Abstract
Induction of phosphorylated extracellular-regulated kinase (pERK) is a reliable molecular readout of learning-dependent neuronal activation. Here, we describe a pERK immunohistochemistry protocol to study the profile of hippocampal neuron activation following exposure to a spatial learning task in a mouse model characterized by cognitive deficits of neurodevelopmental origin. Specifically, we used pERK immunostaining to study neuronal activation following Morris water maze (MWM, a classical hippocampal-dependent learning task) in Engrailed-2 knockout (En2-/-) mice, a model of autism spectrum disorders (ASD). As compared to wild-type (WT) controls, En2-/- mice showed significant spatial learning deficits in the MWM. After MWM, significant differences in the number of pERK-positive neurons were detected in specific hippocampal subfields of En2-/- mice, as compared to WT animals. Thus, our protocol can robustly detect differences in pERK-positive neurons associated to hippocampal-dependent learning impairment in a mouse model of ASD. More generally, our protocol can be applied to investigate the profile of hippocampal neuron activation in both genetic or pharmacological mouse models characterized by cognitive deficits.
Keywords: Neuroscience, Issue 99, Immunohistochemistry, Morris water maze, ERK, hippocampus, cognition, memory, autism
Introduction
Neurodevelopmental disorders include a wide and heterogeneous group of disorders such as Down syndrome, fragile X syndrome (FXS), Rett syndrome, neurofibromatosis, tuberous sclerosis and ASD, in which the development and maturation of the central nervous system (CNS) is disturbed early during the prenatal period1. These developmental brain dysfunctions can cause profound, lifelong effects on motor function, language, learning and memory process. A plethora of genetic and environmental factors have been implicated in the pathogenesis of neurodevelopmental disorders during the last few years2,3. Even if the molecular mechanisms underlying the clinical phenotype remain unknown, the above mentioned findings have allowed the development of several mouse models of these disorders. Learning and memory deficits have been identified in a number of these mouse models such as Tsc1+/-, Tsc2+/-, Nf1+/- and En2-/- mice2,4-7. An important challenge in the field of neurodevelopmental disorders is the identification of cellular and molecular processes underlying memory and learning dysfunction. Selected signaling pathways activated during learning or memory can induce the transcription of specific genes and ultimately lead to de novo protein synthesis. Immediate-early genes (IEGs) activation and protein-dependent synaptic modifications are rapidly induced in brain neurons in response to neuronal activity and behavioral training8,9.
Deficits in signaling pathways involving neurofibromin have been associated with impaired learning in neurodevelopmental disorders. Neurofibromin is the product of the NF1 gene, whose mutation causes neurofibromatosis type 1, a complex genetic syndrome characterized by nervous system tumors, behavioral and motor delays, and cognitive disabilities10. Mice heterozygous for Nf1 deletion restricted to inhibitory neurons show deficits in the early phase of long-term potentiation (LTP), as well as compromised spatial learning in MWM5,11,12. Interestingly, Nf1 deficiency in this mouse model leads to an over-activation of Ras signaling in inhibitory interneurons during learning, resulting in increased ERK phosphorylation and finally in an abnormal enhancement of GABA release from these neurons5.
Based on these findings, the visualization of neuronal activity after behavioral tasks represents a way to reconstruct specific circuits involved in neurodevelopmental diseases. The immunohistochemistry protocol described here aims to assess and quantify hippocampal ERK phosphorylation levels following MWM in an ASD mouse model with cognitive deficits. MWM is widely used to investigate hippocampal dependent spatial learning and memory in rodents13,14. We decide to use ERK phosphorylation as molecular readout of task-dependent hippocampal learning, since ERK was shown to have an essential role in learning and memory formation15. Moreover, the ERK pathway is necessary for experience-dependent plasticity in the developing visual cortex16. Finally, mice lacking one of the two ERK isoforms (ERK2) in the CNS show marked anomalies in cognitive, emotional and social behaviors17, indicating that ERK signaling might play a critical role in the pathogenesis of neurodevelopmental disorders such as ASD.
We used Engrailed 2 knockout (En2-/-) mice as a model of neurodevelopmental disorders. En2-/- mice show anatomical and behavioral “ASD-like” features, including loss of forebrain interneurons18, reduced expression of ASD-related genes19, decreased sociability, and impaired cognitive flexibility6,7,20. Spatial learning and memory defects, such as those detected in MWM, are especially robust in En2-/- mice6,7 and might be relevant to the cognitive impairments observed in ASD patients21. Furthermore, we showed that impaired spatial learning in MWM is associated with reduced neurofibromin expression and increased pERK levels in the hilus of En2-/-adult mice7. Here we present the detailed protocol for the immunohistochemical characterization of pERK following MWM in this ASD mouse model.
Protocol
All experiments were conducted in conformity with the European Community Directive 2010/63/EU and were approved by the Italian Ministry of Health.
1. Animal Care, Housing and Treatment
Perform all experimental protocols using mice in accordance with the guidelines of the respective institutional animal care.
Maintain animals in a 12 hr light/dark cycle with food and water available ad libitum.
House the mice in groups of 2-4, in standard-sized polycarbonate mouse cages with sawdust and bedding material changed weekly.
Use age and sex-matched mice, because susceptibility to disease can vary with age and gender in different mouse models of neurodevelopmental disorders.
Choose an appropriate number of animals to reach statistical significance (a minimum of 10 mice per genotype for the behavioral study and 4 mice per genotype for the immunohistochemistry experiment).
2. Morris Water Maze (MWM)
- Use a Circular Tank (diameter 100 cm, height 50 cm).
- Place some visual cues on the room dividers around the tank.
- Place a 10 cm diameter platform into the tank at a fixed location in the center of one of four imaginary quadrants. Keep the hidden platform in the same quadrant for all trials across all training sessions.
- Fill the pool with tap water until the platform is 1 cm above the water surface. Equilibrate water temperature should be close to 22 °C; this step may take several hours. Add hot water to shorten the time required to equilibrate water temperature.
- Add approximately 1.5-2 L of white, non-toxic paint to make the water opaque.
- Track Acquisition Procedure
- Set up MWM’s tracking system with standard CCD cameras and video boards. Choose a video-tracking system to monitor and analyze several variables such as path length, escape latency, and time spent in each quadrant.
- Divide the arena in four quadrants and adjust detection settings to ensure optimal contrast between animals and background for live tracking.
- Specify the platform region.
- Set the maximum trial time as 60 sec.
- MWM Testing
- Prior to behavior testing, allow mice an adaptation period of 20-25 min in an area where they cannot see the pool or spatial cues.
- Train each mouse for 9 days (two consecutive trials a day, with 20-30 min interval between them).
- Switch on the computer and the camera and start the video-tracking system software.
- Take the mouse by its tail and place it into water, with its head pointed towards the side of pool, starting from one of the four start positions (north, south, east, west). For each mouse, the start position should be different and pseudorandomized across trials.
- Let the mouse swim and search for the platform for a maximum of 60 sec. Once the animal reaches the platform, allow it to stay on the platform for 20 sec.
- If mouse failed to locate the platform within that time, gently guide the mouse to the platform, holding it by its tail and let it to stay there for 20 sec. Once the animal has completed all two trials, dry it off with a towel and put it back into cage.
- Each day of the training period, repeat the same procedure (steps 2.3.4-2.3.6).
- Perform the probe test on day 9 after the last training trail.
- Remove the platform from the pool.
- Let the mouse swim and search for the platform for a maximum of 60 sec.
- Periodically, check the water temperature so that it should be the same (22 °C) and clean out the pool.
3. Preparation of Sections from Perfused Animals
Sacrifice the mice at the end of MWM. Four mice per genotype must be anesthetized prior to euthanization according to local and international guidelines.
Perfuse the animal transcardially (cut the right auriclewith a scissor to allow bleeding and introduce a butterfly cannula into the left ventricle to perfuse) with phosphate buffered saline (PBS) for 4-5 min, followed by 4% paraformaldehyde in PBS for 10-15 min22.
Dissect and post-fix the brains in 4% paraformaldehyde at 4 °C for at least 12 hr.
Place the brains in 50 ml polystyrene conical tubes containing 15 ml of 0.02% Sodium Azide in 0.1 M PBS and store at 4 °C until required for slicing.
Cut and remove the cerebellum before vibratome cutting.
Glue the brain upright with the cutting site, using superglue, onto the holder plate of the vibratome.
Cut the brain into 20-40 µm thick coronal sections, in serial order throughout the dorsal hippocampus, on appropriate vibratome.
Collect vibratome slices with a paint brush and transfer to a 24 multi-well plate containing 1 ml of 0.1 M PBS for each well.
Store sections for short term in PBS or long term in 0.02% Sodium Azide in PBS at 4 ºC (tissue might be used for up to one year).
4. Immunohistochemistry
Transfer 3-5 dorsal hippocampal sections for each mouse, into 24 multi-well plate containing 500 µl of 0.1 M PBS for each well.
Wash the sections contained in each well with 500 µl of 0.1 M PBS (5 min, 3 times with gentle shaking).
Quench endogenous peroxidase activity, by incubating sections in 1% H2O2 in PBS for 20 min with gentle agitation.
Wash sections in 0.1 M PBS with gentle agitation (3 x 5 min).
Permeabilization of tissue: incubate sections for 5 min in 0.2% Triton X-100 in PBS.
During the above steps (4.4; 4.5), make enough blocking buffer (10% Goat Serum and 0.1% Triton-X100 in PBS) for the entire experiment (number of sections x 100 µl x 3 steps).
Note: Use serum from the same species in which the secondary Ab (conjugated to biotin) was made.
Prevent non-specific binding of the antibody, by incubating sections in blocking buffer (100 µl/section) for 1 hr at room temperature (RT) with gentle agitation.
Incubate sections in primary Ab (pERK) diluted in blocking buffer, overnight at 4 °C, with gentle agitation.
Wash sections in 0.1 M PBS with gentle agitation (3 x 5 min).
Incubate with the secondary Ab conjugated with biotin (1:250) diluted in blocking buffer (same as used in step 4.6) for 1 hr at RT with gentle agitation.
Prepare ABC reagent at the same time, because Avidin and biotin require at least 30 min at room temperature to complex.
Prepare the required volume of ABC reagent according to manufacturers’ protocol.
Wash sections in 0.1 M PBS with gentle agitation (3 x 5 min).
Incubate sections for 45 min at RT with ABC Reagent.
Wash sections in 0.1 M PBS with gentle agitation (3 x 5 min).
Prepare DAB working solution, every step involving DAB must be done in the fume hood as DAB is carcinogenic and teratogenic.
Transfer DAB solution to a new plate by plastic transfer pipette.
Place sections in the plate containing DAB working solution and slowly swirl plate by hand, to allow maximum exposure to sections.
Monitor DAB reaction on microscope until adequate signal develops (2-5 min).
Stop the reaction at the desired color intensity by washing the tissue in 0.1 M PBS (3 x 5 min).
Use bleach to deactivate any remaining DAB substrate solution in the fume hood overnight and dispose of it according to laboratory guidelines.
Mount sections on gelatin coated slides by floating in PBS. Then dry them at room temperature at least 3-4 hr.
Dehydrate the sections sequentially in 50%, 70%, 95% and 100% ethanol for 2 min each, and clear in 100% xylene for 5 min.
Coverslip using mounting medium and leave in fume hood to dry overnight.
5. Cell Count
Count pERK-positive cells, three to five sections at the level of the dorsal hippocampus should be analyzed per animal.
Acquire multiple bright-field images from each section at 200X magnification using an appropriate microscope, and then assemble them using an image processing software.
Perform cell counts on tiff-converted mosaic images using the ImageJ free software.
Count pERK-stained cells in the hilus and granule cell layer over two to three counting boxes of 100 × 100 µm each.
Cell densities will be expressed as the number of labeled cells per counting window (100 × 100 µm).
Perform statistical analysis to assess significant difference between genotypes. This can be accomplished with Student's t test or ANOVA followed by appropriate post hoc test using commercial softwares. Set statistical significance level at p <0.05.
Representative Results
The protocol described here was designed to visualize, by immunohistochemistry, the expression of a specific marker of hippocampal neuron activity after MWM in a mouse model of neurodevelopmental disorders. All the experimental data shown herein were taken from our recent work7. Male and female WT and En2-/- age-matched adult littermates (3-5 months old; weight = 25-35 g) obtained from heterozygous matings were used. Twenty-two mice (11 per genotype) were used for the MWM. Eight mice (4 per genotype) were sacrificed at the end of the MWM probe trial and used for the immunohistochemistry experiments.
Assessment of spatial learning deficits in En2-/- mice
To specifically examine the presence of spatial learning deficits in En2-/- mice, we used the MWM test, a classical hippocampal-dependent spatial learning task13,14. On day 1 of training trials, no differences in latency and speed swim have been detected between En2-/- and WT mice, suggesting that both genotypes show similar visual and motor performance. Even if all experimental groups demonstrated the ability to learn during the trails, En2-/- mice reached the goal less efficiently than WT mice starting from the 3rd training day7. On probe trial (day 9), when the hidden platform was removed from the pool, WT mice (Figure 1A) showed a directed path-navigation to the expected target location (dotted black box), while En2-/- mice (Figure 1B) swam more irregularly, displaying a wandering path to the target quadrant (dotted black box). Moreover, during the probe trial WT mice showed closer proximity to platform, as compared to En2-/- mutants (Figure 1C) (one-way ANOVA followed by Holm-Sidak post-hoc test, *p = 0.027, **p = 0.007). The proximity score is considered more sensitive for detecting group differences than traditional measures (time spent and number of crossings in the target quadrant23).
Deregulation of hippocampal ERK phosphorylation in En2-/- mice after MWM task
To examine ERK phosphorylation, we then performed an immunohistochemical staining in hippocampal subfield of WT and En2-/- mice after MWM (Figure 2A). The number of pERK-positive CA3 neurons was significantly lower in En2-/- mice, compared with WT (Student’s t test, p = 0,0108; Figure 2B-C). Conversely, a significantly higher number of pERK-positive neurons was present in hilus of En2-/- mice, as compared with WT (Student’s t test, p = 0,0012; Figure 2B, D). In addition, a significant higher number of pERK-positive neurons was also detected in the granule cell layer (GCL) of the dentate gyrus of En2-/- mice, as compared with WT controls (Student’s t test, p = 0,0021; Figure 2B, E).
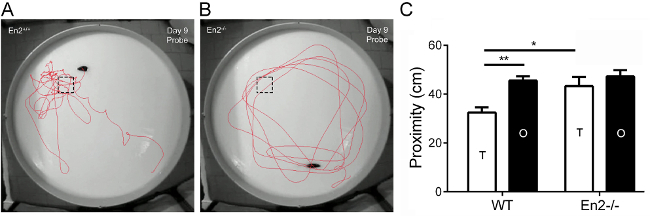 Figure 1: En2-/- mice show impaired spatial learning in the Morris water maze. (A-B) Pictures show WT the path-navigation to the expected platform location (dotted black box), in WT (A) and En2-/- (B) mice during probe trial. (C) Proximity to platform in target and opposite quadrants during probe trial. Abbreviations and symbols: T, target quadrant; OP, quadrant opposite to target; *p <0.05, **p <0.01 (one-way ANOVA followed by Holm-Sidak post-hoc test; n = 11 per genotype). Modified from ref7. Please click here to view a larger version of this figure.
Figure 1: En2-/- mice show impaired spatial learning in the Morris water maze. (A-B) Pictures show WT the path-navigation to the expected platform location (dotted black box), in WT (A) and En2-/- (B) mice during probe trial. (C) Proximity to platform in target and opposite quadrants during probe trial. Abbreviations and symbols: T, target quadrant; OP, quadrant opposite to target; *p <0.05, **p <0.01 (one-way ANOVA followed by Holm-Sidak post-hoc test; n = 11 per genotype). Modified from ref7. Please click here to view a larger version of this figure.
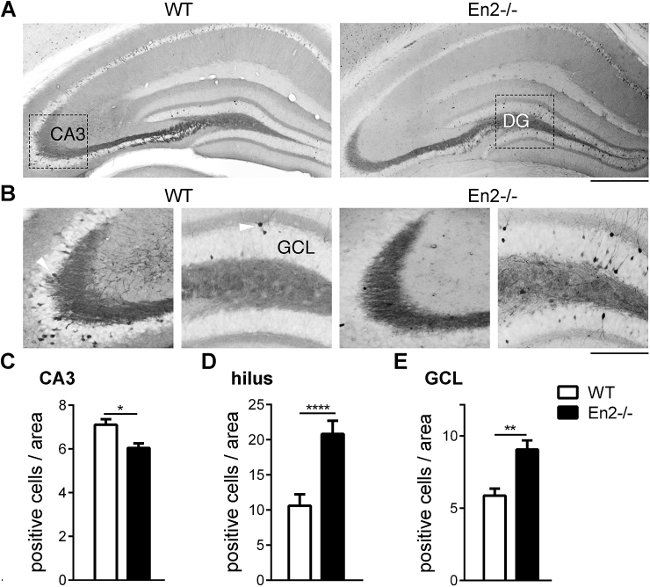 Figure 2: pERK staining in the hippocampus of MWM-trained WT and En2-/- mice. (A) Representative pERK immunostaings on whole dorsal hippocampi. (B) Details of pERK immunostaining in the CA3 and dentate gyrus the same mice as in (A). White arrowheads indicate examples of pERK-positive neurons. Genotypes are as indicated. Abbreviations: CA3, CA3 pyramidal layer; GCL, granule cell layer. Scale bars: 300 µm (A), 150 µm (B). (C-E) Counts of pERK-positive neurons in the CA3 (C), hilus (D) and GCL (E) of MWM-trained WT and En2-/- mice. *p <0.05, **p <0.01, ****p <0.0001 (Student’s t-test; n = 4 per genotype and treatment group). Modified from ref7.
Figure 2: pERK staining in the hippocampus of MWM-trained WT and En2-/- mice. (A) Representative pERK immunostaings on whole dorsal hippocampi. (B) Details of pERK immunostaining in the CA3 and dentate gyrus the same mice as in (A). White arrowheads indicate examples of pERK-positive neurons. Genotypes are as indicated. Abbreviations: CA3, CA3 pyramidal layer; GCL, granule cell layer. Scale bars: 300 µm (A), 150 µm (B). (C-E) Counts of pERK-positive neurons in the CA3 (C), hilus (D) and GCL (E) of MWM-trained WT and En2-/- mice. *p <0.05, **p <0.01, ****p <0.0001 (Student’s t-test; n = 4 per genotype and treatment group). Modified from ref7.
Discussion
Here, we provide a pERK immunohistochemistry protocol for revealing neuronal activation following MWM in En2-/- mice, a mouse model of neurodevelopmental disorders. Decreased levels of pERK were detected in the CA3 subfield of En2-/- mutants compared to WT. Differently from what observed in CA3 subfield, a general increase of pERK-positive neurons was instead detected in both hilus and GCL of En2-/- mice compared to WT. A possible explanation of this opposite trend in ERK phosphorylation levels might rely on subfield-specific mechanisms of pERK induction following MWM.
The protocol contains several critical steps, such as the evaluation of MWM data, brain fixation, choice and calibration of the antibody, washing procedures, signal detection, and cell counting procedure. All these critical steps should be carefully performed to ensure high quality and reproducible results. For example, if the brain is not fixed properly (steps 3.2 and 3.3), the sections will easily tear during the staining procedures. Moreover, sections should not dry out at any point during the washing steps in order to avoid low signal in staining. Lastly, if a pERK antibody different than that reported here (see Materials List) is used, it is important to calibrate both the antibody concentration and incubation time in order to identify the most robust and appropriate conditions. Furthermore, to ensure an accurate and unbiased outcome for the immunohistochemistry experiments, it is critical that both MWM test and cell counting will be done by operators blind to the animal's genotype or pharmacological treatment. pERK positive neurons can be also detected by immunofluorescence staining using secondary antibody conjugated to an appropriate fluorophore. In this regard, it is necessary to omit the quenching of endogenous peroxidases (step 4.3) and use an aqueous mounting media to preserve the fluorescent dyes (step 4.24).
So far, expression of immediate early gene (IEG,) has been extensively used as a molecular signature of task-dependent hippocampal learning24. This has been essentially addressed by c-fos mRNA in situ hybridization or immunohistochemistry. Conversely, few immunohistochemistry studies systematically addressed neuronal activity by addressing ERK phosphorylation in mouse models of neurodevelopmental disorders. Our recent study7 and previous work5 demonstrated as pERK immunostaining is a reliable marker to trace hippocampal neuronal circuits involved in learning and memory. One limitation of this technique is represented by the several critical steps that might increase the variability of the results. Nevertheless, the experimental approach presented here provides researchers with a concise, easy-to-follow outline of how to profile of hippocampal neuron activation in both genetic or pharmacological mouse models characterized by cognitive deficits. More generally, this protocol can also be used to investigate neuronal activity in cognitive behavioral tasks different than MWM, and to map neuronal activation to other brain regions potentially involved in cognitive functions.
Disclosures
The authors have nothing to disclose
Acknowledgments
We wish to thank the administrative staff of CIBIO (University of Trento) and CNR Neuroscience Institute for assistance. Giovanni Provenzano is supported by a post-doctoral fellowship from Fondazione Veronesi (Milan, Italy). This work was funded by the Italian Ministry of University and Research (PRIN 2008 grant # 200894SYW2_002 and PRIN 2010-2011 grant # 2010N8PBAA_002 to Y.B.), University of Trento (CIBIO start-up grant to Y.B.) and Telethon Foundation (grant # GGP13034 to Y.B.).
References
- Castren E, Elgersma Y, Maffei L, Hagerman R. Treatment of neurodevelopmental disorders in adulthood. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:14074–14079. doi: 10.1523/JNEUROSCI.3287-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, et al. Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nature medicine. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/- mice in the absence of cerebral lesions and seizures. Annals of neurology. 2007;62:648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- Cui Y, et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brielmaier J, et al. Autism-relevant social abnormalities and cognitive deficits in engrailed-2 knockout mice. PloS one. 2012;7:e40914. doi: 10.1371/journal.pone.0040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano G, et al. Hippocampal dysregulation of neurofibromin-dependent pathways is associated with impaired spatial learning in engrailed 2 knock-out mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:13281–13288. doi: 10.1523/JNEUROSCI.2894-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in neurons: role of cellular immediate-early genes. Trends in neurosciences. 1989;12:459–462. doi: 10.1016/0166-2236(89)90096-9. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annual review of neuroscience. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Parada LF, Silva AJ, Ratner N. Neurofibromatosis type 1: modeling CNS dysfunction. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:14087–14093. doi: 10.1523/JNEUROSCI.3242-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- Silva AJ, et al. A mouse model for the learning and memory deficits associated with neurofibromatosis type. I. Nature. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- Morris RG, Garrud P, Rawlins JN, O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nature. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- Adams JP, Sweatt JD. Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annual review of pharmacology and toxicology. 2002;42:135–163. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- Di Cristo G, et al. Requirement of ERK activation for visual cortical plasticity. Science. 2001;292:2337–2340. doi: 10.1126/science.1059075. [DOI] [PubMed] [Google Scholar]
- Satoh Y, et al. ERK2 contributes to the control of social behaviors in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:11953–11967. doi: 10.1523/JNEUROSCI.2349-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgado P, et al. Loss of GABAergic neurons in the hippocampus and cerebral cortex of Engrailed-2 null mutant mice: implications for autism spectrum disorders. Experimental neurology. 2013;247:496–505. doi: 10.1016/j.expneurol.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgado P, et al. Transcriptome profiling in engrailed-2 mutant mice reveals common molecular pathways associated with autism spectrum disorders. Molecular autism. 2013;4:51. doi: 10.1186/2040-2392-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheh MA, et al. En2 knockout mice display neurobehavioral and neurochemical alterations relevant to autism spectrum disorder. Brain research. 2006;1116:166–176. doi: 10.1016/j.brainres.2006.07.086. [DOI] [PubMed] [Google Scholar]
- Dawson G, et al. Defining the broader phenotype of autism: genetic, brain, and behavioral perspectives. Development and psychopathology. 2002;14:581–611. doi: 10.1017/s0954579402003103. [DOI] [PubMed] [Google Scholar]
- Gage GJ, et al. Whole animal perfusion fixation for rodents. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]
- Maei HR, Zaslavsky K, Teixeira CM, Frankland PW. What is the Most Sensitive Measure of Water Maze Probe Test Performance. Frontiers in integrative neuroscience. 2009;3:4. doi: 10.3389/neuro.07.004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos and zif268. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


