Abstract
Activation of macrophages triggers the release of pro-inflammatory cytokines leading to inflammation. Numb is a negative regulator of Notch signaling, but the role of Numb in macrophages is not fully understood. In this study, the role of Numb as a regulator of inflammatory responses in macrophages was investigated. Murine bone marrow-derived macrophages, in which expression of Numb was silenced, secreted significantly less TNFα, IL-6 and IL-12 and more IL-10 upon activation by lipopolysaccharide (LPS), a ligand for Toll-like receptor 4 (TLR4), despite increased Notch signaling. The Tnfα mRNA levels both in Numb-deficient and wild-type macrophages were not significantly different, unlike those of Il6 and Il12-p40. In Numb-deficient macrophages, the Tnfα mRNAs were degraded at faster rate, compared to those in control macrophages. Activation of p38 MAPK and NF-κΒ p65 were compromised in activated Numb deficient macrophages. Numb was found to interact with the E3 ubiquitin ligase, Itch, which reportedly regulates p38 MAPK. In addition, blocking the Notch signaling pathway in activated, Numb-deficient macrophages did not further reduce TNFα levels, suggesting a Notch-independent role for Numb. A proteomics approach revealed a novel funciton for Numb in regulating complex signaling cascades downstream of TLRs, partially involving Akt/NF-κB p65/p38 MAPK in macrophages.
Macrophages are among the first immune cells to encounter microorganisms and initiate immune defense. Therefore, they play an important role in immune response. Macrophages can recognize and discriminate pathogens from self-molecules through binding of their surface and intracellular receptors to different exogenous and endogenous ligands. The receptors, known as pattern recognition receptors (PRR), can bind to many essential parts of pathogens, collectively called pathogen-associated molecular patterns (PAMPs). These interactions result in changes in the activation status of transcription factors such as MAPK and NF-κB that govern macrophage activation1,2, leading to production of various antimicrobial molecules such as anti-microbial peptides, cytokines and chemokines3. Unless these immune responses are tightly regulated, damage to host cells may occur.
One pathway that is crucial for the feedback inhibition of LPS signaling is the phosphoinisitide-3 kinase (PI-3K)-Akt/PKB signaling pathway. The Akt family of serine-threonine protein kinases is activated by PI-3K and plays a regulatory role in many cellular functions such as cell proliferation, differentiation, and metabolism. Stimulating macrophages with LPS activates PI-3K and its downstream target, Akt, which suppresses LPS-activated NF-κB and MAPK signaling pathways. This negative feedback loop results in decreased pro-inflammatory cytokine production4,5. All three isoforms of Akt (Akt1, Akt2, Akt3) have been implicated in modulating host immune defense in infection and autoimmunity6.
Evolutionarily conserved, Notch signaling has been reported to regulate development and function of cells in the immune system7,8. The Notch transmembrane receptors (Notch-1, -2, -3, and -4) are activated upon engagement of ligands (Jagged-1 and -2, Delta-1, -3, and -4). After ligand-receptor binding, the receptor undergoes enzymatic cleavage, generating a Notch intracellular domain (NICD) that can translocate to the nucleus. There it functions as a transcriptional co-activator through association with its DNA binding partner, CSL (CBF1, Suppressor of Hairless, LAG-1/RBP-Jκ) and other co-activators such as p300, to regulate transcription of specific target genes. Several target genes have been identified, including transcriptional repressors of the HES family (Hes-1, Hes-2)9.
Numb is a membrane-associated protein that contains an N-terminal phosphotyrosine-binding domain (PTB) and C-terminal proline-rich region (PRR), harboring putative Src homology 3-binding sites and Eps15 homology (EH) regions10. In vitro studies demonstrated that Numb interacted with various intracellular molecules of different signaling pathways, thereby regulating multiple cell functions. It has been extensively studied for its role in the maintenance of several types of neuronal stem cells during fetal development11,12 and in many types of cancers13,14,15. Wilson et al. used Numb-deficient mice to show that Numb was dispensable for the development of hematopoiesis and lymphopoiesis into myeloid and lymphoid cells, respectively16. Numb was also dispensable for T cell proliferation and function17. However, several studies demonstrated that Numb played critical roles in regulating asymmetric T cell division that occurred during development, differentiation, and in response to infection18,19.
The E3 ubiquitin ligase, Itch, a member of the HECT (homologous to the E6-AP C-terminus) family contains a C2 domain, four WW domains, and a HECT ligase domain20. These WW domains are implicated in protein-protein interactions, whereas the HECT domain helps in recruitment of E2 ubiquitin loading enzymes and transfers ubiquitin to their substrates21. Mice deficient in Itch developed a skin-scratching or “itchy” phenotype22 together with severe immune dysregulation, including lymphadenopathy, splenomegaly, and inflammation in the lungs and digestive tract23. In addition, Itch was shown to play a key role in peripheral T-cell tolerance. Disruption of Itch-mediated JunB ubiquitination results in continuous binding of JunB to Il-4 promoter, thereby promoting a strong TH2 response24. Itch was also identified as a negative regulator of homeostasis and hematopoietic stem cell function by regulating the Notch signaling pathway25.
Numb can interact with Itch to antagonize Notch signaling via ubiquitin-mediated degradation of the Notch receptors26,27, raising the possibility that Numb may play a critical role in other types of immune cells in which Notch signaling is involved. Studies from our group and others suggest that, upon stimulation of macrophages, Notch signaling is activated and this activation up-regulates gene expression patterns involved in macrophage activation, including Irf1, Socs1, and H-2A, as well as the production of both pro- and anti-inflammatory cytokines such as TNFα, IL-6, IL-12, and IL-1027,28,29. These studies demonstrated that using γ-secretase inhibitors or Notch1 RNAi to interfere with Notch signaling dampened macrophage activation and abrogated pro-inflammatory cytokine production. In this study, we describe a novel role for Numb in positively regulating TNFα, IL-6, and IL-12 cytokine production. Furthermore, Numb interacts with Itch which, in turn, regulates downstream signaling pathways, including NF-κB p65 and p38 MAPK. Moreover, we utilized a quantitative proteomics approach, combined with analysis of a bioinformatics database to propose that the Numb/Akt1 axis regulates pro-inflammatory cytokines in macrophages.
Materials and Methods
Construction of retrovirus-based shRNA plasmids and generating retrovirus particles for transduction
To construct pMKO.1-shNumb-GFP plasmid, the pMKO.1-GFP retroviral vector was used as a backbone vector (Addgene plasmid #10676, USA). Small hairpin loop-forming double-stranded oligonucleotides containing a Numb-specific sequence (5′- AACCACTTTCACAAGAGAAGG -3′)30, which targets the murine Numb gene, was inserted into pMKO.1-GFP. To obtain retroviral particles containing the plasmids, human embryonic kidney 293T cells (ATCC CRL-3216) were cultured in DMEM supplemented with 10% FBS at a concentration of 4 × 105 cells/ml for 24 hrs and transfected with pMKO.1-shNumb-GFP or pMKO.1-GFP, as a control vector. Cells were maintained in DMEM media supplemented with 10% FBS in 5% CO2 incubator at 37 °C. Two days later, the cell culture supernatants were collected and filtered through 0.45 μm filter and used as retroviral particles for transduction.
Generation of bone marrow-derived macrophages (BMMs) and transduction by retroviral plasmid
C57BL/6 mice were purchased from the National Laboratory Animal Center (Mahidol University, Salaya, Thailand). All procedures involving laboratory animals were reviewed and approved by Chulalongkorn University Institutional Animal Care and Use Committee (CU-IACC protocol review number #001/2555-20/54). All the procedures involving laboratory animals were carried out in accordance with the guidelines issued by CU-IACC. Bone marrow cells were harvested and filtered through 70 μm nylon mesh. The cells were incubated in L929-conditioned medium at a density of 6.25 × 105 cells/mL and maintained in a 5% CO2 incubator at 37 °C. Two days later, cells were harvested and seeded at 1 × 106 cells/well in 12-well tissue culture plates. The supernatant from 293T cell culture containing retroviral particles was mixed with X-tremeGENE HP transfection reagent (Roche, Germany) and added to the BM culture. After 1 hr of incubation in a 5% CO2 incubator at 37 °C, L929-conditioned medium was added to the BM cultures and incubation continued for 24 hrs. The transduction steps were repeated using freshly prepared retroviral partcles. L929-conditioned medium was added to the BM cultures every 3 days during 7-day incubation period. On day 8 after the first transduction, cells were harvested for cell sorting using FACSAria II (BD Biosciences) to obtain GFP+ cells. The sorted cells were seeded at 5 × 104 cells/well in 24-well plates and incubated for 5 days. The cells were harvested and the purity of F4/80+ GFP+ cells was >90%, as determined by flow cytometry.
Overexpression of Numb in macrophage-like cell line RAW264.7
RAW264.7 cells (ATCC TIB-71) were transfected with either pCI-OVA31, as a control, or pCIneoHA mouse Numb65(Addgene plasmid #37012) using FuGENE HD transfection reagent (Promega, USA). After transfection for 24 hrs, RAW264.7 cells were stimulated with 100 ng/mL E. coli LPS (Sigma Aldrich, USA) and protein lysates or culture supernatants were collected for further analysis.
Reagents
BMMs were activated by 100 ng/mL of Escherichia coli lipopolysaccharide (LPS) (Sigma-Aldrich, USA). The γ-secretase inhibitor (GSI), DAPT (Merck, Germany), was used to inhibit Notch signaling as previously described27,29. To inhibit Notch signaling in BMMs, 25 μM of DAPT or DMSO, as vehicle control, was added to the cells for 1 hr prior to stimulation with LPS.
mRNA decay assay
To assess mRNA stability, cells were seeded at 1 × 104 cells/well in 96-well plates and maintained in DMEM supplemented with 10% FBS in 5% CO2 incubator at 37 °C for 24 hrs. The cells were pre-treated with LPS for 1 hr prior to treatment with actinomycin D (Merck, Germany) at the final concentration of 20 μg/mL for indicated time periods. Total RNA was isolated from the cultured cells with TRIzol reagent (Invitrogen, UK) as described in the manufacturer’s instructions and reverse transcribed to cDNA for quantitative real-time PCR analysis.
Co-immunoprecipitation
Briefly, 5 × 105 BMM cells were maintained in complete DMEM supplemented with 10% FBS. Cells were stimulated with 100 ng/ml E. coli-derived LPS for 30 mins. After stimulating, cells were washed and detached using cold PBS. Cell pellets were lysed by vigorous vortexing in 0.5% NP-40 lysis buffer containing protease inhibitors and phosphatase inhibitor (Roche, Germany) and lysis was allowed to continue on ice for 30 minutes. Cells were centrifuged at 4 °C and protein lysates were transferred to new tubes. Protein concentration of each sample was determined using a BCA Assay Protein Assay kit (Pierce, USA). Protein A agarose beads (Cell Signaling Technology, USA) were used for the pre-clearing step of the immunoprecipitation. For immunoprecipitation, 250 ng of anti-IgG or anti-Numb antibody (Cell Signaling Technology, USA) was added to cell lysates and rotated on a rocking platform overnight.
Immunoblotting
After stimulation with LPS at indicated time points, whole cell lysates were extracted using RIPA lysis buffer containing protease inhibitor cocktail and a phosphatase inhibitor (Roche, Germany). Protein concentrations were measured using a BCA Protein Assay kit (Pierce, USA) following the manufacturer’s instructions. Antibodies used in this study were anti-Notch1 (clone C-20), anti-GAPDH (Santa Cruz Biotechnology, USA), anti-ITCH (Epitomics, USA), anti-cleaved Notch1 (Val1744), anti-Numb, anti-phospho NF-κB p65, anti-NF-κB p65, anti-phospho-p38 MAPK, anti-p38 MAPK, anti-phospho-p42/44 MAPK, anti-p42/44 MAPK, anti-phospho-SAP/JNK MAPK, anti-SAP/JNK MAPK, anti-pAkt (Thr308), anti-Akt, HRP-conjugated anti-rabbit IgG (Cell Signaling Technology, USA).
Semi-quantitative RT-PCR (qPCR)
Total RNA was isolated from cultured cells with TRIzol reagent (Invitrogen, UK) as described in the manufacturer’s instructions, and was reverse transcribed to cDNA as described previously28. Primers specific for Numb were designed (forward 5′-ACTACGGCAAAGCTTCAGGA-3′, reverse 5′-TGCATTCCTCTTGACTCATCA-3′), Numb-like (forward 5′-TACGGTTGAATGAGCTGCCA-3′; reverse 5′-AGGCAGAAGTCCCTG TTGTG-3′) and primers for Hes1, Tnfα, IL-6, IL-12p40, IL-10, Ifn-β, Akt1, Akt2, Ticam1, Map3k10, and Gapdh were used as previously described28,29,32,33,34,35. Semi-quantitative real-time PCR was carried out using MJ Mini Personal Thermal Cycler and iQ SYBR Green system (Bio-Rad, USA) following the manufacturer’s instructions. The relative gene expression levels were analyzed by CFX Manager (Bio-Rad, USA) and calculated as described previously36.
ELISA
Cell culture supernatants were collected at indicated times after treatment with specific inhibitor and stimulation with LPS. To assess protein levels of TNFα, IL-6, IL-10, and IL-12, ELISA were performed using LEGEND MAXTM mouse TNFα, IL-6, IL-10, and IL-12p40 ELISA kits (BioLegend, USA) according to the manufacturer’s instructions.
Flow cytometry
For intracellular cytokine staining, cells were treated with 1 μL of GolgiPlugTM (BD Biosciences, USA) according to the manufacturer’s instructions, just before stimulation with LPS. BD Cytofix/CytopermTM kit (BD Biosciences, USA) was used for staining according to the manufacturer’s instructions. At indicated time points, cells were harvested, washed, and incubated with 2.4G2 antibody (BD Biosciences, USA) to block FcγRII/III receptors. Antibodies used in this study: PE-labeled anti-F4/80, biotin-conjugated anti-CD11b, avidin-conjugated PECy5 (BioLegend, USA), PE-labeled anti-MHC class II, biotin-conjugated anti-CD86 (BioLegend, USA), avidin-conjugated ECD (Beckman Coulter, USA), PE-labeled anti-TNFα, PE-labeled anti-IL-6, and biotin-conjugated IL-12p40 and avidin-conjugated PECy5 (BioLegend, USA). The acquired data were analyzed using FlowJo vX (TreeStar, USA).
Protein preparation and proteomics data analysis
BMM containing shRNA plasmids were prepared as previously described. The cells were seeded overnight at 2.5 × 105 cells/well in 12-well plates in L929-conditioned medium. On the next day, cells were stimulated with 100 ng/ml of E. coli LPS (Sigma-Aldrich, USA) for 30 mins and were harvested using RIPA lysis buffer. Cell lysates were stored at −80 °C until use. Protein concentration of the samples were measured by Lowry method37. The absorbance at 750 nm (OD750) was measured and protein concentration was calculated using the bovine serum albumin standard curve. Protein separation by SDS-PAGE, in-gel digestion, and proteomic data analysis were processed as described in Supplementary Materials.
Statistical analysis
All experimental data in this study were presented as the means ± SEM of three independent experiments or from a representative experiment of three independent experiments unless indicated otherwise. The statistical significances of the differences in the experimental data were evaluated using the Student’s t test. A P value of < 0.05 was considered to be significant.
Results
Numb expression is decreased in LPS-stimulated macrophages and silencing Numb increases activation of Notch signaling
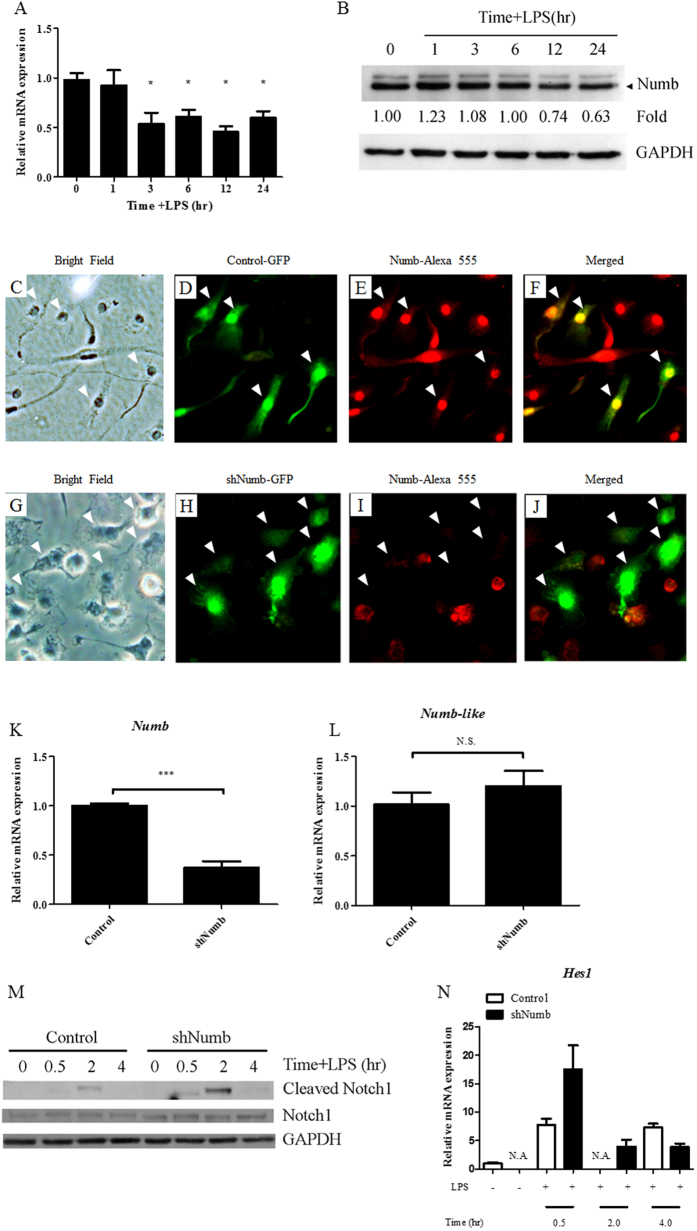
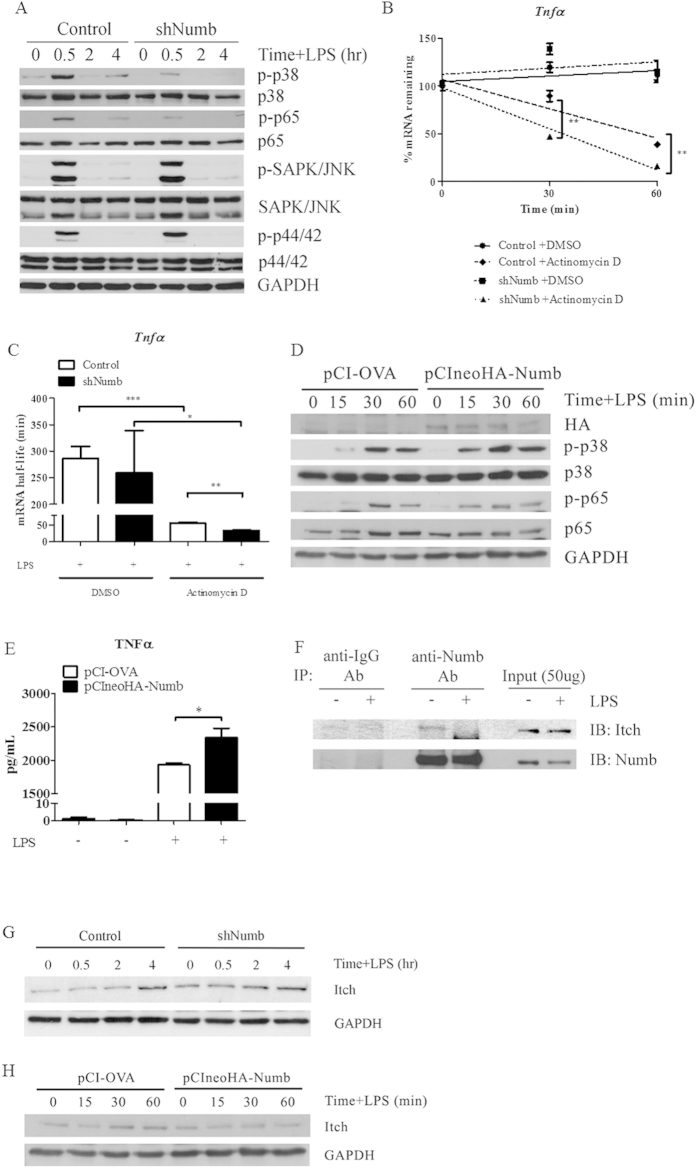
We began our investigation by exploring the expression of Numb in LPS-stimulated macrophages. We found that the expression level of Numb mRNA significantly decreased to approximately one-half and remained at this level after LPS stimulation for 3 hrs and up to 24 hrs (Fig. 1A). For protein expression, we observed a slight increase of Numb at 1 hr post stimulation and a gradual decrease over the next 24 hrs (Fig. 1B and Supplementary Figure 1A). To further investigate the effect of Numb in macrophages, we constructed a retroviral shRNA vector, pMKO.1-shNumb-GFP, introduced it into BMMs and confirmed its target specificity. In BMMs retrovirally transduced with pMKO.1-shNumb-GFP vectors, the expression level of Numb decreased significantly compared to that of the control-transduced macrophages (Fig. 1C–J). To further assess the target specificity of the construct, macrophages containing pMKO.1-shNumb-GFP, or control vector, were sorted to obtain homogenous GFP+ populations. The mRNA level of Numb and its homolog, Numb-like, that has been reported to have redundant functions with Numb in other cell types were measured30,38,39. The expression level of Numb mRNA was reduced to one-half, whereas the level of Numb-like mRNA was not affected (Fig. 1K,L). These results confirmed that the shRNA vector specifically targeted only Numb. Numb has been reported to be a negative regulator of Notch signaling. When we assessed cleaved Notch1 expression (Val1744), and the level of Hes1 mRNA, we observed that both were increased in LPS-activated Numb-silenced macrophages, compared to control cells (Fig. 1M,N). These results confirmed the role of Numb as a negative regulator of Notch signaling, as previously described.
Figure 1. Numb expression in macrophages after LPS stimulation and effects of Numb silencing on Notch signaling.
(A) Bone marrow-derived macrophages were stimulated with LPS (100 ng/mL) and Numb mRNA was measured. (B) Numb protein expression was shown and its intensity was measured as fold-increase compared to unstimulated cells. (C–J) Immunofluorescent staining of macrophages retrovirally infected with control vector (C–F) or shNumb vector (G–J), and stained for Numb (red). White arrows indicate macrophages containing shRNA vector (green). (K–L) Expression level of Numb (K) and Numb-like (L) mRNA from GFP+ macrophages as determined by qPCR. (M) Cleaved Notch1 (Val1744) was detected in LPS-stimulated macrophages by immunoblot. (N) Hes1 mRNA was measured in LPS-stimulated macrophages by qPCR. N.D. = not detectable. Data are the mean ± SEM from three independent experiments, and a representative blot from at least two independent replicates.
Numb is dispensable for differentiation of bone marrow macrophages in vitro
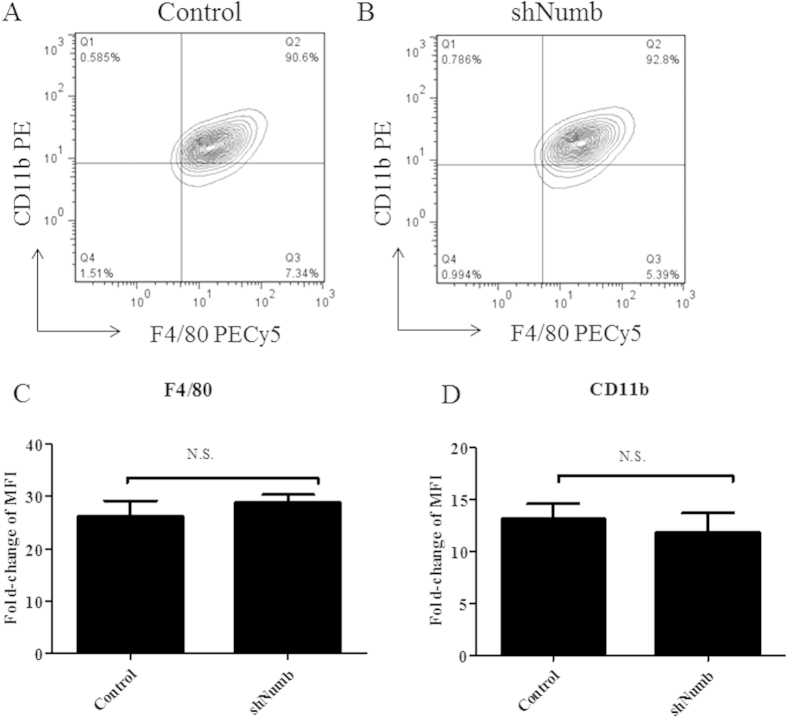
A report by Wilson et al.16 demonstrated that hematopoietic stem cell compartments, including macrophages, developed normally in Numb and Numb-like double-knockout mice. Our system employed an insertion of the retroviral vector in hematopoietic cells from the bone marrow, causing constitutive silencing of Numb. We, therefore, tested whether silencing of Numb affects the development of macrophages from bone marrow hematopoietic stem cells in vitro. After retroviral transduction of pMKO.1-shNumb-GFP or control vectors, bone marrow cells were induced to differentiate to macrophages. Cells containing the vectors were gated for GFP+ and assayed for the expression of two macrophage-specific surface markers, F4/80 and CD11b. Our results showed that the expression levels both of F4/80 and CD11b were comparable between control and Numb-silenced macrophages (Fig. 2A–D), indicating that Numb was dispensable for macrophage differentiation in vitro.
Figure 2. Differentiation of bone marrow cells to macrophages upon silencing of Numb.
(A,B) Expression of macrophage markers, CD11b and F4/80, from GFP+ population of macrophages that were retrovirally infected with control (A) or shNumb (B) vectors was determined by flow cytometry. (C,D) Expression level of F4/80 (C) and CD11b (D) from (A) and (B) were shown as fold-change of mean fluorescence intensity (MFI). Data are the mean ± SEM from three independent experiments. N.S. = not statistically significant.
Numb-deficient macrophages secreted less pro-inflammatory cytokines
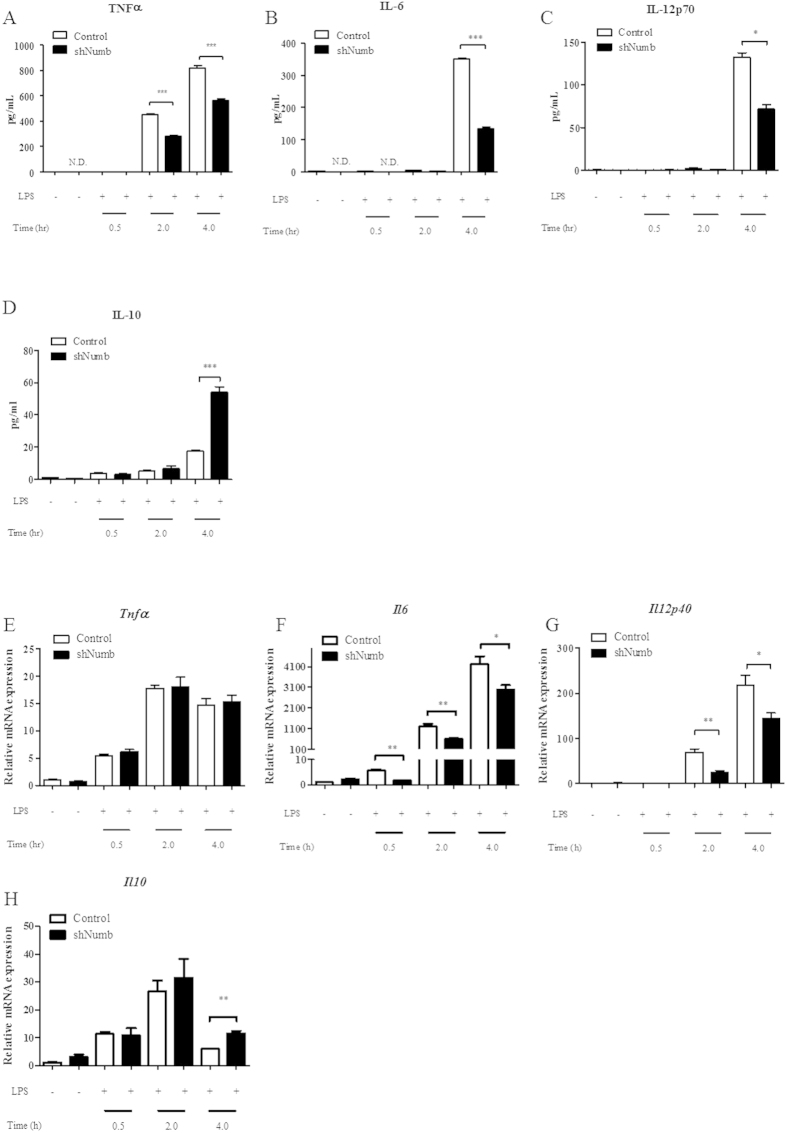
Our group and others reported that abrogating Notch signaling diminishes the ability of macrophages to secrete pro-inflammatory cytokines and perform other effector functions27,28,29,40. Therefore, we hypothesized that pro-inflammatory cytokines under the regulation of Notch signaling would increase in macrophages wherein Numb was silenced. Surprisingly, we found that Numb-silenced macrophages secreted significantly less TNFα, IL-6, and IL-12 after LPS stimulation (Fig. 3A–C). In contrast, the anti-inflammatory cytokine, IL-10, was higher in Numb-silenced macrophages (Fig. 3D). Furthermore, the expression of the costimulatory molecule CD86 decreased significantly (Supplementary Figure 1B) after LPS stimulation, whereas the expression of MHCII was not different between LPS-activated Numb-sufficient and Numb-deficient macrophages (Supplementary Figure 1C). These results suggest that Numb positively regulates pro-inflammatory programs in macrophages. We next investigated cytokine mRNA expression. We observed no difference in the levels of Tnfα between Numb-deficient and control macrophages (Fig. 3E). In contrast, the levels of Il6 and Il12p40 were readily down-regulated in Numb-deficient macrophages at all time points tested (Fig. 3F,G). In addition, the mRNA level of Il10 and Ifnβ was increased at 4 hrs after stimulation (Fig. 3H and data not shown). From these results, we conclude that the expression of Il6, Il12p40 and Il10, but not Tnfα, are regulated by Numb at the transcriptional level.
Figure 3. Expression of cytokines in macrophages lacking Numb after LPS stimulation.
(A–D) Secretion levels of TNFα (A), IL-6 (B), IL-12p70 (C), and IL-10 (D) from control- (open bars) or shNumb-infected macrophages (closed bars) following LPS stimulation were measured by ELISA. (E–H) mRNA levels of Tnfα (E), Il6 (F), Il12p40 (G), and Il10 (H) from control- or shNumb-infected macrophages following LPS stimulation were measured by qPCR. Data are the mean ± SEM from the representative experiment of two independent experiments performed in triplicates.
Numb regulates production of TNFα in a Notch-independent manner
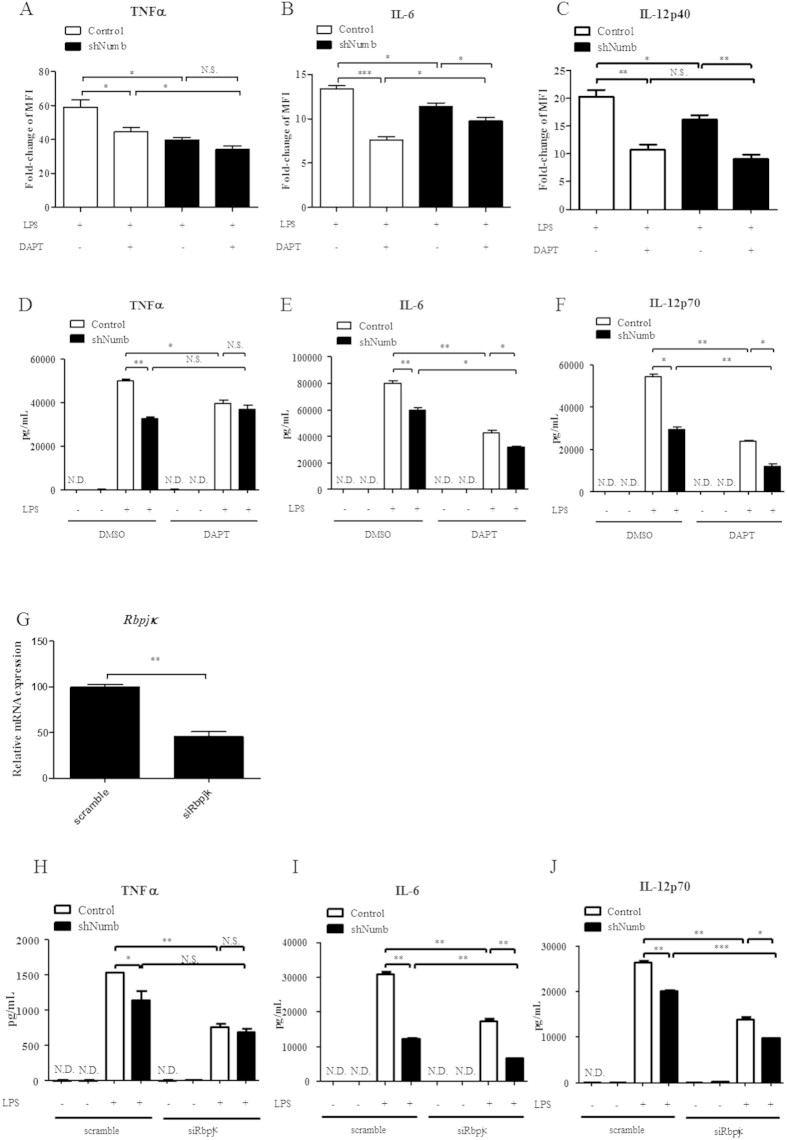
Our earlier observation revealed that Notch signaling was up-regulated in Numb-deficient macrophages following LPS stimulation. Previous reports have also demonstrated that Notch regulates pro-inflammatory cytokines in macrophages, especially IL-6 and IL-12p4027,28,32,40. Therefore, to elucidate the regulatory mechanism downstream of Numb, we used a γ-secretase inhibitor, DAPT, to pharmacologically inhibit Notch signaling in macrophages prior to and during LPS stimulation. As expected, intracellular and secreted TNFα, IL-6, and IL-12p40 protein level in DAPT-treated, Numb-intact macrophages decreased after LPS stimulation (Fig. 4A–F). However, there was not a significant difference in TNFα levels in Numb-silenced macrophages treated with DMSO or DAPT (Fig. 4A,D). In contrast, the levels of intracellular and secreted IL-6 (Fig. 4B,E) and IL-12p40 (Fig. 4C,F) in DAPT-treated, Numb-deficient macrophages were lower than those of the DMSO-treated, Numb-deficient cells. These results suggest that Notch and Numb counter-regulate expression of IL-6 and IL-12p40, but not TNFα. To confirm this, we silenced CSL/RBP-Jκ, which plays a central role in canonical Notch signaling41. The mRNA level of CSL/Rbp-jκ was reduced by ~70% (Fig. 4G). Consistent with the results obtained by the treatment with DAPT, the levels of IL-6 and IL-12p40, but not of TNFα, in Numb-deficient macrophages were further decreased, compared to the those in macrophages transfected with scramble siRNA (Fig. 4H–J). Despite increased activation of Notch signaling in Numb-deficient macrophages, a reduction in TNFα expression that results from silencing Numb, demonstrates TNFα is not under the direct influence of Notch signaling. By contrast, it appears IL-6 and IL-12p40 expression are, at least partially, directly regulated by canonical Notch signaling.
Figure 4. Expression of pro-inflammatory cytokines in macrophages lacking both Numb and Notch signaling after LPS stimulation.
(A–F) GFP+ macrophages containing control (open bars) or shNumb (closed bars) vectors were pretreated with DAPT for 1 hr prior to LPS stimulation. The levels of intracellular TNFα, IL-6, and IL-12 were determined by flow cytometry (A–C) or ELISA (D–F). N.D. = not detectable. (G) Expression level of Rbpjκ from macrophages transduced with siRbpjκ, or scrambled siRNA as control, was measured by qPCR. (H–J) Expression levels of TNFα, IL-6, and IL-12p70 from GFP+ macrophages containing control (open bars) or shNumb (closed bars) vectors which were transfected with scramble siRNA or siRbpjk were measured by ELISA. N.D. = not detectable.
Activation of the NF-κB and p38 MAPK pathways are affected by decreasing Numb
Regulatory mechanisms of TNFα, IL-6, and IL-12p40 expression have been extensively studied and shown to be regulated through different MAPK and NF-κB pathways1,2,42. Therefore, we tested the effect of Numb silencing on activation of MAPK and NF-κB pathways in macrophages. We found that the phosphorylation levels of both NF-κB p65 (RelA) and p38 MAPK were compromised in Numb-deficient macrophages after LPS stimulation, while the phosphorylation levels of ERK1/2 and SAP/JNK MAPKs remained similar between control and Numb-deficient macrophages (Fig. 5A). This selective effect silencing Numb has on p38 MAPK and NF-κB p65 activation following TLR4 stimulation may act to decrease TNFα, IL-6 and IL-12p40. Our previous results showed that the mRNA levels of Tnfα did not differ between Numb-deficient and control macrophages, consistent with several studies that demonstrated Tnfα mRNA is regulated by p38 MAPK, followed by post-transcriptional regulation43,44. Therefore, to address whether Numb exerts an effect on Tnfα mRNA stability, we performed a Tnfα mRNA decay assay, using actinomycin D to inhibit mRNA synthesis. We found that Tnfα mRNA was degraded at a faster rate in activated macrophages wherein Numb was silenced (Fig. 5B,C). On the other hand, we found that the mRNA stability of Il6 was similar in the presence or absence of Numb (Supplementary Figure 2). Taken together, these results suggested that Numb positively regulates phosphorylation of p65 in the NF-κB pathway, possibly facilitating Il6 and Il12p40 mRNA transcription, as well as phosphorylation of p38 in MAPK, which may further mediate post-transcriptional regulation of Tnfα mRNA.
Figure 5. Effect of silencing Numb on activation of MAPK and NF-κB pathways in activated macrophages and the interaction of Numb and Itch.
(A) After stimulation with LPS for indicated times, phosphorylation levels of NF-κB p65, p38, ERK1/2, and JNK MAPK from GFP+ macrophages were analyzed. Data are representative of two independent experiments. (B,C) GFP+ macrophages containing control (open bars) or shNumb (closed bars) vectors were stimulated with LPS for 1 hr prior to treatment with DMSO or with actinomycin D to inhibit mRNA synthesis. The relative amount of remaining Tnfα mRNA was measured by qPCR. Half-life of Tnfα mRNA was calculated using linear regression line equations and shown in (C). Data are the mean ± SEM from representative of two independent experiments performed in triplicates. (D) RAW264.7 cell line was transiently transfected with the control plasmid or pCIneoHA-Numb and stimulated with LPS as indicated. p38 MAPK and NF-κB p65 were detected by immunoblotting. (E) RAW264.7 cell line transfected with the control plasmid or pCIneoHA-Numb were stimulated with LPS for 1 hr. The amount of TNFα was measured by ELISA. (F) Co-immunoprecipitation of endogenous Numb from unstimulated or LPS-stimulated macrophages was analyzed by immunoblotting. (G) Expression of Itch in macrophages containing control or shNumb vector was detected by immunoblotting following LPS stimulation. (H) RAW264.7 cell line transfected with the control plasmid or pCIneoHA-Numb were stimulated with LPS for the times indicated and the level of Itch was detected by immunoblotting.
To complement the silencing approach, an overexpression of Numb in macrophage-like cell line, RAW264.7 was performed. As shown in Fig. 5D, Numb overexpression resulted in increased activation of p38 MAPK and NF-κB p65. Furthermore, increased TNFα production was detected, as expected if Numb functions to regulate TNFα (Fig. 5E).
Numb was reported to interact with Itch to promote ubiquitin-mediated degradation of Notch receptors45, and a recent report demonstrated that Itch functioned as a negative regulator of p38α46. Therefore, we hypothesized that Numb may regulate the activation of p38 MAPK through Itch. To test our hypothesis, we immunoprecipitated Numb and determined its physical interaction with Itch, using immunoblotting techniques. Our result show that Numb and Itch could be found in a complex in resting macrophages, but Itch dissociated from Numb upon LPS stimulation (Fig. 5F). Furthermore, Itch expression was higher in Numb-silenced macrophages, compared to those in control macrophages and, in contrast, Numb overexpression reduced Itch level (Fig. 5G,H). These results suggested that Numb and Itch interact physically and may affect activation of p38 MAPK and NF-κB p65.
Proteomics analysis identifies Akt1 as a potential regulator of pro-inflammatory cytokines in Numb-deficient macrophages
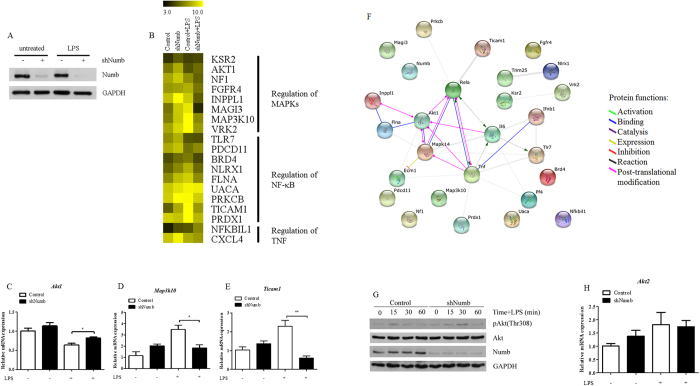
To investigate further the role of Numb in macrophages, we employed a quantitative proteomics approach using Numb-silenced macrophages, or control cells, stimulated with LPS. The expression of Numb was separately confirmed (Fig. 6A) prior to gel loading for SDS-PAGE followed by trypsin digestion. Based on the LC/MS/MS data, we identified 758 proteins that were differentially expressed in the unstimulated or LPS-activated Numb-intact and Numb-deficient macrophages (Supplementary Figure 3). Among these proteins, 551 had previously described functions and so were subjected to analysis for protein-protein interaction using the software STRING v9.147. Gene ontology (GO) enrichments in biological processes of these proteins using the software identified categories of proteins involved in regulating MAPKs, NF-κB signaling pathways and TNF production as represented by a heat map (Fig. 6B). To validate our proteomics results, we performed qPCR to measure Akt1, Map3k10, and Ticam1 in Numb-silenced macrophages and found that their expression pattern correlated well with our proteomics data (Fig. 6C–E).
Figure 6. Proteomic data analysis of proteins from Numb-silenced macrophages.
(A) Expression level of Numb in GFP+ macrophages following LPS stimulation was detected by immunoblotting. (B) Heat map representing expression levels of proteins as identified by GO enrichment in biological process. (C–E) Expression level of Akt1 (C), Map3k10 (D), and Ticam1 (E) in control (open bars) or shNumb (closed bars) macrophages were detected by qPCR. (F) A network of protein-protein interactions generated from proteins in (B) using the software STRING v9.1. (G) Phosphorylation of Akt (Thr308) and total Akt from macrophages containing control or shNumb vector was detected by immunoblotting. (H) Schematic diagram showing how Numb interacts with different proteins and regulates pro-inflammatory cytokines (see text for details). Solid lines depict the confirmed links while the dotted lines depict the potential links.
To further predict whether these proteins were involved in the regulation of pro-inflammatory cytokine production in Numb-deficient macrophages, TNFα, IL-6, IFN-β, Numb, NF-κB p65 (RelA), and p38 MAPK (MAPK14) were included to create the protein-protein interaction network. Remarkably, network analysis revealed that Akt1 interacted both with NF-κB p65 and p38 MAPK, suggesting that Akt1 may play a role in role in the Numb-mediated regulatory network in macrophages (Fig. 6F). Although all three Akt isoforms (Akt1, Akt2, Akt3) share a high degree of homology, in vitro studies showed no difference in the ability of these isoforms to recognize and phosphorylate protein substrates48. We observed increased phosphorylation of Akt in LPS-stimulated macrophages wherein Numb was silenced (Fig. 6G); however, levels of Akt2 in Numb-silenced macrophages and wild-type cells did not differ (Supplementary Figure 4). These results were consistent with studies of macrophages from Akt1 knockout mice, in which TNFα and IL-6 were up-regulated49,50. Taken together, these data report a novel and as-yet-undescribed function for Numb, as a regulator of pro-inflammatory cytokines in macrophages through Numb-Itch interactions as well as through the Akt pathway which affects both p38 MAPK and NF-κB pathways, in addition to its role as a negative regulator of Notch signaling (Fig. 6H).
Discussion
To our knowledge, the function of Numb in activated macrophage has never been documented. Here we describe both Notch-dependent and -independent roles for Numb in macrophage activation. As reported for other cell types, Numb was found to affect diverse signaling pathways in macrophages which might ultimately dictate the outcomes of the inflammatory responses. Our data showed that the amount of activated, intracellular Notch was higher in LPS-stimulated macrophages wherein Numb was silenced, suggesting that Numb functions as a negative regulator of Notch signaling. In addition, we also found that the Notch target, Hes1, was elevated following the activation in Numb-deficient macrophages. These results are consistent with other reports demonstrating that Numb is a negative regulator of Notch signaling in other cell types45,51. Therefore, we can extend this role for Numb as a negative regulator of Notch signaling to include macrophages.
Numb was first reported as a regulator of cell fate determinant by interrupting Notch signaling in sensory organ progenitors of Drosophila melanogaster and in mammalian retinal cells and neurons26,30,52,53. Previous studies using hematopoietic bone marrow cells, demonstrated that activating Notch signaling using immobilized Delta-1 resulted in apoptosis and inhibited monocyte into macrophage differentiation under M-CSF stimulation54. However, our results demonstrate that sustained Notch activity, as a result of interrupting Numb in bone marrow, did not affect monocyte differentiation into macrophages. Consistent with our results, studies using Numb and Numb-like double-knockout mice revealed that bone marrow cells, including macrophages, developed and distributed normally16. When Numb was overexpressed to inhibit Notch signaling in T cells, T cell maturation and development also remained intact55. These results imply that Numb may influence cellular differentiation in a context-dependent manner.
To our surprise, silencing Numb resulted in down-regulation of pro-inflammatory cytokines such as TNFα, IL-6, and IL-12p40, despite activation of Notch signaling. Furthermore, silencing Numb strongly affected the activation of p38 MAPK and NF-κB p65. Recently, Itch was demonstrated to be a negative regulator of NF-κB and p38 MAPK signaling pathways that control pro-inflammatory cytokines, mainly through ubiquitination and degradation of Tak1 and Tab146,56,57. We found that Itch expression increased in LPS-activated, Numb-silenced macrophages, suggesting that Numb and Itch may counter-regulate signaling cascades to control the expression of pro-inflammatory cytokines in macrophages. Additionally, Itch has been identified as a Numb binding partner in order to regulate Notch signaling in other cell types45,58,59. To this end, we demonstrated that Numb interacts with Itch in resting macrophages and they dissociate after LPS stimulation. Therefore, upon LPS stimulation, Itch may be released from Numb to function as a negative regulator of pro-inflammatory response. Understanding the precise means by which Itch and Numb interact will require further investigation.
From our proteomics data, we constructed a network of proteins capable of regulating pro-inflammatory cytokines, in which Akt1 was located at the center and interacted with NF-κB p65 and p38 MAPK. In addition, quantitative proteomics and mRNA analyses revealed that the expression level of Akt1 increased in Numb-deficient macrophages. These results suggested that Akt1 also played a role in regulating pro-inflammatory cytokine production in macrophages. In support of our results, studies in Akt1−/− mice showed that LPS stimulation of Akt1−/− macrophages gave rise to pro-inflammatory phenotypes which included secretion of more TNFα and IL-649,50. There are three Akt isoforms each of which displays distinct tissue distribution and function. Akt1 is required for induction of nitric oxide synthase and endothelial cell function whereas Akt2 is required for insulin-responsive signaling60,61,62. In contrast, Akt3 is involved in brain development and function; however, its function remains largely undefined63,64. We observed an upregulation of Akt1 in Numb-deficient macrophages, whereas Akt2 remained comparable to that of wild-type. In addition, we showed that the phosphorylation level of Akt was higher in LPS-stimulated, Numb-deficient macrophages. Taken together, our results are consistent with a model showing Numb regulates multiple pathways as depicted in Fig. 6H. In this model Numb negatively regulates Notch, Akt1 and Itch, and each pathway further affects defined downstream signaling. Exactly how Numb regulates Akt levels is a subject for further investigation.
Additional Information
How to cite this article: Kueanjinda, P. et al. A Novel Role of Numb as A Regulator of Pro-inflammatory Cytokine Production in Macrophages in Response to Toll-like Receptor 4. Sci. Rep. 5, 12784; doi: 10.1038/srep12784 (2015).
Supplementary Material
Acknowledgments
This work was supported in part by the Fogarty International Research Collaborative Award (NIH, USA), the Special Task Force for Activating Research (STAR) from the centenary academic development project (Chulalongkorn University), the Ratchadaphiseksomphot Endowment Fund of Chulalongkorn University (RES560530144-AS), the Omics Sciences and Bioinformatics Center, and the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund). The authors wish to thank Dr. Lisa M. Minter for critical reading of the manuscript.
Footnotes
Author Contributions P.K. designed, performed and analyzed results of all experiments and wrote the manuscript. S.R. designed and analyzed the proteomics experiments. T.P. designed, analyzed and wrote the main manuscript text. All authors reviewed the manuscript.
References
- Ghosh S., May M. J. & Kopp E. B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annual review of immunology 16, 225–260, 10.1146/annurev.immunol.16.1.225 (1998). [DOI] [PubMed] [Google Scholar]
- Lee J. C. et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372, 739–746, 10.1038/372739a0 (1994). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology 11, 373–384, 10.1038/ni.1863 (2010). [DOI] [PubMed] [Google Scholar]
- Fukao T. et al. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nature immunology 3, 875–881, 10.1038/ni825 (2002). [DOI] [PubMed] [Google Scholar]
- Guha M. & Mackman N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. The Journal of biological chemistry 277, 32124–32132, 10.1074/jbc.M203298200 (2002). [DOI] [PubMed] [Google Scholar]
- Weichhart T. & Saemann M. D. The PI3K/Akt/mTOR pathway in innate immune cells: emerging therapeutic applications. Annals of the rheumatic diseases 67 Suppl 3, iii70–74, 10.1136/ard.2008.098459 (2008). [DOI] [PubMed] [Google Scholar]
- Amiel E. et al. Pivotal Advance: Toll-like receptor regulation of scavenger receptor-A-mediated phagocytosis. Journal of leukocyte biology 85, 595–605, 10.1189/jlb.1008631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M. D. & Lake R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999). [DOI] [PubMed] [Google Scholar]
- Lai E. C. Notch signaling: control of cell communication and cell fate. Development 131, 965–973, 10.1242/dev.01074 (2004). [DOI] [PubMed] [Google Scholar]
- Dho S. E., French M. B., Woods S. A. & McGlade C. J. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. The Journal of biological chemistry 274, 33097–33104 (1999). [DOI] [PubMed] [Google Scholar]
- Zhong W., Feder J. N., Jiang M. M., Jan L. Y. & Jan Y. N. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 17, 43–53, 10.1016/S0896-6273(00)80279-2 (1996). [DOI] [PubMed] [Google Scholar]
- Zhong W., Jiang M. M., Weinmaster G., Jan L. Y. & Jan Y. N. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development 124, 1887–1897 (1997). [DOI] [PubMed] [Google Scholar]
- Rennstam K. et al. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast cancer research and treatment 122, 315–324, 10.1007/s10549-009-0568-x (2010). [DOI] [PubMed] [Google Scholar]
- Sheng W. et al. Cooperation among Numb, MDM2 and p53 in the development and progression of pancreatic cancer. Cell and tissue research 354, 521–532, 10.1007/s00441-013-1679-6 (2013). [DOI] [PubMed] [Google Scholar]
- Flores A. N., McDermott N., Meunier A. & Marignol L. NUMB inhibition of NOTCH signalling as a therapeutic target in prostate cancer. Nature reviews. Urology, 10.1038/nrurol.2014.195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. et al. Normal hemopoiesis and lymphopoiesis in the combined absence of numb and numblike. Journal of immunology 178, 6746–6751, 10.4049/jimmunol.178.11.6746 (2007). [DOI] [PubMed] [Google Scholar]
- Anderson A. C. et al. The Notch regulator Numb links the Notch and TCR signaling pathways. Journal of immunology 174, 890–897 (2005). [DOI] [PubMed] [Google Scholar]
- Aguado R., Martin-Blanco N., Caraballo M. & Canelles M. The endocytic adaptor Numb regulates thymus size by modulating pre-TCR signaling during asymmetric division. Blood 116, 1705–1714, 10.1182/blood-2009-10-246777 (2010). [DOI] [PubMed] [Google Scholar]
- Chang J. T. et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687–1691, 10.1126/science.1139393 (2007). [DOI] [PubMed] [Google Scholar]
- Melino G. et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell death and differentiation 15, 1103–1112, 10.1038/cdd.2008.60 (2008). [DOI] [PubMed] [Google Scholar]
- Hershko A. & Ciechanover A. The ubiquitin system. Annual review of biochemistry 67, 425–479, 10.1146/annurev.biochem.67.1.425 (1998). [DOI] [PubMed] [Google Scholar]
- Perry W. L. et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nature genetics 18, 143–146, 10.1038/ng0298-143 (1998). [DOI] [PubMed] [Google Scholar]
- Hustad C. M. et al. Molecular genetic characterization of six recessive viable alleles of the mouse agouti locus. Genetics 140, 255–265 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D. et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nature immunology 3, 281–287, 10.1038/ni763 (2002). [DOI] [PubMed] [Google Scholar]
- Rathinam C., Matesic L. E. & Flavell R. A. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nature immunology 12, 399–407, 10.1038/ni.2021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Jan L. Y. & Jan Y. N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41 (1996). [DOI] [PubMed] [Google Scholar]
- Monsalve E. et al. Notch1 upregulates LPS-induced macrophage activation by increasing NF-kappaB activity. European journal of immunology 39, 2556–2570, 10.1002/eji.200838722 (2009). [DOI] [PubMed] [Google Scholar]
- Wongchana W. & Palaga T. Direct regulation of interleukin-6 expression by Notch signaling in macrophages. Cellular & molecular immunology 9, 155–162, 10.1038/cmi.2011.36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaga T. et al. Notch signaling is activated by TLR stimulation and regulates macrophage functions. European journal of immunology 38, 174–183, 10.1002/eji.200636999 (2008). [DOI] [PubMed] [Google Scholar]
- Rasin M. R. et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci 10, 819–827, 10.1038/nn1924 (2007). [DOI] [PubMed] [Google Scholar]
- Yoshida A., Nagata T., Uchijima M., Higashi T. & Koide Y. Advantage of gene gun-mediated over intramuscular inoculation of plasmid DNA vaccine in reproducible induction of specific immune responses. Vaccine 18, 1725–1729 (2000). [DOI] [PubMed] [Google Scholar]
- Boonyatecha N., Sangphech N., Wongchana W., Kueanjinda P. & Palaga T. Involvement of Notch signaling pathway in regulating IL-12 expression via c-Rel in activated macrophages. Molecular immunology 51, 255–262, 10.1016/j.molimm.2012.03.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrby M., Evertsson K., Fjallstrom A. K., Svensson A. & Tagerud S. Akt (protein kinase B) isoform phosphorylation and signaling downstream of mTOR (mammalian target of rapamycin) in denervated atrophic and hypertrophic mouse skeletal muscle. Journal of molecular signaling 7, 7, 10.1186/1750-2187-7-7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijneman R. J. et al. Pla2g2a attenuates colon tumorigenesis in azoxymethane-treated C57BL/6 mice; expression studies reveal Pla2g2a target genes and pathways. Cellular oncology : the official journal of the International Society for Cellular Oncology 31, 345–356, 10.3233/CLO-2009-0480 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H., Matsumoto M., Funami K., Akazawa T. & Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nature immunology 4, 161–167, 10.1038/ni886 (2003). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L. & Randall R. J. Protein measurement with the Folin phenol reagent. The Journal of biological chemistry 193, 265–275 (1951). [PubMed] [Google Scholar]
- Petersen P. H., Zou K., Hwang J. K., Jan Y. N. & Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934, 10.1038/nature01124 (2002). [DOI] [PubMed] [Google Scholar]
- Yang J. et al. Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart. Cardiovasc Res 96, 276–285, 10.1093/cvr/cvs250 (2012). [DOI] [PubMed] [Google Scholar]
- Hu X. et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity 29, 691–703, 10.1016/j.immuni.2008.08.016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nature immunology 13, 642–650, 10.1038/ni.2304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. et al. A novel mechanism for TNF-alpha regulation by p38 MAPK: involvement of NF-kappa B with implications for therapy in rheumatoid arthritis. Journal of immunology 173, 6928–6937, 10.4049/jimmunol.173.11.6928 (2004). [DOI] [PubMed] [Google Scholar]
- Liu L. et al. Estrogen-induced nongenomic calcium signaling inhibits lipopolysaccharide-stimulated tumor necrosis factor alpha production in macrophages. PLoS One 8, e83072, 10.1371/journal.pone.0083072 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guma M. et al. Antiinflammatory functions of p38 in mouse models of rheumatoid arthritis: advantages of targeting upstream kinases MKK-3 or MKK-6. Arthritis Rheum 64, 2887–2895, 10.1002/art.34489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. A. & McGlade C. J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. The Journal of biological chemistry 278, 23196–23203, 10.1074/jbc.M302827200 (2003). [DOI] [PubMed] [Google Scholar]
- Theivanthiran B. et al. The E3 ubiquitin ligase Itch inhibits p38alpha signaling and skin inflammation through the ubiquitylation of Tab1. Science signaling 8, ra22, 10.1126/scisignal.2005903 (2015). [DOI] [PubMed] [Google Scholar]
- Franceschini A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research 41, D808–815, 10.1093/nar/gks1094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K. S. et al. Activation of protein kinase B beta and gamma isoforms by insulin in vivo and by 3-phosphoinositide-dependent protein kinase-1 in vitro: comparison with protein kinase B alpha. The Biochemical journal 331 (Pt 1), 299–308 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F. et al. Akt1-mediated regulation of macrophage polarization in a murine model of Staphylococcus aureus pulmonary infection. The Journal of infectious diseases 208, 528–538, 10.1093/infdis/jit177 (2013). [DOI] [PubMed] [Google Scholar]
- Androulidaki A. et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31, 220–231, 10.1016/j.immuni.2009.06.024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frise E., Knoblich J. A., Younger-Shepherd S., Jan L. Y. & Jan Y. N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proceedings of the National Academy of Sciences of the United States of America 93, 11925–11932 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. et al. Numb proteins specify asymmetric cell fates via an endocytosis- and proteasome-independent pathway. Molecular and cellular biology 25, 2899–2909, 10.1128/MCB.25.8.2899-2909.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M. & Raff M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development 130, 2329–2339 (2003). [DOI] [PubMed] [Google Scholar]
- Ohishi K. et al. Monocytes express high amounts of Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1. Blood 95, 2847–2854 (2000). [PubMed] [Google Scholar]
- French M. B. et al. Transgenic expression of numb inhibits notch signaling in immature thymocytes but does not alter T cell fate specification. Journal of immunology 168, 3173–3180 (2002). [DOI] [PubMed] [Google Scholar]
- Ajibade A. A. et al. TAK1 negatively regulates NF-kappaB and p38 MAP kinase activation in Gr-1 + CD11b + neutrophils. Immunity 36, 43–54, 10.1016/j.immuni.2011.12.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N. et al. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nature immunology 12, 1176–1183, 10.1038/ni.2157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. A., Dho S. E., Weinmaster G. & McGlade C. J. Numb regulates post-endocytic trafficking and degradation of Notch1. The Journal of biological chemistry 284, 26427–26438, 10.1074/jbc.M109.014845 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L. et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene 30, 65–76, 10.1038/onc.2010.394 (2011). [DOI] [PubMed] [Google Scholar]
- Yang Z. Z. et al. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. The Journal of biological chemistry 278, 32124–32131, 10.1074/jbc.M302847200 (2003). [DOI] [PubMed] [Google Scholar]
- Cho H., Thorvaldsen J. L., Chu Q., Feng F. & Birnbaum M. J. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. The Journal of biological chemistry 276, 38349–38352, 10.1074/jbc.C100462200 (2001). [DOI] [PubMed] [Google Scholar]
- Cho H. et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 292, 1728–1731, 10.1126/science.292.5522.1728 (2001). [DOI] [PubMed] [Google Scholar]
- Easton R. M. et al. Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Molecular and cellular biology 25, 1869–1878, 10.1128/MCB.25.5.1869-1878.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp O. et al. Essential role of protein kinase B gamma (PKB gamma/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132, 2943–2954, 10.1242/dev.01864 (2005). [DOI] [PubMed] [Google Scholar]
- Kansaku A., Hirabayashi S., Mori H., Fujiwara N., Kawata A., Ikeda M., Rokukawa C., Kurihara H. & Hata Y. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene Aug 24;25(37):5071-84. doi:10.1038/sj.onc.1209468 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.