Abstract
The genus Nocardiopsis is a widespread group within the phylum Actinobacteria and has been isolated from various salty environments worldwide. However, little is known about whether biogeography affects Nocardiopsis distribution in various hypersaline environments. Such information is essential for understanding the ecology of Nocardiopsis. Here we analyzed 16S rRNA, gyrB, rpoB and sodA genes of 78 Nocardiopsis strains isolated from hypersaline environments in Yunnan and Xinjiang Provinces of western China. The obtained Nocardiopsis strains were classified into five operational taxonomic units, each comprising location-specific phylo- and genotypes. Statistical analyses showed that spatial distance and environmental factors substantially influenced Nocardiopsis distribution in hypersaline environments: the former had stronger influence at large spatial scales, whereas the latter was more influential at small spatial scales.
The study of biogeography addresses spatial patterns of species and ecosystems in geographic space or through geological time1. It is widely accepted that macro-organism populations have many similar characteristics and common ancestry and thus possess different geographical distributions due to geographical isolation2. In contrast, debate has been lasting on microbial biogeography in that microorganisms have great potential for global dispersal and adaptability to diverse environments3. However, numerous studies recently reported bio-geographic patterns for microbial distributions4,5.
Microbial biogeography is mainly ascribed to two factors: the environmental heterogeneity and spatial distance6, either of which alone cannot entirely account for geographic distribution patterns of some widespread bacterial groups7,8. However, little is known about the relative importance of these two factors to shaping microbial biogeography in saline and hypersaline environments. Theoretically, saline and hypersaline environments are suitable for studying impact of environmental effects on bacterial biogeography because of strong environmental selective pressures in such environments9. Previous studies have shown that spatial distance and environmental factors contribute differently to shaping microbial biogeography in saline environments with respect to certain microbial groups10.
As one group of widespread Actinobacteria, Nocardiopsis species have drawn extensive attention from microbial ecologists due to their capacity to produce compounds of potential biotechnological relevance11,12,13. Most characterized Nocardiopsis strains were mainly recovered from various salty habitats, such as marine environments, deserts, alkaline or hypersaline soils14,15,16. In these salty environments, Nocardiopsis spp. underwent a wide range of environmental pressures and thus developed distinct genetic and metabolic features among different habitats11,12,13. So in order to understand the underlying reasons for the endemicity of Nocardiopsis species in various hypersaline environments, it is imperative to know 1) whether geographic isolation affects Nocardiopsis distribution in hypersaline environments and 2) relative importance of spatial distance and environmental factors to shaping Nocardiopsis distribution in (hyper) saline environments.
Investigation on the biogeography of Nocardiopsis spp. requires a detailed taxonomic classification of Nocardiopsis strains from different habitats. High phylogenetic resolution can be achieved by multilocus sequence analyses, which combine phylogenies of 16S rRNA and functional housekeeping genes such as gyrB, rpoB and sodA17,18,19. Previous studies have shown that 16S rRNA gene together with gyrB, rpoB and sodA genes could provide better phylogenetic resolution20,21,22,23 than one single gene. Here, we applied a multilocus phylogenetic analysis of 16S rRNA, gyrB, rpoB and sodA genes to investigate the biogeographic patterns of Nocardiopsis strains retrieved from the hypersaline environments in Xinjiang and Yunnan Provinces of western China and assessed their correlations with spatial distance and environmental factors.
Results
Geochemistry differentiations of the sampled sediments
In Xinjiang Province, Qijiaojing Salt Lake is 140 km away from Aydingkol Salt Lake; while in Yunnan Province, Heijing Salt Mine is about 560 km away from Jiangcheng Salt Mine. The sampling sites of Xinjiang Province are about 4300 km away from those of Yunnan Province (Table S1). The Heijing and Jiangcheng salt mines are under subtropical monsoon climate. The salt ores in the two salt mines are rich in sodium chloride as well as potassium chloride24. In Xinjiang Province, due to strong evaporation, most of surface areas of Aydingkol and Qijiaojing salt lakes have been highly mineralized, containing abundant alkaline rock salt (e.g., Glauber’s salt, glauberite, gypsum, sodium chloride) but low concentration of potassium salt25.
Principle component analysis (PCA) showed that the Yunnan sampling sites were different from that of Xinjiang with respect to environmental factors: the sediment chemistry of the Aydingkol and Qijiaojing salt lakes was different from that of the Heijing and Jiangcheng salt mines: the former possesses higher salinity, pH and concentrations of Ca2+, Mg2+ and Mn2+ but lower concentrations of trace elements (e.g. K+, Cu2+, Zn2+) and total phosphorus than the latter (cumulative contribution value = 91.5%, Fig. 1). In addition, the sampling sites within one region (Yunnan or Xinjiang) were different from each other on the basis of climate types, geographic distances, and geochemistry factors (Tables S1 & S2, Fig. 1).
Figure 1.
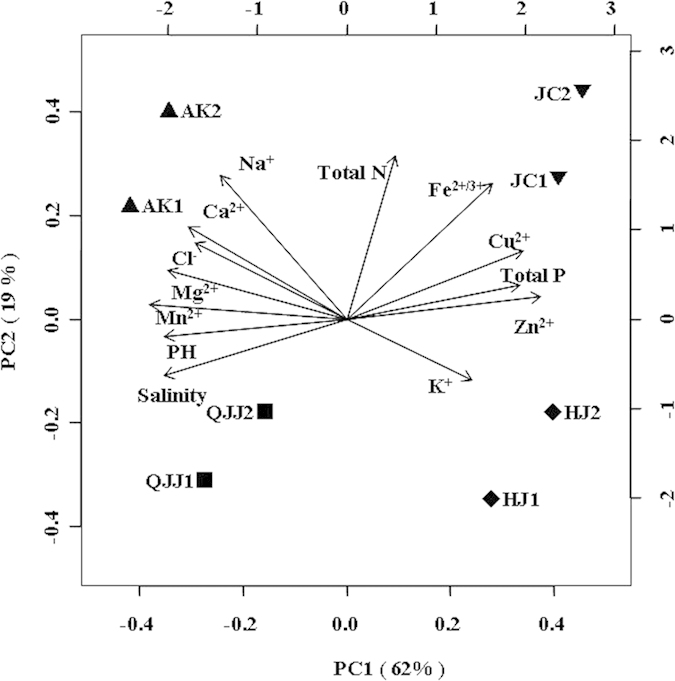
(A) PCA map showing the 8 sampling sites and their correlation with trace elements. Note: pH, Cl–, Ca2+, Mg2+, K+, Na+, Fe2+, Mn2+, Cu2+, Zn2+, total N (nitrogen) and total P (phosphorus) were used to evaluate the influence of each variable. The longer the arrow, the greater the influence; the smaller the angle between two arrows, the closer their correlation. solid squares (■), upright (▲) and inverse (▼) triangles, and diamonds (♦) denote the Qijiaojing (QJJ), Aydingkol (AK), Jiangcheng (JC) and Heijing (HJ) sampling sites, respectively.
Phylogenic analysis of the 16S rRNA, gyrB, rpoB and sodA genes
A total of 78 Nocardiopsis strains were retrieved and subjected to phenotypic characterization as well as OTU identification26. The obtained Nocardiopsis strains belonged to five OTUs (N. dassonvillei, N. aegyptia, N. terrea, N. quinghaiensis, and N. xinjiangensis) (Table S3). Each of the identified OTUs covered more than eight strains and contained at least one strain from a sampling site (Table S3). The multi-locus sequence typing (MLST) phylogeny showed endemism of Nocardiopsis strains: each endemic sequence type (ST) was specific to a site or a region (Table 1 and Fig. 2 & Fig. S1E, Bootstrap value >80%). A total of 34 STs (Table S4) were identified, with either region containing 17 STs and each sampling site including at least 8 STs (Table 1 & Table S5).
Table 1. Geographic information for sampling sites and numbers of sequence types specific to a sampling site, a habitat, and a region.
| Sampling Sites | Latitude (N) and Longitude (E) | STs of concatenated sequences (Bootstrap values>80%) |
|---|---|---|
| Yunnan region | 17 | |
| HJ1 | 25° 22.366′ N, 101° 44.568′ E | 5 |
| HJ2 | 25° 23.567′ N, 101° 45.105′ E | 3 |
| JC1 | 22° 35.069′ N, 101° 50.034′ E | 4 |
| JC2 | 22° 36.069′ N, 101° 52.034′ E | 5 |
| Xinjiang region | 17 | |
| AK1 | 42° 29′ N, 89° 22′ E | 4 |
| AK2 | 42° 29′ N, 89° 23′ E | 5 |
| QJJ1 | 43° 26′ N, 91° 38′ E | 6 |
| QJJ2 | 43° 26′ N, 91° 38.6′ E | 2 |
QJJ denotes Qijiaojing sampling sites, AK denotes Aydingkol sampling sites, JC denotes Jiangcheng sampling sites, and HJ denotes Heijing sampling sites.
Figure 2. Maximum Likelihood based Phylogenetic tree of concatenated sequences of 16S rRNA, gyrB, rpoB and sodA, showing endemism for five Nocardiopsis OTUs.
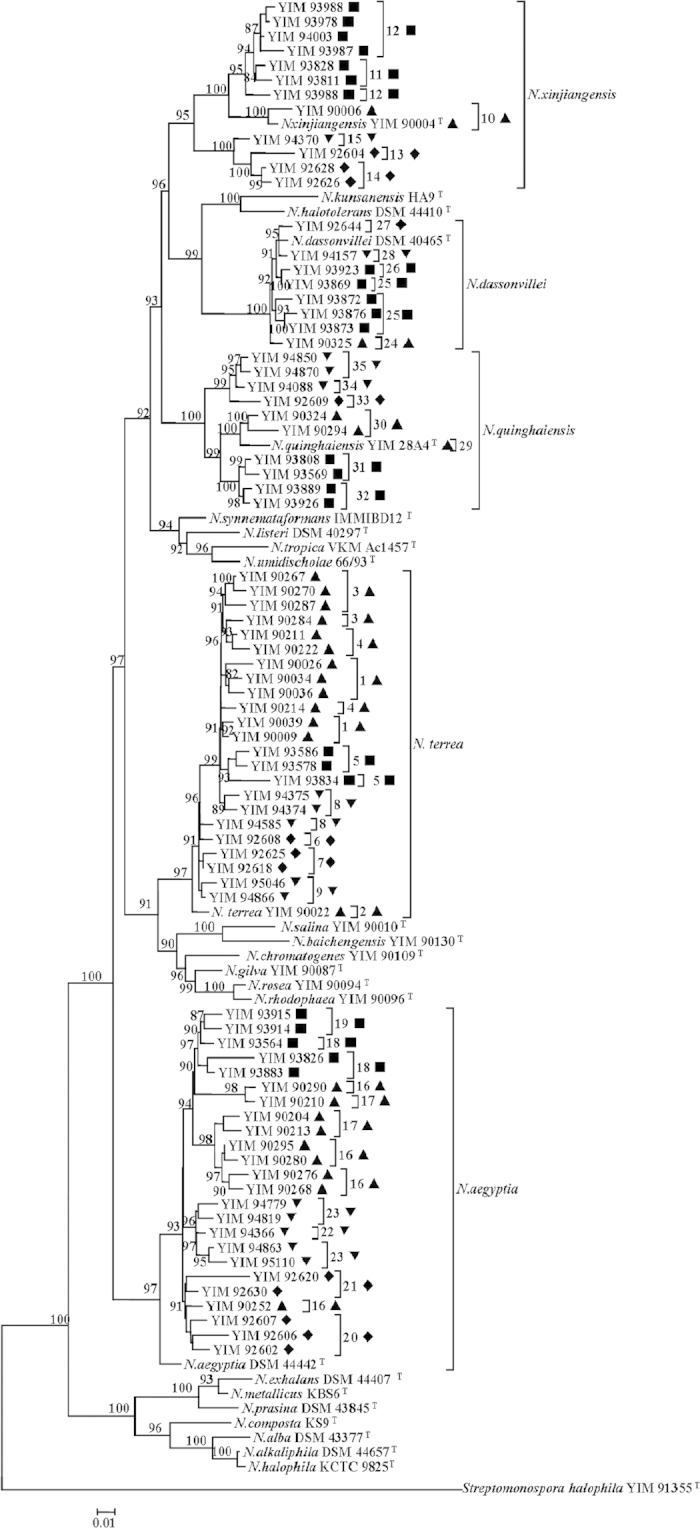
Bar, 0.05, five nucleotide substitutions per 100 nt. Bootstrap values are shown as percentage of 1000 replicates, and only the bootstrap values above 50% are shown. Solid squares (■), upright triangles (▲), inverse triangles (▼) and diamonds (♦) indicate Nocardiopsis strains from Qijiaojing (QJJ) and Aydingkol (AK) sampling sites of Xinjiang Province and Jiangcheng (JC) and Heijing (HJ) sampling sites of Yunnan Province, respectively. The 34 STs are marked in the clades, and each ST is supported by high bootstrap value (>80%).
Effects of spatial distance and environmental factors on the geographic patterns of Nocardiopsis strains
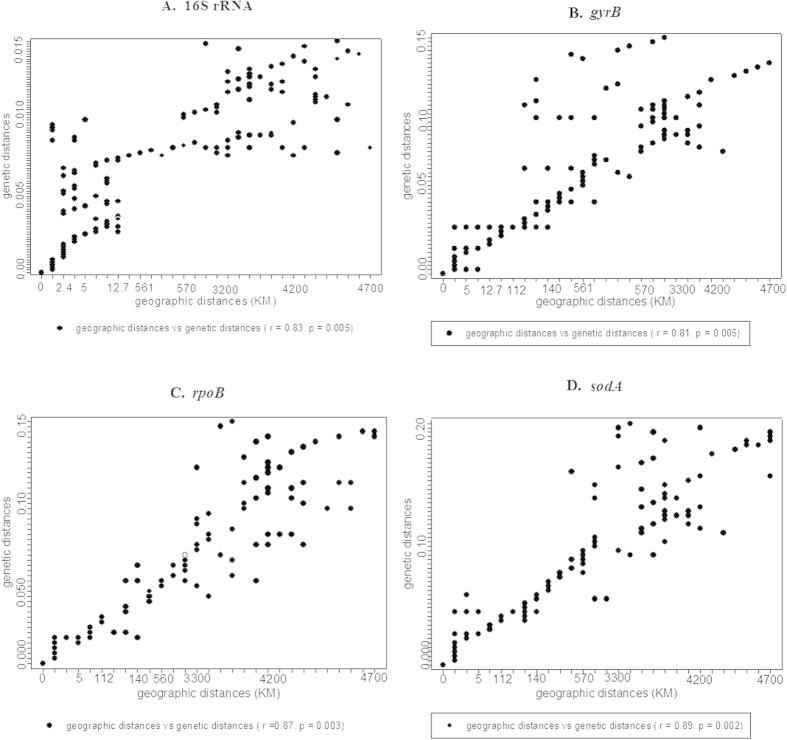
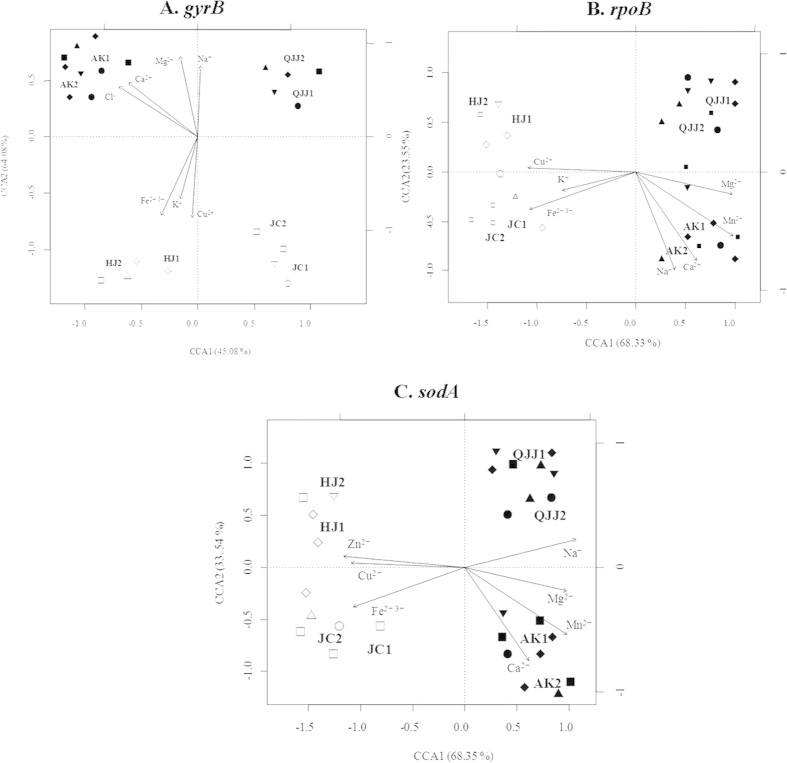
MLST of four housekeeping genes revealed significant correlations of gene sequences with geographic distance (16S rRNA: r = 0.83, p = 0.005; gyrB: r = 0.81, p = 0.005; rpoB: r = 0.87, p = 0.003; sodA: r = 0.89, p =0.002) (Fig. 3). In addition, the closely related phylo- or genotypes were present within a very small scale (<100 km) but not at distant locations (>100 km) (Fig. 3). Mantel test (r > 0.5, P < 0.05) and canonical correlation analysis showed that the differentiations of endemic genotypes of gyrB, rpoB and sodA genes were significantly correlated with the geochemistry variations of sediments from eight habitats between Yunnan and Xinjiang Provinces (Table 2; Fig. 4).
Figure 3. Mantel correlation between genetic distances of the four genes and geographic distance for the five identified Nocardiopsis OTUs.
(A–D) panels indicate the correlation between geographic distance and genetic distances of 16S rRNA gene (r = 0.83, p = 0.005), gyrB (r = 0.81, p = 0.005), rpoB (r = 0.87, p = 0.003) and sodA (r = 0.89, p = 0.002) genes, respectively.
Table 2. Simple Mantel test for genotypes of five Nocardiopsis OTUs in this study.
| environmental factor |
gyrB genotypes |
rpoB genotypes |
sodA genotypes |
|||
|---|---|---|---|---|---|---|
| r | p | r | p | r | P | |
| Cl− (ppm) | 0.536 | 0.046 | 0.594 | 0.047 | 0.087 | 0.486 |
| Ca2+ (ppm) | 0.636 | 0.026 | 0.694 | 0.047 | 0.587 | 0.046 |
| Mg2+ (ppm) | 0.736 | 0.028 | 0.540 | 0.032 | 0.527 | 0.035 |
| Na+ (ppm) | 0.739 | 0.033 | 0.506 | 0.046 | 0.579 | 0.0036 |
| Fe2/3+ (ppm) | 0.531 | 0.054 | 0.497 | 0.055 | 0.578 | 0.019 |
| Mn2+ (ppm) | 0.089 | 0.334 | 0.645 | 0.035 | 0.537 | 0.044 |
| Cu2+ (ppm) | 0.601 | 0.043 | 0.590 | 0.038 | 0.502 | 0.042 |
| Zn2+ (ppm) | 0.245 | 0.113 | 0.322 | 0.051 | 0.643 | 0.038 |
r2 is the correlation value; positive or negative values reflect the type of relationship between the two matrices, while p is the probability associated with r2. P values are significant if P is <0.05 (boldface).
Figure 4.
Canonical correlation analysis of linear relationships between geochemistry variations, and differentiations of endemic genotypes among the eight sampling sites (A–C) panels are for gyrB, rpoB, and sodA genes, respectively). In Nocardiopsis xinjiangensis, inverse solid (▼) and open (▽) triangles and upright solid triangles (▲) denote NXX, NXY and NDX phylotypes/genotypes from Xinjiang sites, respectively; upright open triangles denote NDY phylotypes/genotypes from Yunnan sites; in Nocardiopsis quinghaiensis, solid (●) and open (◯) circles denote NQX and NQY phylotypes/genotypes from Xinjiang and Yunnan sites, respectively; in Nocardiopsis aegyptia, Solid (■) and open (□) squares denote NAX and NAY phylotypes/genotypes from Xinjiang and Yunnan sites, respectively; in Nocardiopsis terrae, solid (◆) and open (◇) diamonds denote NTX and NTY phylotypes/genotypes from Xinjiang and Yunnan sites, respectively.
The Z-test of three functional housekeeping gene (gyrB, rpoB and sodA) transcripts by optimum ‘positive selection’ models (M2a and M8) showed that evolutions of gyrB, rpoB and sodA genes in the 78 Nocardiopsis strains in this study and 24 type strains from other habitats were partially under positive environmental selection (ω2 1 and ω8 1, respectively, Table S6). Mutations of seven, five and five residues of the B subunit of DNA gyrase, the β subunit of RNA polymerase and the A subunit of superoxide dismutase, respectively, were positively influenced by environmental forces (Table S6). The predicted molecular function ontology of gyrB, rpoB and sodA gene transcripts (Table S7) showed that seven N-terminus residues of the B subunit of DNA gyrase had high potential for binding magnesium ion, ATP or integrating with nucleotides, five N-terminus residues of the β subunit of RNA polymerase had high potential of binding cations or rifampicin, and five N-terminus residues of the A subunit of superoxide dismutase had high potential of binding magnesium, iron, or copper ions.
Discussion
Geographic patterns and endemism of Nocardiopsis genotypes within an OTU
Our study supports previously detected biogeographical patterns among Nocardiopsis, being consistent with the fact that some Nocardiopsis species have been exclusively isolated from certain habitats to date27,28,29,30,31. The patchy distribution of Nocardiopsis among species and endemic patterns within one species (Fig. 2 & Fig. S1E) indicated that biogeography may influence microbial distribution within a species but may not function among species within the genus at a large geographic scale. The observed endemic distribution of Nocardiopsis strains was consistent with previous studies about other microbial groups32,33,34,35,36,37. For example, a patchy geographic distribution was found for the bacterial isolates within a homogeneous background (sulfate-reducing sediments from four continents32. Similarly, crenarchaeal assemblages in mesophilic soil habitats were distributed in mosaic patterns of different phylotypes6,36. Likewise, individual genotypes of purple non-sulfur bacterium Rhodopseudomonas palustris were detected only locally and exhibited patchy distribution at 10-m or even 1-m scales33.
Relative importance of spatial distance and environmental factors upon endemism of Nocardiopsis strains
The biogeographic distribution of Nocardiopsis could be ascribed to spatial distance and environmental factors. However, little is known about the relative importance of spatial distance and environmental factors on the distributional patterns of the five known Nocardiopsis species. In this study, the impact of spatial distance upon endemicity of Nocardiopsis in Yunnan or Xinjiang could be validated by the fact that the closely related phylo- or genotypes was present within a very small scale (<100 km) but not at distant (>100 km) locations (r > 0.80 p < =0.005, Fig. 3). This indicated that spatial distance significantly contribute to the observed biogeographic patterns of Nocardiopsis strains at a large scale, which was consistent with some previous studies6,38. Previously, spatial distance together with genetic drift or physical isolation was proposed to lead to microbial population endemism at a large scale38,39. Our data suggest that the spatial distance notably resulted in differentiations of Nocardiopsis strains between regions (>100 km, Fig. 3).
Our study suggested that both environmental parameters and spatial distance played a role. However, environmental parameters apparently rather influence microbial endemism at the local scale. In the present study, the genetic differentiations of gyrB, rpoB and sodA genes of the retrieved Nocardiopsis strains significantly corresponded to heterogeneities of some cations or anions in the sediments of the studied sampling sites within a habitat (Table 2 and Fig. 4). This observation was consistent with some previous studies, in which environmental factors rather than spatial distance were shown to cause bacterial variation at a local scale (within 1 km)38,39. Previous studies indicated that Na+, Mg2+, Ca2+, Mn2+ and Fe2+/3+ ions were significant in influencing bacterial biogeography at the species (97% OTU) or subspecies (99%) levels40. The Na+ ions were important to some halophilic bacteria or alkaliphilic bacteria as they replaced protons and coupled ion to cope with the high external pH, rather than increasing the electric potential difference across the cytoplasmic membrane41. Mg2+ was a chaotropic agent and a limiting factor in the diversity of microbes in the hypersaline environment39. In addition, Cu2+, Ca2+, Mn2+ and Fe2+/3+ ions were important regulators of some extremozymes in Nocardiopsis genus, for example, xylanases, alpha amylases, thermoalklotolerant β-1,3-glucanases and cellulases42,43. Thus, it is reasonable to observe the significant influence of environmental factors on the biogeographic distribution of Nocardiopsis strains.
Environmental factors influenced some functional genes important for bacterial survival more significantly than 16S rRNA gene. For example, the phylogenies of gyrB, rpoB and sodA genes of the obtained Nocardiopsis strains showed more visible endemic clusters within one habitat or one region than the highly conserved 16S rRNA gene (Fig. 2 & Fig. S1, Table 1). This observation could be ascribed to the fact that some residues of the three functional housekeeping genes could be subjected to mutation due to cation binding (P > 90%, Table S6 and Table S7; reliability >70%), which led to the catalytic regulation functions of their corresponding enzymes44,45,46,47,48.
In summary, Nocardiopsis spp. in hypersaline environments possessed geographic distribution patterns. Spatial distance and environmental factors influenced the biogeography distribution of Nocardiopsis at large and local scales, respectively.
Material and Methods
Site description and sample collection
In this study, two salt mines (Heijing and Jiangcheng) from Yunnan Province and two salt lakes (Aydingkol and Qijiaojing) from Xinjiang Province of western China were selected (Table 1). Two sites each were sampled at the Heijing saline mine (HJ1, an abandoned salt mine; HJ2, a natural hypersaline spring) and the Jiangcheng salt mine (JC1, an abandoned salt mine; JC2 site, a natural hypersaline spring), respectively. Two (AK1 and AK2) and two sites (QJJ1 and QJJ2) were sampled at Aydingkol and Qijiaojing salt lakes, respectively. At each selected sampling site, sediments were sampled at the 10–30 cm depth and collected into sterile 50 ml sterile Falcon centrifuge tubes. GPS coordinates were recorded at each sampling point with a portable meter in the field and were subsequently imported into Map-Source according to the manufacturer’s instructions to measure the geographic distances among the sites. The samples for microbial cultivation and geochemistry measurement were stored at 4 °C in the field and during transportation.
Geochemistry measurements
The pH and salinity of the sampled sediments were measured with portable meters after sediments being dissolved into distilled water. The concentrations of major cations and trace elements in sediments from nine sampling sites were measured by flame atomic absorption spectrometry (HITACHI Z-2310). Total nitrogen of the sediment samples was determined by the semi-micro-Kjeldahl method49, and total phosphorus of the sediment samples was determined by the alkali fusion–Mo-Sb Anti-spectrophotometric method49. Principle component analysis (PCA) of the studied sediment samples was performed with the use of the R program50.
Isolation of Nocardiopsis strains
The sediment samples (2 g, wet weight) were dispersed into 18 ml sterilized physiological saline water (con. 0.70%, w/v, equal to bacterial cell physiological salinity) and were incubated at 30 °C for 30 min with shaking at 150 rpm. The resulting slurry was serially diluted with sterilized physiological saline water (NaCl con. 0.70%, w/v). Aliquots (0.2 ml) of each dilution were spread onto petri dishes containing three different media: cellulose-casein multi-salt medium and modified ISP 4 and ISP 5 media51. All the agar plates were supplemented with 5% (w/v) NaCl and potassium dichromate (15 mg/L)51. The petri dishes were incubated at 37 °C for 4–6 weeks. Based on the morphologic characteristics of Nocardiopsis spp. described previously27, colonies were picked and checked by light microscopy (BH-2; Olympus). Candidate strains were purified on inorganic salts-starch agar supplemented with 5% (w/v) NaCl27 and cultivated using the ISP4 medium (Difco Laboratories, Detroit, Mich) at 37 °C for four weeks29. Genomic DNA of the obtained strains was extracted and 16S rRNA genes were PCR amplified19. PCR amplification of gyrB, rpoB and sodA genes was performed according to the methods described previously19. The amplified PCR products were purified using a TaKaRa DNA fragment purification kit (Ver. 2.0) and were sequenced using an ABI 3100 automated sequencer with primers of four genes (16S rRNA, 27f and 1525r; gyrB, UP-1F and UP-2R; rpoB, MF and MR; sodA, Z205 and Z212)19 at Shanghai Sangon Biotech (Shanghai, China). The 16S rRNA gene sequences obtained from the candidate strains were compared with reference taxa via the EzTaxon-e database52. The sequences similarity levels were calculated between the candidate strains and their related Nocardiopsis taxa in the EzTaxon-e database52.
Phylogenetic analysis of isolated Nocardiopsis strains
Multiple alignments and genetic distance calculations were carried out by using CLUSTAL_X53 after retrieving the reference sequences of Nocardiopsis type strains from the EzTaxon-e database. The pair-wise similarities between Nocardiopsis strains were calculated by the software package MEGA 4.054. OTU classification was performed using DOTUR appliying a 98.5% 16S rRNA sequence similarity cut-off26. The 98.5% identity of 16S rRNA gene sequences corresponded to 70% of DNA-DNA relatedness, which was widely used as the cutoff value for species definition in prokaryotes26. Reference sequences were retrieved from NCBI (National Center for Biotechnology Informatics, http://www.ncbi.nlm.nih.gov) with BLAST (Basic Local Alignment Search Tool, http://blast.ncbi.nlm.nih.gov/Blast.cgi).
After designation of OTUs, phylogenies of the four investigated genes (16S rRNA, gyrB, rpoB and sodA) were constructed by using PhyML 3.055 with maximum-likelihood56. Bootstrap analysis was used to evaluate the stability of tree topology by resampling 1000 times57. Subsequently, a cluster within an OTU of the 16S rRNA gene phylogenetic trees was defined as a phylotype. Plus, a cluster within an OTU of the gyrB, rpoB and sodA gene phylogenies was nominated as one genotype.
In order to differentiate between OTUs, T-test was performed to analyze pair-wise divergences of genetic distances among different strains using the Vegan package of the R software version 3.0.2. In order to study the biogeographic pattern of Nocardiopsis, 16S rRNA, gyrB, rpoB and sodA gene sequences were assigned with allele numbers and multi-locus sequence types (STs) of concatenated sequences according to the multi-locus sequence typing (MLST) web site (www.mlst.net). Phylogenies of the concatenated sequences of four investigated genes were constructed with Bayesian inference55 by using the PhyML 1.8.3 software with maximum-likelihood method56 and Mr Bayes-3.1.255. Bootstrap analysis was used to evaluate the stability of tree topology by resampling 1000 times57.
Biostatistic and bioinformatic analyses on the biogeographic patterns of Nocardiopsis strains
In order to assess the impact of spatial distances on Nocardiopsis strains’ dispersal, the correlations between Nei’s unbiased genetic distances of the four genes (16S rRNA, gyrB, rpoB and sodA) and their corresponding geographic distances were analyzed using Mantel tests implemented in the NTSYS package58. Additionally, the relationship between differentiation in sediments geochemistry and variations of endemic genotypes of gyrB, rpoB and sodA among eight sampling sites were analyzed by simple Mantel test and Canonical Correlation Analysis (CCA) with the R program50. The maximum-likelihood method of Yang59, implemented in the codeml program from the PAML package, was applied to analyze the effects of environmental forces on adaptive evolution of Nocardiopsis strains59,60. Six models were used to detect positive environmental selection upon evolution of the Nocardiopsis strains. Each model allows for various dN/dS ratios ω among sites, including the simplest model (M0 or one-ratio model), the ‘nearly neutral’ model (Mla), the positive selection model (M2a), the discrete Model M3, Model M7 (β), and the optimum positively selective Model M8. In addition, the protein prediction server (https://www.predictprotein.org)61 was used to map the residues under positive environmental selection to molecular function ontology of three proteins (the B subunit of DNA gyrase, the β subunit of RNA polymerase and A subunit of superoxide dismutase).
Additional Information
How to cite this article: He, S.-T. et al. Biogeography of Nocardiopsis strains from hypersaline environments of Yunnan and Xinjiang Provinces, western China. Sci. Rep. 5, 13323; doi: 10.1038/srep13323 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 30870005 and 41422208). The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the research group no RGP-205. W.-J. Li was also supported by ‘Hundred Talents Program’ of the Chinese Academy of Sciences and Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
Footnotes
Author Contributions S.H., X.Z. and W.L. conceived and designed the experiments; S.H., X.Z., H.J. and W.L. analyzed the data. All of the authors assisted in writing the manuscript, discussed the results and commented on the manuscript.
References
- O’Malley M. A. The nineteenth century roots of ‘everything is everywhere’. Nat. Rev. Microbiol. 5, 647–651 (2007). [DOI] [PubMed] [Google Scholar]
- Leibold M. A. et al. The Meta community concept: a framework for multi-scale community ecology. Ecol. Lett. 7, 601–613 (2004). [Google Scholar]
- Baas-Becking L. G. M. Geobiologie of Inleiding to de Milieukunde. Van Stockum & Zoon, The Hague (1934).
- Casamayor E. O. et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 73, 338–348 (2002). [DOI] [PubMed] [Google Scholar]
- Campbell L., Nolla H. A. & Vaulot D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limonol. Oceanog 39, 955–961 (1994). [Google Scholar]
- Martiny J. B. H. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbol 4, 102–112 (2006). [DOI] [PubMed] [Google Scholar]
- Margos G. et al. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 105, 8730–8735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. R., Goericke R. & Chisholm S. W. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Eco. Prog. Ser. 116, 259–275 (1995). [Google Scholar]
- Pagaling E. et al. Microbial Biogeography of Six Salt Lakes in Inner Mongolia, China, and a Salt Lake in Argentina. Appl. Environ. Microbiol. 75, 5750–5760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M. et al. Genetic diversity and biogeography of haloalkaliphilic sulphur-oxidizing bacteria belonging to the genus Thioalkalivibrio. FEMS Microbiol. Ecol. 56, 95–101 (2006). [DOI] [PubMed] [Google Scholar]
- Tian S. Z. et al. Isolation and characterization of new p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agric. Food. Chem. 61, 3006–3012 (2013). [DOI] [PubMed] [Google Scholar]
- Zitouni A. et al. Nocardiopsis and Saccharothrix genera in Saharan soils in Algeria: isolation, biological activities and partial characterization of antibiotics. Res. Microbiol. 156, 984–993 (2005). [DOI] [PubMed] [Google Scholar]
- Engelhardt K. et al. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 76, 4969–4976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola J. S. et al. Isolation of toxigenic Nocardiopsis strains from indoor environments and description of two new Nocardiopsis species, N. exhalans sp. nov. and N. umidischolae sp. nov. Appl. Environ. Microbiol 67, 4293–4304 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tai A. M. & Ruan J.-S. Nocardiopsis halophila sp. Nov., a new halophilic actinomycete isolated from soil. Int. J. Sys. Bacteriol 3, 474–478 (1994). [Google Scholar]
- Yassin A. F. et al. Description of Nocardiopsis synnemataformans sp. nov., elevation of Nocardiopsis alba subsp. prasina to Nocardiopsis prasina comb. nov., and designation of Nocardiopsis antarctica and Nocardiopsis alborubida as later subjective synonyms of Nocardiopsis dassonvillei. Int. J. Syst. Bacteriol. 47, 983–988 (1997). [DOI] [PubMed] [Google Scholar]
- Huang W. M. Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 30, 79–107 (1996). [DOI] [PubMed] [Google Scholar]
- Kim K. S. et al. Use of rpoB sequences for phylogenetic study of Mycoplasma species. FEMS Microbiol. Lett. 226, 299–305 (2003). [DOI] [PubMed] [Google Scholar]
- Yang L.-L., Zhi X.-Y. & Li W.-J. Phylogenetic analysis of Nocardiopsis species based on gyrB, rpoB and sodA gene sequences. Acta. Microbiologica. Sinica 47, 951–955 (2007). [PubMed] [Google Scholar]
- Lee S. H. et al. Differentiation of Borrelia burgdorferi sensu lato on the basis of RNA polymerase gene (rpoB) sequences. J. Clin. Microbiol. 38, 2557–2562 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K. S. et al. Application of RNA polymerase beta-subunit gene (rpoB) sequences for the molecular differentiation of Legionella species. J. Clin. Microbiol. 40, 2653–2658 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. J., Lee S. H. & Lyu M. A. Identification of Mycobacterial Species by Comparative Sequence Analysis of the RNA Polymerase Gene (rpoB). J. Clin. Microbiol. 37, 1714–1720 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K. S. et al. RNA polymerase beta-subunit gene (rpoB) sequence analysis for the identification of Bacteroides spp. Clin. Microbiol. Infect. 13, 48–54 (2007). [DOI] [PubMed] [Google Scholar]
- Wu J.-Y. et al. Diversity of Actinobacterial community in saline sediments from Yunnan and Xinjiang, China. Extremophiles 13, 623–632 (2009). [DOI] [PubMed] [Google Scholar]
- Yu G., Harrison S. P. & Xue B. Lake status records from China: Data Base Documentation. Max-Planck-Institute for Biogeochemistry Report, 4, 100–103 (2001). [Google Scholar]
- Stackebrandt E. & Goebel B. M. Taxonomic Note: A Place for DNA-DNA Reassociation and16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849 (1994). [Google Scholar]
- Evtushenko L. et al. Nocardiopsis tropica sp. nov., Nocardiopsis trehalosi sp. nov., nom. rev. and Nocardiopsis dassonvillei subsp. albirubida subsp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 50, 73–81 (2000). [DOI] [PubMed] [Google Scholar]
- Sabry S. A. et al. Nocardiopsis aegyptia sp. Nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 54, 453–456 (2004). [DOI] [PubMed] [Google Scholar]
- Li M.-G. et al. Nocardiopsis xinjiangensis sp. Nov., a halophilic actinomycete isolated from a saline soil sample in China. Int. J. Syst. Evol. Microbiol. 53, 317–321 (2003). [DOI] [PubMed] [Google Scholar]
- Chen Y.-G. et al. Nocardiopsis quinghaiensis sp. Nov., isolated from saline soil in China. Int. J. Syst. Evol. Microbiol. 58, 699–705 (2008). [DOI] [PubMed] [Google Scholar]
- Chen Y.-G. et al. Nocardiopsis terrae sp. nov., a halophilic actinomycete isolated from saline soil. Antonie. Van. Leeuwenhoek 98, 31–38 (2010). [DOI] [PubMed] [Google Scholar]
- Perez-Jimenez J. R. & Kerkhof L. J. Phylogeography of sulfatereducing bacteria among disturbed sediments, disclosed by analysis of the dissimilatory sulfite reductase genes (dsrAB). Appl. Environ. Microbiol. 71, 1004–1011 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent S. J., Gucker C. L., Oda Y. & Forney L. J. Spatial distribution of Rhodopseudomonas palustris ecotypes on a local scale. Appl. Environ. Microbiol. 69, 5192–5197 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R., Bienhold C., Ramette A. & Harder J. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME. J 4, 159–170 (2010). [DOI] [PubMed] [Google Scholar]
- Papke R. T., Niels B., Ramsing N. B., Bateson M. M. & Ward D. M. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5, 650–659 (2003). [DOI] [PubMed] [Google Scholar]
- Escobar-Páramo P., Ghosh S. & DiRuggiero J. Evidence for genetic drift in the diversification of a geographically isolated population of the hyperthermophilic archaen Pyrococcus. Mol. Biol. Evol. 22, 2297–2303 (2005). [DOI] [PubMed] [Google Scholar]
- Cho J. C. & Tiedje J. M. Biogeography and Degree of Endemicity of Fluorescent Pseudomonas Strains in Soil. Appl. Environ. Microbiol. 66, 5448–5456 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y. et al. Differences in soil bacterial diversity: driven by contemporary disturbances or historical contingencies? ISME. J 2, 254–264 (2008). [DOI] [PubMed] [Google Scholar]
- Rosselló-Mora R. et al. Metabolic evidence for biogeographic isolation of the extremophilic bacterium Salinibacter ruber. ISME. J 2, 242–253 (2008). [DOI] [PubMed] [Google Scholar]
- Skulachev V. P. et al. Bacterial energetics at high pH: what happens to the H cycle when the extracellular concentration decreases? Novar. Found. Symp. 221, 200–217 (1999). [DOI] [PubMed] [Google Scholar]
- Hallsworth J. E. et al. Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ. Microbiol. 9, 801–813 (2007). [DOI] [PubMed] [Google Scholar]
- Stamford T. L., Stamford N. P., Coelho L. C. & Araújo J. M. Production and characterization of a thermo-stable alpha-amylase from Nocardiopsis sp. endophyte of yam bean. Bioresour. Technol. 76, 137–141 (2001). [DOI] [PubMed] [Google Scholar]
- Saratale G. D. & Oh S. E. Production of thermotolerant and alkalotolerant cellulolytic enzymes by isolated Nocardiopsis sp. KNU. Biodegradation. 22, 905–919 (2011). [DOI] [PubMed] [Google Scholar]
- Ofran Y. & Rost B. Interaction sites identified from sequence. Bioinformatics 23, 13–16 (2007). [DOI] [PubMed] [Google Scholar]
- Hamp T. et al. Homology-based inference sets the bar high for protein function prediction. BMC. Microbiol. 14, S3–S7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrat F. et al. Reduction of Fe(III) ions complexed to physiological ligands by lipoyl dehydrogenase and other flavoenzymes in vitro: implications for an enzymatic reduction of Fe (III) ions of the labile iron pool. J. Biol. Chem. 278, 46403–46413 (2003). [DOI] [PubMed] [Google Scholar]
- Bruno-Bárcena J. M., Andrus J. M., Libby S. L., Klaenhammer T. R. & Hassan H. M. Expression of a Heterologous Manganese Superoxide Dismutase Gene in Intestinal Lactobacilli Provides Protection against Hydrogen Peroxide Toxicity. Appl. Environ. Microbiol. 70, 4702–4710 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrusa J. M., Bowen S. W., Klaenhammer T. R. & Hassan H. M. Molecular characterization and functional analysis of the manganese-containing superoxide dismutase gene (sodA) from Streptococcus thermophilus AO54. Arch. Biochem. Biophys 420, 103–113 (2003). [DOI] [PubMed] [Google Scholar]
- Yakimov M. et al. Microbial community of a saline mud volcano at San Biagio-Belpasso, Mt. Etna (Italy). Environ. Microbiol. 4, 249–256 (2002). [DOI] [PubMed] [Google Scholar]
- Venables W. N. & Ripley B. D. Modern Applied Statistics with S. Springer-Verlag, New York (2002). [Google Scholar]
- Tang S.-K. et al. Haloactinospora alba gen. nov., sp. Nov., a halophilic filamentous actinomycete of the family Nocardiopsaceae. Int. J. Syst. Evol. Microbiol 58, 2075–2080 (2008). [DOI] [PubMed] [Google Scholar]
- Kim O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S RRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids. Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
- David B. J. & Anderson W. F. A Fast Algorithm for Rendering Space-Filling Molecule Pictures. J. Mol. Graph. 6, 219–220 (1988). [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Condence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- Fitch W. M. Towards defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971). [Google Scholar]
- Yang Z. PAML a program package for phylogenetic analysis by maximum likelihood. Comput. Appli. Biosci 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- Yang Z., Wong W. S. & Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118 (2005). [DOI] [PubMed] [Google Scholar]
- Rost B. Protein secondary structure prediction continues to rise. J. Struct. Biol. 134, 204–218 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.