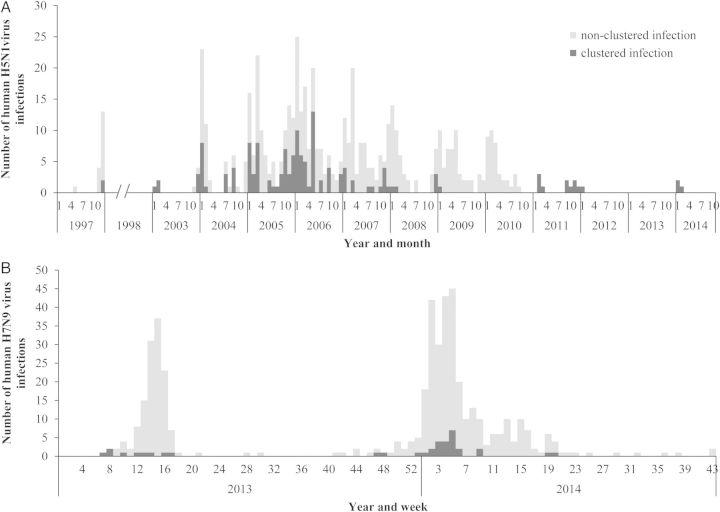
We obtained data on 720 human H5N1 cases that have been reported globally since the first identified cases of human infection in Hong Kong SAR in 1997 up until 2 November 2014 (Table 1). This included 688 laboratory-confirmed cases, 27 probable cases, and 5 suspected cases. We also obtained data on 457 laboratory-confirmed H7N9 cases and 3 suspected case reported worldwide since the first case was confirmed on 31 March 2013 in China up until 2 November 2014 (Table 1). Among these cases, 55 H5N1 clusters and 16 H7N9 clusters were identified (Table 1; Supplementary Table 1). The date of illness onset of clustered H5N1 cases peaked annually in the northern-hemisphere winter, at the same time as sporadic H5N1 cases, with the exception that no clustered H5N1 cases were reported in 2010 and 2013 (Figure 1, panel A). Clustered H7N9 cases were reported both in spring 2013, winter 2013–2014 and spring 2014, concomitant with peaks in the reporting of sporadic H7N9 cases (Figure 1, panel B). Clustered H5N1 cases were reported from the majority of countries in which human H5N1 cases have been detected (11/16, 69%). Clustered H7N9 cases have been reported from 35% (6/17) of the Chinese provinces from which H7N9 cases have been reported (Supplementary Figures 1 and 2). A higher proportion of H5N1 infections occurred in clusters (all reported infections - 144/720; 20%, laboratory confirmed cases only - 112/688; 16%) compared to H7N9 infections (all reported infections - 34/460; 8%, laboratory confirmed cases only - 33/457; 7%).
Table 1.
Number of Human H5N1 and H7N9 Cases and Clusters by Country/Province
| Country/Province | Total No. of Cases | No. of Clusters | n/N (%) of Cases Occurring in Clusters |
|---|---|---|---|
| H5N1 cases and clusters | |||
| Azerbaijan | 9 | 2 | 9/9 (100) |
| Bangladesh | 7 | 0 | 0/7 (0) |
| Cambodia | 58 | 3 | 7/58 (12) |
| Canada | 1 | 0 | 0/1 (0) |
| China, mainland | 47 | 3 | 6/47 (13) |
| China, Hong Kong SAR | 23 | 2 | 5/23 (22) |
| Djibouti | 1 | 0 | 0/1 (0) |
| Egypt | 178 | 4 | 9/178 (5) |
| Indonesia | 208 | 20 | 52/208 (25) |
| Iraq | 3 | 1 | 2/3 (67) |
| Laos PDR | 2 | 0 | 0/2 (0) |
| Myanmar | 1 | 0 | 0/1 (0) |
| Nigeria | 2 | 1 | 2/2 (100) |
| Pakistan | 4 | 1 | 4/4 (100) |
| Thailand | 28 | 3 | 8/28 (29) |
| Turkey | 12 | 3 | 8/12 (67) |
| Vietnam | 136 | 12 | 32/136 (24) |
| Total | 720 | 55 | 144/720 (20) |
| H7N9 cases and clusters | |||
| Anhui, China | 18 | 0 | 0/18 (0) |
| Beijing, China | 5 | 0 | 0/5 (0) |
| Fujian, China | 22 | 0 | 0/22 (0) |
| Guangdong, China | 110 | 4 | 7/110 (6) |
| Guangxi, China | 2 | 0 | 1/2 (50) |
| Hebei, China | 1 | 0 | 0/1 (0) |
| Henan, China | 4 | 0 | 0/4 (0) |
| Hunan, China | 24 | 2 | 4/24 (17) |
| Jilin, China | 2 | 0 | 0/2 (0) |
| Jiangsu, China | 57 | 1 | 2/57 (4) |
| Jiangxi, China | 8 | 0 | 0/8 (0) |
| Shandong, China | 5 | 2 | 4/5 (80) |
| Shanghai, China | 42 | 2 | 5/42 (12) |
| Xinjiang, China | 4 | 0 | 0/4 (0) |
| Zhejiang, China | 141 | 5 | 13/141 (9) |
| Hong Kong SAR, China | 10 | 0 | 0/10 (0) |
| Taiwan, China | 4 | 0 | 0/4 (0) |
| Malaysia | 1 | 0 | 0/1 (0) |
| Total | 460 | 16 | 36/460 (8) |
Figure 1.
Epidemic curve of sporadic and clustered human cases with H5N1 and H7N9 virus infection (as of 2 November 2014). A, Number of sporadic and clustered human cases with H5N1virus infection by month of illness onset. B, Number of sporadic and clustered human cases with H7N9 virus infection by week of illness onset. Note for A, When the date of illness onset is missing, the earliest date among the date of hospitalization, date of outcome, and date of World Health Organization (WHO) report is used. The month of illness onset for 23 cases in total 720 cases are missing and excluded from this epidemic curve: 21 cases of Indonesia in 2009 and 2 cases of Turkey in 2006.
The index case in each cluster is essentially detected in the same way as sporadic cases. Therefore, in order to examine whether the individual characteristics of cluster cases are systematically different from other cases, we compared secondary cluster cases with the combined group of sporadic cases and the index case of each cluster (sporadic/index cases). Twenty-two mild cases of infection with H7N9 viruses were identified through sentinel surveillance of influenza-like-illness (ILI). To assess the potential impact of these ILI cases on the results, we conducted the analysis with and without these 22 cases. Table 2 shows a comparison of the demographic characteristics and outcomes of sporadic/index cases vs secondary clustered cases of H7N9 and H5N1. The average age of sporadic/index cases of H7N9 was higher than secondary cases (median age 59 years vs 31; P < .001). There was no statistically significant difference in the age of sporadic/index H5N1 cases vs secondary H5N1 cases (P = .126). The CFR of sporadic/index H7N9 cases (40%) was higher than the CFR of secondary H7N9 cases (25%) but the difference was not statistically significant. There was also no statistically significant difference in the hospitalization ratio of sporadic/index H7N9 cases vs secondary H7N9 cases. However, 92% of sporadic/index H7N9 cases were classified as severe compared to 55% of secondary cases (P < .001), and 71% of sporadic/index H7N9 cases required admission to intensive care compared to only 33% of secondary cases (P = .007). Data on intensive care unit admission and severity were not available for H5N1 cases, but there was no statistically significant difference in the case fatality or hospitalization ratio of sporadic/index H5N1 cases vs secondary H5N1 cases, and hospitalization ratio of sporadic/index cases (99.7%) is only slightly higher than that of secondary cases (95.8%) (P = .011). A sensitivity analysis of excluding the 22 H7N9 cases identified through sentinel ILI surveillance sites did not change the findings (Supplementary Table 2).
Table 2.
Demographic Characteristics and Outcomes of Sporadic and Clustered Cases of H5N1 and H7N9 Virus Infection
| Characteristics | H5N1 Cases |
H7N9 Cases |
||||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 720) | Cluster Secondary Cases (n = 89) | Sporadic Cases or Cluster Index Cases (n = 631) | P Valuea | Total (n = 460) | Cluster Secondary Cases (n = 20) | Sporadic Cases or Cluster Index Cases (n = 440) | P Valuea | |
| Age | ||||||||
| Median (range) | 18 (0.3, 86) | 16 (0.3, 51) | 18 (0.7, 86) | .126 | 58 (0.4, 91) | 31 (3, 87) | 59 (0.4, 91) | <.001 |
| Age group | ||||||||
| 0–9 | 232 (33.4%) | 31 (35.2%) | 201 (33.1%) | 25 (5.4%) | 7 (35.0%) | 18 (4.1%) | ||
| 10–19 | 138 (19.9%) | 20 (22.7%) | 118 (19.4%) | 4 (0.9%) | 0 (0.0%) | 4 (0.9%) | ||
| 20–29 | 150 (21.6%) | 23 (26.1%) | 127 (20.9%) | 18 (3.9%) | 2 (10.0%) | 16 (3.6%) | ||
| 30–39 | 110 (15.8%) | 12 (13.6%) | 98 (16.1%) | 61 (13.3%) | 3 (15.0%) | 58 (13.2%) | ||
| 40–49 | 34 (4.9%) | 1 (1.1%) | 33 (5.4%) | 44 (9.6%) | 1 (5.0%) | 43 (9.8%) | ||
| 50–59 | 20 (2.9%) | 1 (1.1%) | 19 (3.1%) | 91 (19.8%) | 4 (20.0%) | 87 (19.8%) | ||
| ≥60 | 11 (1.6%) | 0 (0.0%) | 11 (1.8%) | 217 (47.2%) | 3 (15.0%) | 214 (48.6%) | ||
| Unknown | 25 | 1 | 24 | 0 | 0 | 0 | ||
| Gender | ||||||||
| Female | 370 (53.1%) | 42 (47.2%) | 328 (53.9%) | .256 | 145 (31.5%) | 9 (45.0%) | 136 (30.9%) | .219 |
| Male | 327 (46.9%) | 47 (52.8%) | 280 (46.1%) | 315 (68.5%) | 11 (55.0%) | 304 (69.1%) | ||
| Unknown | 23 | 0 | 23 | 0 | 0 | 0 | ||
| Outcome | ||||||||
| Death | 426 (59.8%) | 45 (54.2%) | 381 (61.6%) | .285 | 180 (40.6%) | 5 (27.8%) | 175 (41.2%) | .331 |
| Survive | 286 (40.2%) | 38 (45.8%) | 248 (39.4%) | 263 (59.4%) | 13 (72.2%) | 250 (58.8%) | ||
| Unknown | 8 | 6 | 2 | 17 | 2 | 15 | ||
| Severity | ||||||||
| Hospitalization | 638 (99.2%) | 68 (95.8%) | 570 (99.7%) | .011 | 436 (99.5%) | 20 (100.0%) | 416 (99.5%) | 1 |
| Unknown | 77 | 18 | 59 | 22 | 0 | 22 | ||
| Severe cases | NA | NA | NA | 407 (90.2%) | 11 (55.0%) | 396 (91.9%) | <.001 | |
| Unknown | NA | NA | NA | 9 | 0 | 9 | ||
| ICU admission | NA | NA | NA | 197 (68.6%) | 5 (33.3%) | 192 (70.6%) | .007 | |
| Unknown | NA | NA | NA | 173 | 5 | 168 | ||
| Exposure history | ||||||||
| Any exposure to poultry | 576 (94.4%) | 65 (87.8%) | 511 (95.3%) | .015 | 302 (80.5%) | 12 (63.2%) | 290 (81.5%) | .069 |
| Unknown | 110 | 15 | 95 | 85 | 1 | 84 | ||
| Occupational exposure to live poultry | 13 (9.8%) | 0 (0.0%) | 13 (12.3%) | .070 | 28 (6.1%) | 0 (0.0%) | 28 (6.4%) | .625 |
| Unknown | 587 | 62 | 525 | 3 | 0 | 3 | ||
| Visit LBMs | 72 (52.6%) | 4 (21.1%) | 68 (57.6%) | .005 | 219 (59.8%) | 6 (30.0%) | 213 (61.6%) | .008 |
| Unknown | 583 | 70 | 513 | 94 | 0 | 94 | ||
| Exposure to sick or dead poultry | 417 (89.7%) | 52 (83.9%) | 365 (90.6%) | .117 | 10 (2.8%) | 2 (10.0%) | 8 (2.4%) | .102 |
| Unknown | 255 | 27 | 228 | 102 | 0 | 102 | ||
| Exposure to backyard poultry | 158 (67.2%) | 28 (63.6%) | 130 (68.1%) | .596 | 92 (30.9%) | 8 (40.0%) | 84 (30.2%) | .452 |
| Unknown | 485 | 45 | 440 | 162 | 0 | 162 | ||
| Human case contact | 47 (29.4%) | 45 (91.8%) | 2 (1.8%) | <.001 | 24 (9.9%) | 19 (95.0%) | 5 (2.2%) | <.001 |
| Unknown | 560 | 40 | 520 | 217 | 0 | 217 | ||
Abbreviations: ICU, intensive care unit; LBM, live bird market; NA, not available.
a The Mann–Whitney test is used for median age, whereas the Fisher exact test is used for other variables.
A history of exposure to poultry prior to onset was common for all groups (Table 2). For both H5N1 and H7N9, and for various types of poultry exposure, poultry exposure was less commonly reported in secondary cases than in sporadic/index cases (except exposure to sick or dead poultry and backyard poultry for H7N9 cases); however, most of these differences were not statistically significant except any exposure to poultry for H5N1 cases (P = .015), visiting live bird markets for H5N1 cases (P = .005), and visiting live bird markets for H7N9 cases (P = .008). A history of exposure to sick or dead poultry was more common for H5N1 cases compared to H7N9 cases, as would be expected given the highly pathogenic phenotype of H5N1 in poultry and the low pathogenic phenotype of H7N9. Where data were available, prior exposure to poultry or poultry markets was reported for 97.2% (518/533) of H5N1 sporadic/index cases and 85.0% (307/361) of H7N9 sporadic/index cases. All index cases of H7N9 clusters and 96% (49/51) index cases of H5N1 clusters reported poultry exposure. If this exposure history is taken as evidence that infection was acquired from the natural reservoir, then using the approach of Cauchemez et al, the reproduction number has an upper limit of the 95% confidence interval of 0.12 for H5N1 and 0.27 for H7N9 (Supplementary Tables 3 and 4) [13]. These estimates were not very sensitive to the method used (R = 1-F or R = 1-G), the proportion of cases detected, or case-to-case variation in infectiousness.
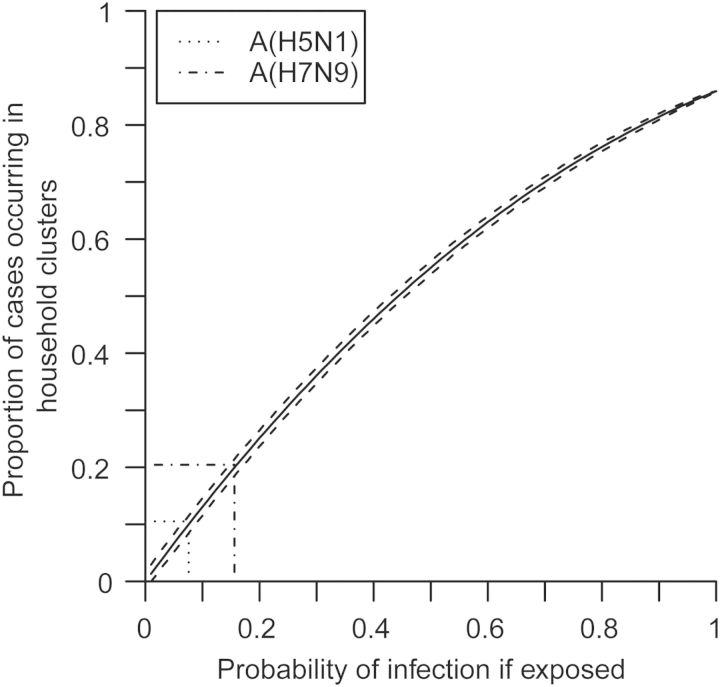
The observed average cluster size was 2.62 (standard deviation [SD] 1.21) for H5N1 and 2.25 (SD 0.58) for H7N9 (Table 3). For those clusters where full data on close contacts was available, the RR of infection in blood-related contacts of cases compared to unrelated contacts was 8.96 for H5N1 (95% confidence interval (CI), 1.30, 61.86) and 0.80 for H7N9 (95% CI, .32, 1.97). The difference in RR was statistically significant (P = .03). If equal susceptibility of every individual is assumed, the observed proportion of cases occurring in clusters is consistent with an estimated probability of infection given exposure of 15.6% (95% CI, 14.4, 16.8) for H5N1 and 7.6% (95% CI, 6.3, 10.5) for H7N9 (Figure 2).
Table 3.
Size and Blood Relationships of H5N1 and H7N9 Clusters
| Characteristic | H5N1 Clusters |
H7N9 Clusters |
||
|---|---|---|---|---|
| No. of cases per cluster | ||||
| Median (range) | 2 (2,8) | 2 (2,4) | ||
| Average (SD) | 2.618 (1.209) | 2.25 (0.577) | ||
| No. of Clusters (%) | No. of Infections (%) | No. of Clusters (%) | No. of Infections (%) | |
| 2 cases | 37 (67.3) | 74 (51.4) | 13 (81.3) | 26 (72.2) |
| 3 cases | 10 (18.2) | 27 (20.8) | 2 (12.5) | 6 (16.7) |
| 4 cases | 5 (9.1) | 20 (13.9) | 1 (6.3) | 4 (11.1) |
| 5 cases | 1 (1.8) | 10 (3.5) | 0 (0.0) | 0 (0.0) |
| 6 cases | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 7 cases | 1 (1.8) | 7 (4.9) | 0 (0.0) | 0 (0.0) |
| 8 cases | 1 (1.8) | 8 (5.6) | 0 (0.0) | 0 (0.0) |
| Total | 55 (100.0) | 144 (100.0) | 16 (100.0) | 36 (100.0) |
| Secondary cases in related and unrelated close contacts of index casesa | ||||
| Blood-relative contact infected | 28 | 7 | ||
| Blood-relative contact not infected | 38 | 23 | ||
| Unrelated contact infected | 0 | 7 | ||
| Unrelated contact not infected | 19 | 17 | ||
| Relative risk (95% CI) | 8.96 (1.30, 61.86)b | 0.80 (0.33, 1.97) | ||
Abbreviations: CI, confidence interval; SD, standard deviation.
a Refers to blood relationship with the cluster index case. Index cases of the clusters are excluded. A blood-relative relationship was defined as parent-offspring, siblings, grandparent-grandchild, and uncle/aunt-niece/nephew. An unrelated contact was defined as spouse, healthcare worker, son/daughter-in-law, parent-in-law, and other unrelated household member.
b 1 is added in each cell to calculate the relative risk since there is a zero cell.
Figure 2.
Proportion of cases occurring in household clusters by the probability of infection given exposure.
DISCUSSION
We have assembled a comprehensive dataset of human infections with the 2 most important zoonotic influenza viruses currently infecting humans and undertaken the largest comparative analysis and detailed analysis of clustered and nonclustered cases. This was conducted in order to inform estimates of the severity and transmissibility of infection, and the susceptibility of the population. Our analysis provides important new insights into the epidemiology of both H5N1 and H7N9, demonstrating the value of comparative analyses such as this.
We have shown that secondary cases of H7N9 occurring in clusters are markedly younger than sporadic/index cases and are less severe. This suggests an important case ascertainment bias, with sporadic cases detected through routine surveillance or through the healthcare system being biased towards older and more severe cases. This in turn indicates that a large number of mild cases in younger people are likely not being detected, a finding that is supported by the analysis of Yu et al and by serological studies [5, 14–16]. In contrast, we did not find any similar evidence of case-ascertainment bias for H5N1. This suggests that sporadic/index H5N1 cases are representative of all cases, and therefore that the majority of H5N1 cases are probably severe. This finding helps to resolve the ongoing debate about the virulence and transmissibility of H5N1, and, in line with the interpretations of H5N1 seroepidemiological studies by Toner et al and Kerkhove et al, suggests that large numbers of mild or asymptomatic H5N1 infections have not occurred [6, 17].
We also show that clusters of H5N1 cases are more common than clusters of H7N9 cases and a higher proportion of all H5N1 cases are clustered cases than H7N9 cases. As our analysis demonstrates (Figure 2), this could be the result of a higher risk of H5N1 infection if exposed compared to H7N9, but we argue that this is an unlikely explanation given the much faster rate of accumulation of human H7N9 cases (457 confirmed cases in less than 2 year) compared to H5N1 (688 confirmed cases in the decade since its reemergence in 2003) despite the much greater extent of the H5N1 epizootic. An alternative explanation is that the clustering rates are similar for H5N1 and H7N9 but that H5N1 clusters are more easily detected than H7N9 clusters. Although this is plausible given the greater severity of H5N1 cases compared to H7N9 cases, it is not consistent with the very active response to the emergence of H7N9 in 2013, and the more widespread availability of molecular diagnostic methods during H7N9 emergence. A third explanation is that person to person transmission is more common for H5N1 than H7N9, but we found no evidence to support this in our analysis of the reproduction number or average cluster size. A fourth explanation is that the greater clustering of H5N1 cases reflects familial susceptibility. This hypothesis is supported by our finding of an increased RR of secondary infection in blood relatives of H5N1 index cases, which was not found in blood relatives of H7N9 cases. The differences in familial RR are consistent with other analyses [12, 18]. Mouse models clearly demonstrate a strong genetic effect on susceptibility to a range of influenza viruses and the severity of influenza infection [19–21]. Many candidate genes for susceptibility to severe influenza have been proposed based on understanding of the pathogenesis and immune evasion strategies of influenza virus and information gained from mice model and other in-vitro studies [22–25]. However, relatively few human studies have systematically evaluated the influence of genetic polymorphisms on susceptibility and disease severity in influenza infections [26–35]. The work by Wang Z et al is the only human genetic study in H7N9 infection [35]. They identified the rs12252-C genotype that compromises the interferon-induced transmembrane 3 (IFITM3) gene function as a primary genetic correlate of severe A(H7N9) pneumonia, which corroborates previous findings in pandemic influenza A(H1N1)pdm09 infections [30, 33].
As we only have access to public resources for H7N9 and H5N1 cases outside of China, Viet Nam, and Azerbaijan, the major limitation of our analysis is the availability and quality of data on exposure variables, clinical outcomes, and household contacts. First, we were only able to obtain the crude outcomes data of deaths and hospitalizations for H5N1 cases. Therefore, we are not able to assess if there are differences in severity of secondary and sporadic/index H5N1 cases other than death and hospitalization, where no difference was observed Previous work by Wang C et al found that a history of smoking was associated with a reduced risk of hospitalization [36]. However, as smoking and preexisting health status were not available in the current study, we could not assess these factors. Second, our analysis of familial RR was limited to an analysis only of clusters where full data were available [29% (16/55) H5N1 clusters and 69% (11/16) H7N9 clusters]. A more powerful analysis of familial RR could be conducted if better data were available, and we recommend that data on the number of close contacts, their relationship with the index case, and the outcome of health surveillance is routinely recorded and centralized for all public health investigations of human cases of avian influenza. Third, data on various exposure history variables were missing for 15%–82% H5N1 cases, whereas various exposure history variables were missing for 1%–47% H7N9 cases. The simple estimators proposed by Cauchemez et al to determine the average number of persons infected by a human case (R) and the case detection rate are dependent on good data on exposure to the animal reservoir or to other human cases [13]. In summary, our data are only inclusive of what has been reported and in some cases publically available, and this might lead to over or underestimates of findings mentioned above.
CONCLUSION
The data suggest that the severity and average age of laboratory-confirmed H7N9 cases is biased upward, compared to all symptomatic H7N9 cases, by the under-ascertainment of young, mild cases, whereas this is not true for H5N1. This suggests quite different risk profiles for these 2 viruses. H5N1 infection causes a severe disease, but there may be quite restricted population susceptibility, perhaps genetically based. The long-term and widespread circulation of H5N1 with relatively few human cases and without the emergence of a human adapted strain supports the existence of strong, but as yet unidentified, biological barriers to transmission and adaptation. In contrast, the data indicate there may be a large number of undetected mild cases of H7N9, suggesting more widespread human susceptibility to H7N9 than to H5N1. Greater population susceptibility and a higher number of cases increase the opportunities for adaptive evolution, whereas mild cases make the detection and monitoring of virus changes more challenging [37]. This suggests the potential pandemic risk is greater for H7N9 than H5N1.
Evaluating the threat posed by H7N9 and H5N1 requires improvements in the standardization, collation and reporting of basic epidemiological and clinical data, and a formal test of the hypothesis of host genetic susceptibility to H5N1 infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Author contributions. H. Y. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: H. Y., P. W. H., B. J. C. Acquisition of data: Y. Q., E. C., L. G., J. O., T. H. N., T. N. D., V. G., L. F., P. W., H. J., X. R., Z. P., S. L., M. L., J. Z., S. L., S. H., R. H. Analysis and interpretation of data: Y. Q., P. W. H., T. K. T., B. J. C., H. Y. Drafting of the manuscript: P. W. H., Y. Q., B. J. C., H. Y. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: T. K. T., B. J. C. Study supervision: H. Y., J. J. F., G. F. G., G. M. L.
Additional contributions. We thank staff members at county-, district-, prefecture-, and provincial- level Chinese Centers for Disease Control and Preventions (CDCs) at the provinces where human H7N9 and H5N1 cases occurred for providing assistance with field investigation, administration, and data collection.
Financial support. This study was funded by the US National Institutes of Health (Comprehensive International Program for Research on AIDS grant U19 AI51915), grants from the Ministry of Science and Technology, China (2012 ZX10004-201), the Harvard Center for Communicable Disease Dynamics from the National Institute of General Medical Sciences (grant U54 GM088558), and the Area of Excellence Scheme of the Hong Kong University Grants Committee (grant no. AoE/M-12/06). P. W. H. is funded by the Wellcome Trust (grants 089276/Z/09/Z and 089276/B/09/Z) and the EU FP7 project PREPARE (602525).
Disclaimer. The funding bodies had no role in the study design, data collection and analysis, preparation of the manuscript, or the decision to publish. The views expressed are those of the authors and do not necessarily represent the policy of China CDC or the institutions with which the authors are affiliated.
Potential conflicts of interest. G. M. L. has received consulting honoraria from Janssen Pharmaceuticals. B. J. C. reports receipt of research funding from MedImmune Inc. and Sanofi Pasteur, and consults for Crucell NV. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Influenza at the Human-Animal Interface. Monthly Risk Assessment Summary. Available at: http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_6January2015.pdf?ua=1 Accessed 8 January 2015.
- 2.World Health Organization. WHO risk assessment of human infection with avian influenza A(H7N9) virus. Available at: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/riskassessment_h7n9_2Oct14.pdf?ua=1 Accessed 8 January 2015.
- 3.Cowling BJ, Jin L, Lau EH, et al. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 2013; 382:129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Influenza Risk Assessment Tool (IRAT). Available at: http://www.cdc.gov/flu/pandemic-resources/tools/risk-assessment.htm Accessed 8 January 2015.
- 5.Yu H, Cowling BJ, Feng L, et al. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 2013; 382:138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toner ES, Adalja AA, Nuzzo JB, Inglesby TV, Henderson DA, Burke DS. Assessment of serosurveys for H5N1. Clin Infect Dis 2013; 56:1206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TT, Parides MK, Palese P. Seroevidence for H5N1 influenza infections in humans: meta-analysis. Science 2012; 335:1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuen KY, Chan PK, Peiris M, et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 1998; 351:467–71. [DOI] [PubMed] [Google Scholar]
- 9.Summary of human infection with highly pathogenic avian influenza A (H5N1) virus reported to WHO, January 2003-March 2009: cluster-associated cases. Wkly Epidemiol Rec 2010; 85:13–20. [PubMed] [Google Scholar]
- 10.Li Q, Zhou L, Zhou M, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 2014; 370:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi X, Qian YH, Bao CJ, et al. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ 2013; 347:f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horby P, Sudoyo H, Viprakasit V, et al. What is the evidence of a role for host genetics in susceptibility to influenza A/H5N1? Epidemiol Infect 2010; 138:1550–8. [DOI] [PubMed] [Google Scholar]
- 13.Cauchemez S, Epperson S, Biggerstaff M, Swerdlow D, Finelli L, Ferguson NM. Using routine surveillance data to estimate the epidemic potential of emerging zoonoses: application to the emergence of US swine origin influenza A H3N2v virus. PLoS Med 2013; 10:e1001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma MJ, Ma GY, Yang XX, et al. Avian influenza A(H7N9) virus antibodies in close contacts of infected persons, China, 2013–2014. Emerg Infect Dis 2015; 21:709–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Chen Y, Cui D, et al. Avian-origin influenza A(H7N9) infection in influenza A(H7N9)-affected areas of China: a serological study. J Infect Dis 2014; 209:265–9. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Fang S, Lu X, et al. Seroprevalence to avian influenza A(H7N9) virus among poultry workers and the general population in southern China: a longitudinal study. Clin Infect Dis 2014; 59:e76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Kerkhove MD, Riley S, Lipsitch M, et al. Comment on "Seroevidence for H5N1 influenza infections in humans: meta-analysis". Science 2012; 336:1506; author reply. [DOI] [PubMed] [Google Scholar]
- 18.Aditama TY, Samaan G, Kusriastuti R, et al. Risk factors for cluster outbreaks of avian influenza A H5N1 infection, Indonesia. Clin Infect Dis 2011; 53:1237–44. [DOI] [PubMed] [Google Scholar]
- 19.Horby P, Nguyen NY, Dunstan SJ, Baillie JK. The role of host genetics in susceptibility to influenza: a systematic review. PloS One 2012; 7:e33180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horby P, Nguyen NY, Dunstan SJ, Baillie JK. An updated systematic review of the role of host genetics in susceptibility to influenza. Influenza Other Respi Viruses 2013; 7(suppl 2):37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin TY, Brass AL. Host genetic determinants of influenza pathogenicity. Curr Opin Virol 2013; 3:531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juno J, Fowke KR, Keynan Y. Immunogenetic factors associated with severe respiratory illness caused by zoonotic H1N1 and H5N1 influenza viruses. Clin Dev Immunol 2012; doi:10.1155/2012/797180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mubareka S, Palese P. Human genes and influenza. J Infect Dis 2008; 197:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trammell RA, Toth LA. Genetic susceptibility and resistance to influenza infection and disease in humans and mice. Expert Rev Mol Diagn 2008; 8:515–29. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Katz JM, Gwinn M, Dowling NF, Khoury MJ. Systems-based candidate genes for human response to influenza infection. Infect Genet Evol 2009; 9:1148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keynan Y, Juno J, Meyers A, et al. Chemokine receptor 5Δ32 allele in patients with severe pandemic (H1N1) 2009. Emerg Infect Dis 2010; 16:1621–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan JF, To KK, Tse H, et al. The lower serum immunoglobulin G2 level in severe cases than in mild cases of pandemic H1N1 2009 influenza is associated with cytokine dysregulation. Clin Vaccine Immunol 2011; 18:305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferdinands JM, Denison AM, Dowling NF, et al. A pilot study of host genetic variants associated with influenza-associated deaths among children and young adults. Emerg Infect Dis 2011; 17:2294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonopoulou A, Baziaka F, Tsaganos T, et al. Role of tumor necrosis factor gene single nucleotide polymorphisms in the natural course of 2009 influenza A H1N1 virus infection. Int J Infect Dis 2012; 16:e204–8. [DOI] [PubMed] [Google Scholar]
- 30.Everitt AR, Clare S, Pertel T, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012; 484:519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuniga J, Buendia-Roldan I, Zhao Y, et al. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J 2012; 39:604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hertz T, Oshansky CM, Roddam PL, et al. HLA targeting efficiency correlates with human T-cell response magnitude and with mortality from influenza A infection. Proc Natl Acad Sci U S A 2013; 110:13492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang YH, Zhao Y, Li N, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun 2013; 4:1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.To KK, Zhou J, Song YQ, et al. Surfactant protein B gene polymorphism is associated with severe influenza. Chest 2014; 145:1237–43. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Zhang A, Wan Y, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A 2014; 111:769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Yu H, Horby PW, et al. Comparison of patients hospitalized with influenza A subtypes H7N9, H5N1, and 2009 pandemic H1N1. Clin Infect Dis 2014; 58:1095–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature 2003; 426:658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.