Abstract
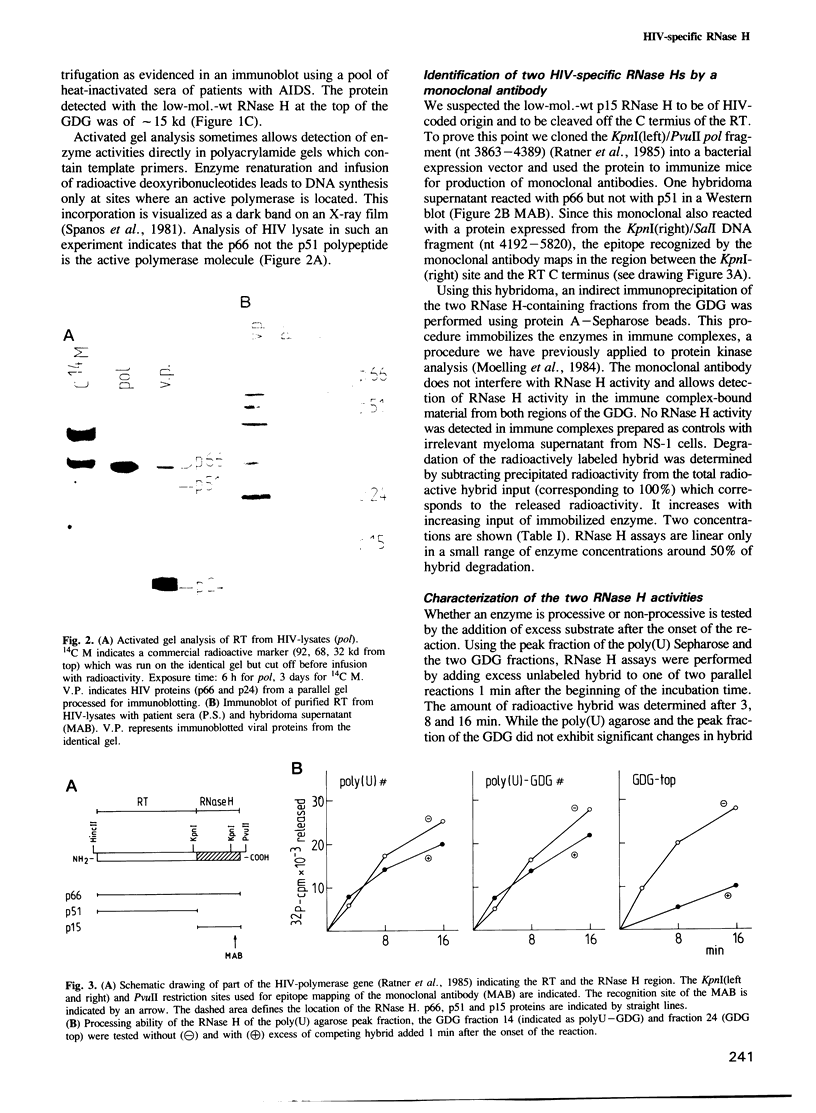
Human immune deficiency virus (HIV) replicates by conversion of the RNA genome into the double-stranded DNA provirus. The reverse transcriptase is not the only enzymatic function crucial in DNA-provirus synthesis. A viral-coded RNase H activity which specifically degrades RNA in RNA-DNA hybrids has been shown to be essential as well. Here we demonstrate that the HIV-reverse transcriptase which consists of a two-polypeptide complex, p66 and p51, copurifies with an RNase H activity which exhibits properties of a processive exonuclease. Only the p66 molecule, not p51, is active as polymerase as evidenced by activated gel analysis. p66 exhibits RNase H activity when precipitated as immune complex by a monoclonal antibody raised against a bacterially expressed carboxy-terminal portion of p66. The monoclonal antibody which does not interfere with enzyme activity also precipitates a second population of molecules with RNase H activity which is of low mol. wt, p15. This RNase H appears therefore to be derived from the carboxy terminus of p66 during processing to the p51 polypeptide. It exhibits low template-binding ability and is of a non-processing mode of action which may be due to the absence of the reverse transcriptase domain. These results lend experimental support to the hypothesis that the RNase H gene maps at the carboxy terminus of the reverse transcriptase. Since both RNase H populations are virus-coded they may be essential for retrovirus replication in general and useful targets for chemotherapeutic agents.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Cathala G., Rech J., Huet J., Jeanteur P. Isolation and characterization of two types of ribonucleases H in Krebs II ascites cells. J Biol Chem. 1979 Aug 10;254(15):7353–7361. [PubMed] [Google Scholar]
- Feinberg M. B., Jarrett R. F., Aldovini A., Gallo R. C., Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986 Sep 12;46(6):807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- Friedrich R., Moelling K. Effect of viral RNase H on the avian sarcoma viral genome during early transcription in vitro. J Virol. 1979 Sep;31(3):630–638. doi: 10.1128/jvi.31.3.630-638.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard G. F. Multiple RNase H activities in mammalian type C retravirus lysates. J Virol. 1978 Apr;26(1):16–28. doi: 10.1128/jvi.26.1.16-28.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D., Quinn T., Hippenmeyer P. J., Oroszlan S. Structural characterization of the avian retrovirus reverse transcriptase and endonuclease domains. J Biol Chem. 1985 Jul 15;260(14):8243–8249. [PubMed] [Google Scholar]
- Hansen J., Schulze T., Moelling K. RNase H activity associated with bacterially expressed reverse transcriptase of human T-cell lymphotropic virus III/lymphadenopathy-associated virus. J Biol Chem. 1987 Sep 15;262(26):12393–12396. [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Schaber M. D., Skalka A. M., Ganguly K., Wong-Staal F., Reddy E. P. HTLV-III gag protein is processed in yeast cells by the virus pol-protease. Science. 1986 Mar 28;231(4745):1580–1584. doi: 10.1126/science.2420008. [DOI] [PubMed] [Google Scholar]
- Lai M. H., Verma I. M. Reverse transcriptase of RNA tumor viruses. V. In vitro proteolysis of reverse transcriptase from avian myeloblastosis virus and isolation of a polypeptide manifesting only RNase H activity. J Virol. 1978 Feb;25(2):652–663. doi: 10.1128/jvi.25.2.652-663.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoote M. M., Coligan J. E., Folks T. M., Fauci A. S., Martin M. A., Venkatesan S. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J Virol. 1986 Nov;60(2):771–775. doi: 10.1128/jvi.60.2.771-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelling K. Characterization of reverse transcriptase and RNase H from friend-murine leukemia virus. Virology. 1974 Nov;62(1):46–59. doi: 10.1016/0042-6822(74)90302-x. [DOI] [PubMed] [Google Scholar]
- Moelling K. Further characterization of the Friend murine leukemia virus reverse transcriptase-RNase H complex. J Virol. 1976 May;18(2):418–425. doi: 10.1128/jvi.18.2.418-425.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelling K., Heimann B., Beimling P., Rapp U. R., Sander T. Serine- and threonine-specific protein kinase activities of purified gag-mil and gag-raf proteins. Nature. 1984 Dec 6;312(5994):558–561. doi: 10.1038/312558a0. [DOI] [PubMed] [Google Scholar]
- Mölling K., Bolognesi D. P., Bauer H., Büsen W., Plassmann H. W., Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971 Dec 22;234(51):240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Roewekamp W., Sekeris C. E. Purification and characteristics of hybridase (ribonuclease H) from rat-liver cytosol. Eur J Biochem. 1974 Apr 1;43(2):405–413. doi: 10.1111/j.1432-1033.1974.tb03426.x. [DOI] [PubMed] [Google Scholar]
- Sarngadharan M. G., Leis J. P., Gallo R. C. Isolation and characterization of a ribonuclease from human leukemic blood cells specific for ribonucleic acid of ribonucleic acid-deoxyribonucleic acid hybrid molecules. J Biol Chem. 1975 Jan 25;250(2):365–373. [PubMed] [Google Scholar]
- Sodroski J., Patarca R., Rosen C., Wong-Staal F., Haseltine W. Location of the trans-activating region on the genome of human T-cell lymphotropic virus type III. Science. 1985 Jul 5;229(4708):74–77. doi: 10.1126/science.2990041. [DOI] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]