Abstract
Bacterial small regulatory RNAs (sRNAs) are commonly known to repress gene expression by base pairing to target mRNAs. In many cases, sRNAs base pair with and sequester mRNA ribosome-binding sites, resulting in translational repression and accelerated transcript decay. In contrast, a growing number of examples of translational activation and mRNA stabilization by sRNAs have now been documented. A given sRNA often employs a conserved region to interact with and regulate both repressed and activated targets. However, the mechanisms underlying activation differ substantially from repression. Base pairing resulting in target activation can involve sRNA interactions with the 5′ untranslated region (UTR), the coding sequence or the 3′ UTR of the target mRNAs. Frequently, the activities of protein factors such as cellular ribonucleases and the RNA chaperone Hfq are required for activation. Bacterial sRNAs, including those that function as activators, frequently control stress response pathways or virulence-associated functions required for immediate responses to changing environments. This review aims to summarize recent advances in knowledge regarding target mRNA activation by bacterial sRNAs, highlighting the molecular mechanisms and biological relevance of regulation.
Keywords: sRNA, Hfq, RNase E, anti-antisense, degradation interference, sponge RNA
This review discusses recent advances in understanding of positive regulation of gene expression by small RNAs in bacteria.
Graphical Abstract Figure.
This review discusses recent advances in understanding of positive regulation of gene expression by small RNAs in bacteria.
INTRODUCTION
Regulation of gene expression at the level of mRNA translation or stability has been discovered in all domains of life. Altering the abundance of mature mRNAs constitutes an important layer of gene control and an effective means to rapidly change the translational output of a cell. In bacteria, archaea and eukaryotes, small riboregulators have been shown to mediate post-transcriptional mechanisms of gene regulation. In bacteria, small regulatory RNAs (sRNAs) regulate target mRNAs through sequence-specific base pairing (Waters and Storz 2009). The sRNAs typically interact with the 5′ untranslated region (UTR) of target mRNAs and directly modulate ribosome binding. The most commonly described consequence of sRNA-mediated regulation is repression resulting from inhibition of translation initiation, enhancement of target mRNA decay or both. Nevertheless, sRNAs have also been documented to activate gene expression. In this review, we focus on bacterial sRNAs that activate translation or enhance stability of one or more mRNA targets, and discuss the molecular mechanisms of post-transcriptional activation as well as the physiological outcomes of regulation, where known.
In contrast to their microRNA counterparts from eukaryotes (Ha and Kim 2014), bacterial sRNAs are heterogeneous in size (∼50 to 250 nts) and structure. The largest class of sRNAs associates with the RNA chaperone, Hfq. Hfq belongs to the extensive group of Sm-like (LSm) proteins which are common in eukaryotes and archaea (Vogel and Luisi 2011). Approximately, 50% of all bacterial species, including sundry pathogens, encode a homolog of Hfq (Valentin-Hansen, Eriksen and Udesen 2004; Chao and Vogel 2010; Finn et al. 2014). At a mechanistic level, Hfq acts as a ‘molecular matchmaker’ for sRNA-target mRNA interactions and is frequently required to protect sRNAs from cellular ribonucleases such as RNase E (Moll et al. 2003). Active Hfq assembles into a homohexameric ring structure with one RNA-binding site on each side of the ring. The individual sides of the hexamer are designated proximal and distal with respect to N- and C-termini of the protomers. The proximal side is required for binding the U-rich sequences contained in the transcriptional terminator located at the 3′ end of most sRNAs (Otaka et al. 2011; Sauer and Weichenrieder 2011; Kovach et al. 2014), while the distal side preferentially associates with polyA and AAN (A, A, any nucleotide) motifs in mRNAs (Link, Valentin-Hansen and Brennan 2009; Robinson et al. 2014). In Salmonella, Hfq associates with ∼1250 mRNAs many of which possess Rho-independent terminators at their 3′ ends (Chao et al. 2012). In some cases, the 3′ UTRs of mRNAs can function as regulatory RNAs themselves (Davis and Waldor 2007; Chao et al. 2012; Guo et al. 2014; Miyakoshi, Chao and Vogel 2015b; Papenfort et al. 2015), a function which might be facilitated by Hfq binding to the sequences within their intrinsic terminators.
Mutation of the hfq gene typically results in pleiotropic phenotypes affecting virulence gene expression and general stress response pathways (Chao and Vogel 2010; Papenfort and Vogel 2010). In Salmonella enterica and Escherichia coli, deletion of hfq altered the expression of ∼20% of all genes (Guisbert et al. 2007; Sittka et al. 2008; Ansong et al. 2009), including upregulated as well as downregulated genes. While many of these effects could be indirect (e.g. because loss of Hfq affects levels of a transcription factor), there are clearly many cases where loss of Hfq more directly impacts mRNA levels via defects in regulation by sRNAs. For example, increased gene expression in the absence of Hfq might reflect loss of repression by an sRNA, whereas reduced mRNA levels could be caused by loss of an activating sRNA function. It is important to note that a given sRNA can function as a repressor for one or more target mRNAs, while activating others. In most cases, the same sRNA sequences are involved in interactions that cause activation and repression of different mRNAs, suggesting that the molecular information determining the regulatory outcome is contained within the target mRNA.
Here, we focus on bacterial sRNAs that activate target mRNAs via interactions with the 5′ UTR, coding sequence or 3′ UTR of transcripts. These interactions can change transcript stability or modulate translation initiation. We consider the protein factors involved in sRNA-mediated regulation and outline the physiological conditions demanding post-transcriptional target activation through sRNAs. Finally, we provide a view on alternative possibilities for target activation including more indirect mechanisms such as base pairing with other non-coding RNAs or the titration of RNA-binding proteins.
ACTIVATION AT THE 5′ UTR
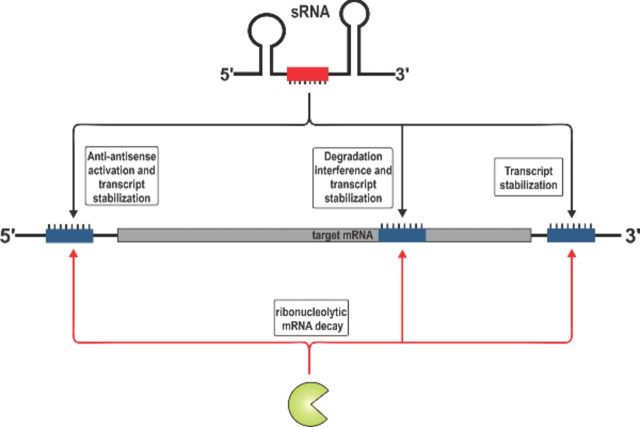
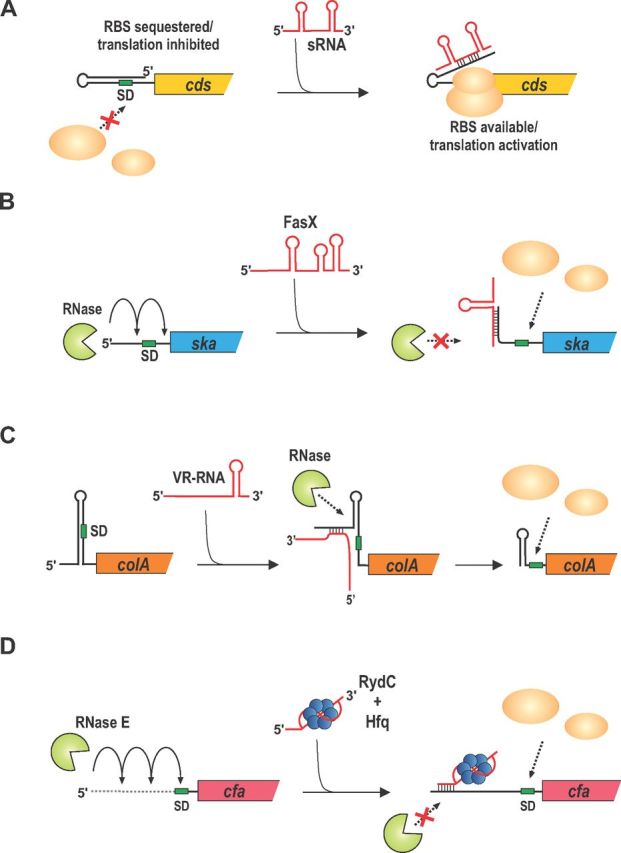
Binding sites within the 5′ UTR of mRNAs are frequently involved in target activation by sRNAs. Two general mechanisms involving activation via 5′ UTR binding sites have been reported. The first is the so-called anti-antisense mechanism, which involves unfolding of an intrinsic translation inhibitory structure in the target transcript (Fig. 1A). For this mechanism, the primary outcome of activation is enhancement of translation via improved mRNA accessibility to ribosomes. The second general mechanism of activation is interference with ribonucleolytic decay (Fig. 1B–D). These mechanisms stand in contrast with repressive mechanisms (De Lay, Schu and Gottesman 2013), which typi-cally involve base pairing interactions encompassing sequences within a window extending from the Shine–Dalgarno (SD) sequence of the mRNA to the fifth codon in the open reading frame (ORF) to directly interfere with ribosome binding (Bouvier et al. 2008). Base pairing interactions upstream (Sharma et al. 2007; Desnoyers and Masse 2012) or downstream (Pfeiffer et al. 2009) of this window can also have a repressive effect on target gene expression. Below, we describe specific examples of sRNAs that activate expression of target genes by anti-antisense control and decay interference mechanisms.
Figure 1.
Mechanisms of target activation at the 5′ end. (A) General scheme of anti-antisense regulation for activation of mRNA translation. An mRNA with a long 5′ UTR forms a translation-inhibitory hairpin in the absence of any sRNA activator. The sRNA forms base pairing interactions that prevent formation of the inhibitory structure, thus allowing ribosomes to access the SD sequence and initiate translation. (B) In group A Streptococcus, FasX sRNA base pairs with sequences in the 5′ UTR of ska mRNA to prevent RNase E-mediated degradation. Increased stability of ska mRNA promotes enhanced translation and production of the encoded streptokinase. (C) In C. perfringens, the colA mRNA forms a translation-inhibitory structure. The VR-RNA base pairs with colA mRNA and promotes processing by an unknown RNase. Processed colA mRNA forms an alternative structure at the 5′ end that stabilizes it and allows enhanced production of the encoded collagenase. (D) In E. coli and Salmonella, the cfa mRNA is inherently unstable due to RNase E-dependent degradation. The RydC sRNA base pairs with an RNase E-sensitive site, stabilizing the cfa mRNA and promoting synthesis of the encoded cyclopropane fatty acid synthase.
Anti-antisense control
Anti-antisense regulation in Gram-positive pathogens
Virulence of Staphylococcus aureus is strictly dependent on synthesis of the ∼500-nt sRNA regulator RNAIII, which is regulated by growth phase and cell density through quorum sensing (Novick and Geisinger 2008). The anti-antisense type of target activation was first discovered for RNAIII activation of hla, which encodes the S. aureus α-toxin (Morfeldt et al. 1995). Base pairing with RNAIII prevents the formation of a translation-inhibitory structure in the hla mRNA; thus, promoting ribosome binding and increased translation initiation (e.g. as illustrated in Fig. 1A). The RNAIII–hla mRNA interaction involves sequences in the 5′ end of RNAIII (Morfeldt et al. 1995). In contrast, RNAIII interactions with other mRNAs that are targets of repression, including the rot, coa and spa mRNAs, utilize sequences in the conserved 3′ end of RNAIII. For these repressed targets, base pairing with RNAIII is frequently coupled to cleavage by the double-strand-specific ribonuclease RNase III, linking translation inhibition to mRNA decay (Romilly et al. 2012). Therefore, recognition of RNAIII–mRNA complexes by RNase III might play a role in determining whether a given transcript is activated or repressed. The role of Hfq in RNAIII activity is currently a matter of debate. Although RNAIII binds Hfq, several studies suggest that Hfq is not relevant for RNAIII-mediated regulation in vivo (Huntzinger et al. 2005; Bohn, Rigoulay and Bouloc 2007; Boisset et al. 2007).
RNAIII is one of only a few ‘dual function’ RNA regulators (Vanderpool, Balasubramanian and Lloyd 2011) since it not only carries out base pairing-dependent post-transcriptional regulation of mRNA targets, but also harbors an ORF (hld) encoding δ-hemolysin (Morfeldt et al. 1995). RNAIII base pairing with another activated target, map mRNA (encoding a surface adhesion protein), is predicted to involve the stem-loop 4 region of RNAIII (Liu et al. 2011), which partially overlaps the hld ORF, suggesting that RNAIII binding to map mRNA could interfere with translation of RNAIII and δ-hemolysin production. It is currently unclear how translation of hld influences the base pairing capabilities of RNAIII, but it is interesting to note that not all RNAIII homologs encode Hld protein (Benito et al. 1998).
Anti-antisense regulation has also been shown to control virulence-related genes in other Gram-positive bacteria. In the human pathogen Listeria monocytogenes, the Rli27 sRNA activates one of two isoforms (that differ in the length of the 5′ UTR) of the lmo0514 transcript through direct interaction. The lmo0514 gene is controlled by two promoters, one giving rise to a transcript with a short 28-nt 5′ UTR and the other producing an mRNA with a 234-nt leader sequence. Extended base pairing of Rli27 with positions –151 to –129 (numbering relative to the start codon) of the long lmo0514 transcript prevents formation of an inhibitory structure in the long 5′ UTR and increases protein production. Activation of lmo0514 by Rli27 is most significant when L. monocytogenes replicates inside host cells, suggesting a potential role for this sRNA in pathogenesis (Quereda et al. 2014).
Activation of the rpoS mRNA
The rpoS mRNA from E. coli is one of the most studied transcripts in the field of bacterial RNA biology. The rpoS gene encodes the stationary sigma factor, σS, which controls transcription initiation of numerous genes with roles in bacterial stress responses and virulence (Battesti, Majdalani and Gottesman 2011). The unusually long (567 nt in E. coli) 5′ UTR of rpoS is conserved in numerous Gram-negative species (Soper and Woodson 2008) and serves as a binding site for at least three independent sRNAs: DsrA, RprA and ArcZ. In the absence of these sRNAs, the rpoS 5′ UTR folds into a hairpin that sequesters the ribosome-binding site (RBS) into a double-stranded secondary structure, inhibiting translation initiation (Fig. 2). Each individual sRNA (DsrA, RprA or ArcZ) binds to a specific site within the rpoS 5′ UTR and sequesters sequences that would otherwise participate in forming the translation-inhibitory structure (Fig. 1A; Mika and Hengge 2014). Thus, sRNA base pairing to the rpoS mRNA relieves translational inhibition and allows increased synthesis of σS.
Figure 2.
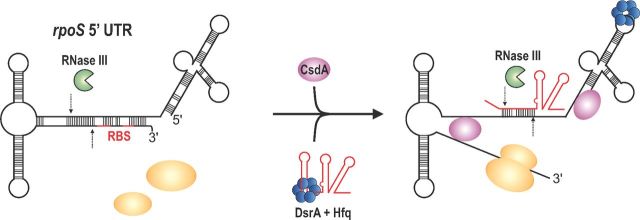
Anti-antisense activation of rpoS by DsrA. In E. coli and related species, the long 5′UTR of the rpoS mRNA forms a complex secondary structure that blocks RBS and thereby inhibits ribosome binding. In addition, this RNA structure is subject to RNase III-mediated cleavage. Together with Hfq, DsrA can base pair with the rpoS 5′ UTR inducing structural rearrangements in the RNA allowing ribosome binding. Interaction with DsrA also creates an alternative RNase III cleavage site in the distal part of the stem-loop structure. At low temperatures, DsrA also requires the DEAD box helicase CsdA for rpoS activation.
Studies of sRNA-mediated activation of rpoS translation, in particular by E. coli DsrA (Lease, Cusick and Belfort 1998; Majdalani et al. 1998), have provided valuable insight into the role played by the RNA chaperone Hfq. DsrA requires Hfq for post-transcriptional activation of rpoS (Sledjeski, Whitman and Zhang 2001). These data suggest that Hfq binding to the rpoS 5′ UTR facilitates subsequent sRNA binding to rpoS mRNA (Soper and Woodson 2008). Hfq binding itself remodels the rpoS 5′ UTR into a compact tertiary structure that positions the translation-inhibitory stem of the rpoS transcript on the proximal side of Hfq, which is known to bind sRNAs (Peng et al. 2014). The conformational changes in rpoS mRNA that occur upon Hfq binding lead to partial opening of the inhibitory stem loop (Muffler, Fischer and Hengge-Aronis 1996; Soper, Doxzen and Woodson 2011; Hammerle et al. 2013); base pairing interactions with sRNAs further relieve formation of the inhibitory structure and promote accessibility of the rpoS RBS and increased translation initiation (Fig. 2). In addition to Hfq, other proteins also affect the translation of rpoS. The major endoribonuclease in enteric species, RNase E, actively degrades the rpoS mRNA and sRNA binding seems to inhibit this process (McCullen et al. 2010). Furthermore, double-stranded sequence elements in the rpoS 5′ UTR are substrates for RNase III. Notably, one of the RNase III cleavage sites is located close to the RBS of rpoS, and nucleolytic processing could reduce translation initiation. Base pairing with DsrA leads to an alternative processing event in the DsrA–rpoS duplex reducing cleavage close to the SD sequence, which indirectly promotes translation initiation (Resch et al. 2008). Concomitant decay of rpoS and DsrA by RNase III inactivates DsrA for further rounds of target regulation, which may be important for controlling DsrA levels and activity (Fig. 2).
Each of the sRNAs that activates rpoS is produced in response to a different stress signal, and the activities of the sRNAs enhance σS production to enable a response that promotes physiological adaptation or resistance to stress. Low temperatures (25°C) increase both the synthesis and stability of DsrA (Repoila and Gottesman 2001, 2003). At low temperatures, DsrA not only requires the activity of Hfq but also the DEAD box helicase CsdA for full activation of rpoS (Resch et al. 2010). Temperature control is also relevant for DsrA activity in the spirochete, Borrelia burgdorferi, causative agent of Lyme disease. At ∼300 nt, Borrelia DsrA is significantly longer than DsrA in enterobacteria and shares very little sequence similarity. However, it mimics the anti-antisense mechanism of rpoS activation in E. coli (Lybecker and Samuels 2007) and also requires an RNA-binding protein similar to Hfq (Lybecker et al. 2010). DsrA may also play a role in acid resistance (Lease et al. 2004), a phenotype that has also been reported for ArcZ and RprA and might relate to their common ability to improve stress resistance through rpoS activation (Gaida et al. 2013; Bak et al. 2014).
DsrA has been reported to regulate two targets in addition to rpoS, most prominently hns, for which it acts as a repressor (Lease, Cusick and Belfort 1998; Majdalani et al. 1998). H-NS is a global transcriptional regulator that controls hundreds of genes. Continuous overexpression of DsrA causes pleiotropic phenotypes including mucoid colony morphology, which is induced by DsrA-mediated reduction of H-NS levels and reduced repression of the H-NS target gene rcsA, encoding a positive regulator of colanic acid capsule expression (Sledjeski and Gottesman 1995). DsrA has also recently been implicated in repression of mreB mRNA, encoding the cell shape regulator MreB (Cayrol et al. 2015). The physiological relevance of putative DsrA-dependent control of cell shape is currently unclear.
Two other sRNAs that activate rpoS, RprA and ArcZ, both bind the rpoS 5′ UTR at positions overlapping the DsrA-binding site and both require Hfq for activity (Majdalani, Hernandez and Gottesman 2002; Soper et al. 2010). The rprA (RpoS regulator RNA) gene was discovered in a screen for post-transcriptional regulators of rpoS (Majdalani et al. 2001) and two additional target mRNAs (both repressed by RprA) have been discovered since then. One of the targets, the csgD mRNA, encodes a major transcriptional regulator of curli fiber and cellulose production in E. coli suggesting that the biological function of RprA might relate to biofilm formation (Jorgensen et al. 2012; Mika et al. 2012). Indeed, the second RprA target, ydaM mRNA, encodes a diguanylate cyclase which functions in concert with the transcriptional regulator MlrA to activate the csgD promoter (Mika and Hengge 2014). Intriguingly, transcription of ydaM itself is controlled by σS making RprA a central regulator of a three-step regulatory cascade: it activates σS production, which in turn leads to increased ydaM mRNA, the protein product of which promotes csgD transcription through increased cyclic di-GMP levels. Both csgD and ydaM are repressed targets, so RprA acts as an activator (through upregulation of rpoS) and repressor of biofilm production at the same time. This network arrangement might be driven by the global function of σS (Weber et al. 2005). By activating rpoS and repressing ydaM and csgD, RprA enhances certain phenotypes associated with σS activity (e.g. general stress response) and represses others, such as curli fiber and cellulose production. This specialization of σS activity through RprA is further supported by the transcriptional input parameters that drive RprA expression. The RcsC/RcsD/RcsB phosphorelay system (Majdalani, Hernandez and Gottesman 2002) and the CpxAR two-component system (Vogt et al. 2014) both activate RprA expression upon insults to the cell envelope. Membrane damage might well require the activity of the σS controlled stress response machinery but does not necessarily support the energy demanding production of curli fiber and cellulose.
ArcZ is one of the rare sRNAs that is post-transcriptionally processed. In Salmonella, overexpression of ArcZ causes pleiotropic phenotypes probably due to competition with other sRNAs for Hfq binding (Papenfort et al. 2009). Besides upregulation of rpoS, ArcZ negatively controls the tpx, sdaCB, STM3216, eptB and fhlDC mRNAs (Papenfort et al. 2009; De Lay and Gottesman 2012; Moon et al. 2013). How all these targets tie together physiologically is currently unclear. One clue might come from transcriptional regulation of arcZ. The ArcB/ArcA two-component system inhibits ArcZ expression under conditions of low oxygen. ArcB activity is controlled by the redox state of the respiratory chain and therefore the energy status of the cell (Malpica et al. 2004). In addition, the arcZ gene is localized in cis and antisense to arcB and acts to repress arcB mRNA levels (Mandin and Gottesman 2010). Repression of arcB by ArcZ therefore reinforces production of σS by a dual mechanism. First, ArcB phosphorylates and activates ArcA, which acts as a transcriptional repressor of rpoS. Secondly, there is some evidence suggesting that ArcB can phosphorylate RssB, a proteolytic targeting factor that labels σS for degradation by the ClpXP protease (Mika and Hengge 2005), though the impact of RssB phosphorylation on in vivo activity remains enigmatic. Mutation of arcZ affects biofilm formation in Salmonella and E. coli (Monteiro et al. 2012; Mika and Hengge 2014), further underscoring the diverse and interesting physiological functions associated with this sRNA.
Others: RyhB, Qrrs and PhrS
Since the discovery of the anti-antisense mechanism for RNAIII pairing with hla and the large body of information that has come from studies of the rpoS 5′ UTR, several other mRNAs have been reported to be activated by a similar mechanism. The iron starvation induced RyhB from E. coli and other enterobacteria negatively regulates many genes that influence iron homeostasis in the cell (Salvail and Masse 2012). In contrast, RyhB activates translation of shiA mRNA through an anti-antisense mechanism (Prevost et al. 2007). The shiA mRNA encodes a permease that imports shikimate, a precursor for siderophore production. In the uropathogenic E. coli strain CFT073, ryhB-deficient cells display reduced siderophore production which compromises bladder colonization in a murine model of urinary tract infection (Porcheron et al. 2014).
The Qrr sRNAs are best known for their role in quorum sensing in Vibrio species (Ng and Bassler 2009). These sRNAs belong to the group of ‘sibling sRNAs’ which come in more than one copy per genome (Caswell, Oglesby-Sherrouse and Murphy 2014). In Vibrio harveyi, Qrr1–5 directly regulate ∼20 mRNAs via base pairing interactions (Shao et al. 2013). Moreover, Qrr-mediated repression of the master transcriptional regulator of high-cell-density behavior, luxR (hapR in V. cholerae) (Lenz et al. 2004) and activation of the mRNA encoding for the low-cell-density transcription factor, AphA (Rutherford et al. 2011), probably accounts for most of the phenotypes associated with these sRNAs. Activation of aphA involves an anti-antisense mechanism requiring base pairing to a specific sequence in the first stem loop of the Qrr sRNAs which is present in Qrr2–4 but not in Qrr1 (Shao and Bassler 2012). Activation of aphA by Qrr sRNAs has a large impact on the quorum-sensing dynamics in Vibrio as base pairing with the first Qrr stem loop facilitates degradation of the sRNAs and transition to high-cell-density behaviors (Feng et al. 2015). In V. cholerae, the Qrr sRNAs also activate the vca0939 mRNA through an anti-antisense mechanism (Hammer and Bassler 2007). The vca0939 gene encodes a GGDEF protein involved in cyclic-di-GMP synthesis and activation of vca0939 by the Qrr sRNAs promotes biofilm formation in V. cholerae (Zhao et al. 2013).
A variation on the canonical anti-antisense theme has been discovered in Pseudomonas aeruginosa. Here, the PhrS sRNA activates translation of the pqsR mRNA by interaction with a sequence in the 5′ UTR of an upstream open reading frame (uORF) that precedes the coding sequence of pqsR. In the absence of PhrS, the SD sequence of the uORF is sequestered in a secondary structure. PhrS pairing with the 5′ UTR enhances translation of the uORF, which in turn facilitates translation of pqsR (Sonnleitner et al. 2011). The PqsR protein encodes a critical activator of pyocyanin production, a redox-active pigment and a toxin of P. aeruginosa (Diggle et al. 2006). Production of the PhrS sRNA is increased in stationary phase and requires the oxygen responsive transcriptional regulator ANR (Sonnleitner, Abdou and Haas 2009; Sonnleitner et al. 2011). Therefore, PhrS in concert with ANR connects oxygen concentrations with virulence factor production in P. aeruginosa.
mRNA stabilization through sRNA binding at the 5′ UTR
Activation by FasX RNA in Streptococcus
The ska gene, encoding streptokinase A (SKA) is highly regulated at transcriptional (Levin and Wessels 1998) and post-transcriptional (Kreikemeyer et al. 2001; Ramirez-Pena et al. 2010) levels, reflecting its important role in the pathogenesis of group A Streptococcus (GAS) (Svensson et al. 2002; Sun et al. 2004; McArthur et al. 2008). SKA activates host plasminogen into plasmin (Svensson et al. 2002), which is a protease that dissolves the fibrin meshwork in a blood clot. SKA thus likely plays an important role in the transition from localized infection to systemic spread of GAS. The ∼200-nt FasX sRNA is an important post-transcriptional activator of ska mRNA (Kreikemeyer et al. 2001; Ramirez-Pena et al. 2010). FasX base pairs with 9 nts at the extreme 5′ end of the ska mRNA (within the 32-nt 5′ UTR), and this interaction stabilizes the ska mRNA (Fig. 1B) (Ramirez-Pena et al. 2010). In the absence of FasX, SKA activity falls to only 12% of the levels seen in wild-type (fasX+) cells, demonstrating the crucial role this sRNA plays in activating synthesis of SKA. The underlying mechanism of enhanced ska mRNA stability upon binding FasX has not yet been discovered. Neither CvfA (an endoribonuclease) nor PNPase (an exoribonuclease) play a role in turnover of ska mRNA (Ramirez-Pena et al. 2010) and other nucleases involved in ska mRNA stability have not been identified. Moreover, the role of translation in ska mRNA stability is not understood. It is possible that FasX could remodel the ska mRNA 5′ UTR to directly enhance translation and therefore mRNA stability. Alternatively, FasX could inhibit ska mRNA decay mediated by as-yet-unidentified ribonucleases. Additional experiments are required to discern between these two alternative mechanisms.
In addition to ska, FasX was shown to regulate another virulence determinant of GAS, namely, the collagen-binding pili (Liu et al. 2012). In this case, the outcome of base pairing between FasX and its target mRNA is translational repression and mRNA degradation. The pilus biogenesis locus is comprised of five genes (cpa, sipA1, tee1, srtC1 and 111) encoded on a single transcript. The polycistronic mRNA has only a 13-nt 5′ UTR and FasX base pairs with sequences overlapping the RBS of the first gene, cpa. This base pairing interaction inhibits cpa translation and promotes turnover of the entire transcript (Liu et al. 2012). The fasX mutant adheres more tightly than the wild-type strain to epithelial cells in tissue culture, consistent with the idea that FasX has a repressive effect on biogenesis of pili, an important adhesion factor.
Transcription of fasX is controlled by the products of genes encoded upstream, fasBCA, which have homology to sensor kinases (fasBC) and a response regulator (fasA) (Kreikemeyer et al. 2001). The environmental signals or conditions required for induction of fasX transcription are not yet known. Nevertheless, the current data strongly suggest that FasX plays a critical role in the regulation of GAS virulence.
Activation by Clostridium VR-RNA
An sRNA regulator also controls synthesis of a key virulence factor in the Gram-positive spore-forming pathogen Clostridium perfringens, the causative agent of gas gangrene and food poisoning in humans. The synthesis of the 386-nt VR-RNA in C. perfringens is controlled by a key two-component system, VirR–VirS, that acts as a global regulator of virulence in this organism (Shimizu et al. 1994; Okumura et al. 2008; Ohtani et al. 2010). VR-RNA has been shown to regulate expression of several genes encoding virulence factors and metabolic functions (Shimizu et al. 2002; Kawsar et al. 2004; Yuan et al. 2012), consistent with the idea that this sRNA plays a crucial role in C. perfringens physiology and pathogenesis. VR-RNA has a positive effect on the stability of colA mRNA, which encodes collagenase, a collagen-degrading toxin produced by C. perfringens (Obana et al. 2010). The colA mRNA has a long 5′ UTR that folds into a stem-loop structure that masks the SD sequence and inhibits translation (Fig. 1C) (Obana, Nomura and Nakamura 2013). This RNA structure has an internal single-stranded bulge that interacts with VR-RNA and prevents formation of the translation-inhibitory structure. The VR-RNA–colA mRNA interaction promotes processing within the 5′ UTR to remove the upstream portion of the stem-loop. Processed colA mRNA is therefore more accessible to ribosomes, which in turn increases mRNA stability (Fig. 1C) (Obana et al. 2010; Obana, Nomura and Nakamura 2013). Thus, for this example of sRNA-mediated gene activation, mRNA stability is enhanced via a processing event that is dependent on sRNA–mRNA base pairing interactions and consequentially improved translation of the processed mRNA.
Activation by inhibiting nucleolytic decay
Many mechanisms of mRNA activation primarily serve to increase the rates of translation initiation by increasing accessibility of an mRNA's RBS. These include the anti-antisense type mechanisms (Fig. 1A) and the mRNA processing-dependent mechanism carried out by VR-RNA (Fig. 1C). In contrast, there are now a few examples of sRNA-mediated activation that work directly at the level of modulating mRNA susceptibility to degradation by ribonucleases. One example is the RydC sRNA, an Hfq-binding sRNA with an unusual pseudoknot structure (Antal et al. 2005) found in a few enteric species including Escherichia, Salmonella, Citrobacter, Enterobacter, Klebsiella and Shigella (Fröhlich et al. 2013). This pseudoknot structure was recently confirmed in the crystal structure of RydC in complex with Hfq (Dimastrogiovanni et al. 2014). Here, the 3′ end of RydC interacts with the proximal face of Hfq via the U-rich sequences of the Rho-independent terminator, while sequences in the 5′ region of RydC interact with Hfq residues on the lateral surface of the hexamer.
RydC carries out base pairing-dependent activation of cfa mRNA (Fig. 1D). Inducing synthesis of RydC causes rapid accumulation of cfa mRNA and subsequently increased levels of Cfa protein (Fröhlich et al. 2013). The cfa gene has two promoters (Wang and Cronan 1994; Kim et al. 2005), which produce cfa transcripts with different 5′ UTRs. A σ70-dependent promoter yields a cfa transcript with a long 210-nt 5′ UTR. A shorter transcript with a 33-nt 5′ UTR is produced from a σS-dependent promoter (Cronan 2002). RydC activates only the longer transcript because the base pairing site for RydC is contained only within the longer cfa 5′ UTR (Fröhlich et al. 2013). RydC positively regulates cfa by base pairing with an RNase E cleavage site in the cfa 5′ UTR and inhibiting RNase E-dependent decay. In a bacterial host where RNase E is inactivated, basal levels of the long cfa transcript increase in a RydC-independent manner, and in this host RydC provides no further enhancement of cfa levels (Fröhlich et al. 2013). Moreover, in wild-type cells, the RydC-mediated increase in cfa mRNA levels is not dependent on cfa translation. Thus, these data indicate that RydC and cfa mRNA constitute an sRNA–mRNA pair where regulation does not rely on changes in the efficiency of mRNA translation.
In addition to regulation of cfa, RydC has also been reported to exert post-transcriptional control of an ABC transporter of unknown substrate specificity, yejABEF (Antal et al. 2005) and the curli gene transcription factor, csgD (Bordeau and Felden 2014). For both of these targets, RydC has a repressive effect on mRNA translation or stability. The physiological signals controlling RydC synthesis remain a mystery, and thus there is currently no clear biological rationale for RydC function. Nevertheless, it is clear that RydC represents yet another example of a bacterial sRNA regulator that carries out both negative and positive regulatory effects.
Activation by base pairing within the mRNA-coding sequence
Activation by the sugar stress RNA SgrS
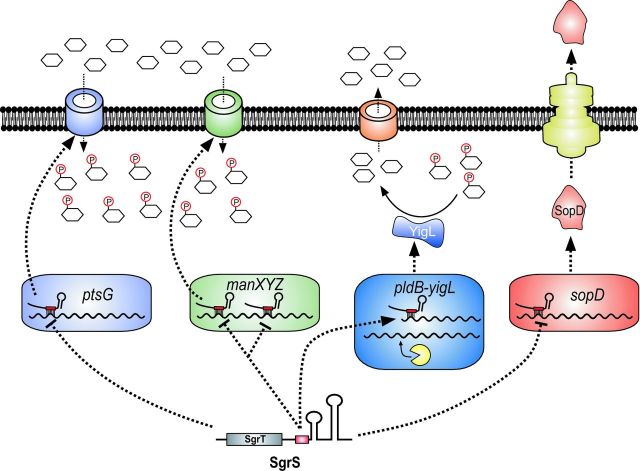
The sRNA SgrS has served as a valuable model for uncovering novel mechanistic aspects of sRNA-based regulation as well as understanding how sRNAs participate in controlling bacterial physiology during stress. SgrS is ∼220 nt long and is found in E. coli, Salmonella and other enteric species (Zhang et al. 2003; Vanderpool and Gottesman 2004; Horler and Vanderpool 2009). SgrS, like RNAIII, is a dual-function RNA regulator (Wadler and Vanderpool 2007; Balasubramanian and Vanderpool 2013). It acts by Hfq-dependent base pairing to post-transcriptionally regulate mRNA targets and produces a small (∼40 aa) protein, SgrT (Fig. 3). SgrS has thus far been demonstrated to regulate four mRNA targets. Three of these targets, ptsG (Vanderpool and Gottesman 2004; Kawamoto et al. 2006), manXYZ (Rice and Vanderpool 2011; Rice, Balasubramanian and Vanderpool 2012) and sopD (Papenfort et al. 2012) are repressed by SgrS at the level of translation and mRNA stability (Fig. 3). The fourth target of SgrS is yigL (Papenfort et al. 2013), encoding a haloacid dehalogenase-like phosphatase (Mustelin 2007). The yigL gene is encoded in a bicistronic operon with the upstream gene pldB. SgrS mediates a ‘discoordinate’ mechanism of regulation of this operon, as it activates only yigL and not pldB (Papenfort et al. 2013). The pldB–yigL mRNA is processed by RNase E, yielding an mRNA species truncated within pldB (‘pldB–yigL) (Papenfort et al. 2013). This processing is SgrS-independent, and is a prerequisite for SgrS activation of yigL. SgrS base pairs with a site in the 3′ region of the pldB-coding sequence and stabilizes the ‘pldB-yigL mRNA. Enhanced stability is independent of the translation of yigL and is due to SgrS sequestration of an RNase E recognition sequence (Papenfort et al. 2013).
Figure 3.
SgrS regulates carbon metabolism and virulence. SgrS represses three mRNA targets, ptsG, manXYZ and sopD, and activates one mRNA target, yigL mRNA. Activation of yigL depends on base pairing of SgrS with the pldB ORF preventing mRNA degradation through RNase E. The result of these activities is inhibition of sugar uptake (via repression of ptsG and manXYZ) and activation of sugar efflux (via activation of yigL), which promotes recovery from glucose-phosphate stress.
At the physiological level, SgrS is needed under conditions known as glucose-phosphate stress (Vanderpool and Gottesman 2004; Vanderpool 2007); these are conditions in which intracellular sugar-phosphate concentrations increase due to mutations in glycolytic genes or exposure of cells to non-metabolizable glucose analogs (Kimata et al. 2001; Morita et al. 2003; Vanderpool and Gottesman 2004). The repressed targets ptsG and manXYZ encode sugar transporters of the phosphoenolpyruvate phosphotransferase system and SgrS-dependent repression of these genes reduces new synthesis of transporters in order to limit further accumulation of sugar phosphates. The benefit to the cell of SgrS activation of YigL synthesis is enhanced capacity for dephosphorylation of accumulated sugar phosphates so that they can be pumped out of the cell cytoplasm. Collectively, base pairing-dependent regulation by SgrS allows cells to inhibit uptake and promote efflux of sugars under conditions where their metabolism is inhibited. Interestingly, the virulence factor encoding sopD mRNA is a repressed target of SgrS in Salmonella (Papenfort et al. 2012), suggesting that SgrS links carbon metabolism and virulence in this important pathogen (Papenfort and Vogel 2014).
Target stabilization by long antisense RNAs
Most trans-acting sRNAs are dedicated non-coding regulators that repress and activate gene expression through a variety of mechanisms. In a recent study from the dental caries pathogen Streptococcus mutans, Liu et al. (2015) demonstrated that the 5′ UTR of the mRNA irvA can act similarly through base pairing with the coding sequence of the gbpC mRNA. RNA-duplex formation with irvA protects gbpC from degradation by RNase J2, which is one of two (the other being RNase J1) major ribonucleases of most firmicutes (Condon 2010; Lehnik-Habrink et al. 2012). Activation of gbpC by irvA has also important physiological consequences, as GbpC is a central factor for cell aggregation and biofilm formation in S. mutans (Idone et al. 2003).
Base pairing-mediated regulation by a long RNA is also relevant during bacteriophage infection of the cyanobacterium Prochlorococcus MED4. Recent studies revealed an operon encoding three proteins of unknown function, and a ribosomal protein-encoding operon along with corresponding long antisense RNAs transcribed from the strand opposite the protein-coding genes (Steglich et al. 2008) that were all induced upon phage infection (Lindell et al. 2007; Steglich et al. 2008; Stazic, Lindell and Steglich 2011). In vitro analyses of Prochlorococcus mRNAs and antisense RNAs demonstrated that the protein-coding mRNAs are susceptible to RNase E-mediated degradation. The antisense RNAs base pair with the mRNAs across their whole length, altering their secondary structures and protecting them from RNase E-mediated decay (Stazic, Lindell and Steglich 2011). A comprehensive study comparing the transcriptomes of two Prochlorococcus strains found evidence for antisense transcription across ∼75% of all genes (Voigt et al. 2014), suggesting that RNA-based mechanisms of regulation depending on interactions within mRNA-coding sequences may be widespread in these organisms.
Activation by base pairing at the 3′ end of target mRNAs
Most bacterial small RNAs characterized thus far are encoded ‘in trans’ to the genes encoding their targets and are dependent on the RNA chaperone Hfq for activity. The 105-nt GadY sRNA differs from other sRNAs with respect to both of these characteristics. GadY is encoded in the gadX–gadW intergenic region and is cis-antisense to a region just outside the 3′ end of gadX (Fig. 4) (Opdyke, Kang and Storz 2004). This bicistronic operon encodes two transcription factors, GadX and GadW, which are involved in a complex regulatory circuit controlling the glutamate-dependent acid resistance system in E. coli (Ma et al. 2002). An initial study characterizing GadY demonstrated that induction of GadY synthesis activates gadX, increasing gadX mRNA levels (Opdyke, Kang and Storz 2004). Because GadY is encoded on the opposite strand from its target, the sRNA and mRNA target are fully complementary. Extensive base pairing between GadY and the gadX–gadW mRNA promotes processing within the complementary region by RNase III, a double-strand specific ribonuclease, and other as yet unidentified nucleases in vivo (Fig. 4) (Opdyke et al. 2011). The processing event yields separate gadX and gadW transcripts that accumulate to higher levels than the unprocessed transcript, presumably because the processed fragments are more stable (Opdyke et al. 2011). The molecular mechanism underlying the differential stability of full length versus processed gadX–gadW mRNA has not yet been uncovered.
Figure 4.
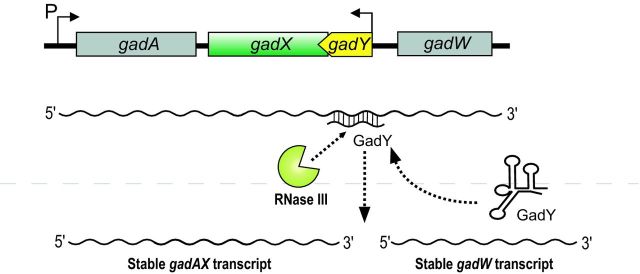
Regulation at the 3′ end through GadY. The GadY sRNA is encoded in cis and antisense to gadX mRNA. GadY base pairing interaction with the gadX–gadW mRNA promotes RNase III-dependent processing, yielding individual gadX and gadW transcripts that are more stable than the full-length mRNA.
Indirect mechanisms of target activation
The CsrA regulon
Post-transcriptional activation of transcripts can also occur indirectly, e.g. through mechanisms that do not require direct base pairing of sRNAs with their targets. In these cases, globally acting RNA-binding proteins, such as Hfq and CsrA often play important roles. Orthologs of CsrA, a global translational regulator in E. coli and Salmonella, have been discovered in many bacteria and some species encode multiple paralogous regulators (Lapouge et al. 2008). CsrA-like proteins typically bind to GGA-rich elements in RNAs. This sequence resembles the canonical SD sequence (AGGAGGU) of bacterial mRNAs and CsrA binding to the SD typically results in reduced translation and subsequent mRNA decay (Romeo, Vakulskas and Babitzke 2013). In few cases, CsrA activates target mRNA translation. For example, interaction of CsrA with the fhlDC mRNA in E. coli protects the transcript from degradation by RNase E (Yakhnin et al. 2013) by a mechanism that is independent of any auxiliary sRNAs. Similarly, CsrA also has a positive effect on the moaA mRNA, which is regulated by a molybdenum cofactor controlled riboswitch (Patterson-Fortin et al. 2013).
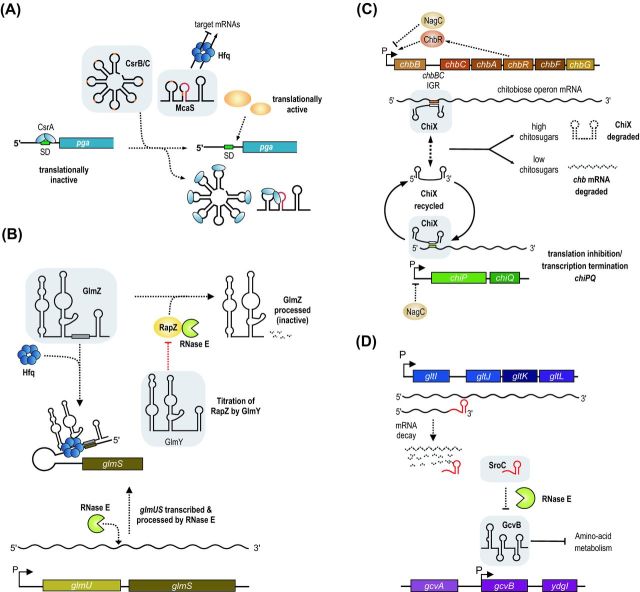
Key regulators of CsrA function are the CsrB-like sRNAs (Babitzke and Romeo 2007). These sRNAs contain multiple high-affinity binding sites for CsrA and act to titrate the protein away from mRNA targets (Duss et al. 2014). Therefore, these sRNAs indirectly activate CsrA-repressed genes by antagonizing CsrA activity (Fig. 5A). Similar to CsrA, genes encoding CsrB-like sRNAs are often found in multiple copies per genome and serve redundant functions. Due to the global regulatory function of CsrA, mutation of the genes encoding CsrB-like sRNAs often cause complex phenotypic changes including pathogenicity (Romeo, Vakulskas and Babitzke 2013). For example, CsrA inhibits virulence gene expression in Salmonella through interaction with the mRNA encoding the master virulence regulator, HilD. Induction of the CsrB and CsrC sRNAs can titrate CsrA from hilD and promote virulence (Martinez et al. 2011).
Figure 5.
Indirect mechanisms of activation by bacterial sRNAs. (A) Titration of a repressor. CsrB, CsrC and McaS bind the translational regulatory protein CsrA to inhibit its activity and relieve repression of CsrA target genes. (B) Decoy RNAs. GlmY acts as a decoy for the active sRNA regulator GlmZ and binds the adaptor protein RapZ, which would otherwise promote RNase E-dependent processing and inactivation of GlmZ. In the presence of GlmY, GlmZ is able to accumulate and activate glmS to promote synthesis of GlcN6P synthase. (C and D) Sponge RNAs. (C) In the presence of chitosugars, chbBCARFG mRNA is produced and sequences in the intergenic region act as a sponge for ChiX sRNA, sequestering it away from another target, chiP mRNA, encoding the chitoporin. The net effect is activation of chitoporin synthesis in the presence of chitosugars. (D) SroC sRNA is a decay product of polycistronic gltIJKL mRNA and base pairs with GcvB sRNA to promote its turnover by RNase E. The activity of SroC results in activation of GcvB-repressed targets.
In E. coli and Salmonella, transcription of CsrB-like sRNAs is controlled by the BarA/UvrY two component system and its orthologs in other species (Seyll and Van Melderen 2013). In many cases, the specific signals for activation of this signaling pathway are currently unknown; however, metabolites of central carbon utilization pathways such as short-chain fatty acids, formate or acetate seem to play a role (Takeuchi et al. 2009; Chavez et al. 2010). The expression of CsrB-like sRNAs is also controlled at the post-transcriptional level: the E. coli CsrD protein can bind the CsrB/C sRNAs and target them for degradation by RNase E (Suzuki et al. 2006). This regulation is part of a feedback loop, since CsrA also inhibits the production of CsrD (Jonas et al. 2008).
A recent addition to the list of the CsrA-binding RNAs is the McaS sRNA from E. coli. McaS is different from the CsrB-like sRNAs as it can act to titrate CsrA but also as a base pairing sRNA through Hfq (Fig. 5A). Hfq- and base pairing-dependent regulation results in activation of fhlD mRNA through an anti-antisense mechanism (Fig. 1A) and repression of the csgD mRNA (Jorgensen et al. 2012; Thomason et al. 2012). In addition, McaS also activates expression of the pgaA gene, encoding a porin required for the export of the adhesin PGA (poly-beta-1,6-N-acetyl-d-glucosamine) and therefore biofilm formation (Itoh et al. 2008). The pgaABCD 5′ UTR harbors six CsrA binding sites, two of which overlap the SD sequence and the start codon. Accordingly, CsrA inhibits translation of the pgaA mRNA (Wang et al. 2005). The McaS sRNA was discovered to titrate the CsrA protein via two exposed GGA motifs and thereby indirectly activate the expression of many CsrA-regulated target genes, including pgaABCD (Fig. 5A) (Jorgensen et al. 2013). McaS is the first sRNA discovered to impact both Hfq and CsrA regulons. Given the relatively relaxed parameters for CsrA binding (one or more GGA motifs), it can be expected that more sRNAs known to regulate mRNAs through Hfq also function as CsrA antagonists. CsrA also regulates the hfq mRNA (Baker et al. 2007), suggesting that these two major post-transcriptional networks could be more connected than previously anticipated, although additional experiments will be required to support this hypothesis.
Molecular mimicry through GlmZ and GlmY
The GlmY sRNA from E. coli and related enterobacteria functions as a sort of decoy RNA. Overexpression of GlmY activates the glmS mRNA (Urban et al. 2007), the protein product of which is glucosamine-6-phosphate (GlcN6P) synthase, an important enzyme for cell envelope synthesis. The glmS mRNA is part of the bicistronic glmUS transcript, which is controlled by the NagC transcriptional regulator in response to exogenous amino sugars (Plumbridge 1995). RNase E cleaves the glmUS transcript at the stop codon of glmU (Kalamorz et al. 2007), which leaves a translationally inactive glmS mRNA due to an intrinsic inhibitory structure within its 5′ UTR (Fig. 5B). Translation repression can be alleviated by base pairing of the GlmZ sRNA to the 5′ UTR of glmS-activating gene expression through an anti-antisense mechanism (Reichenbach et al. 2008; Urban and Vogel 2008). GlmZ and GlmY are highly related sRNAs but only GlmZ binds Hfq and base pairs with glmS mRNA. So, how does GlmY activate glmS? The key to answering this question was the identification of the RNA-binding protein, RapZ (a.k.a. YhbJ). RapZ is a specialized adapter protein that binds GlmZ and targets it for degradation by RNase E (Fig. 5B). Recognition of GlmZ by RapZ is directed by a RNA element conserved between GlmZ and GlmY. At high levels, GlmY can act as a molecular mimic and titrate the RapZ protein from GlmZ (Fig. 5B) (Gopel et al. 2013). In addition, GlmY is also regulated through polyadenylation by PAPI (encoded by the pcnB gene) (Reichenbach et al. 2008; Urban and Vogel 2008).
Two overlapping promoters control transcription of glmZ and glmY. In both cases, recognition sequences for σ70- and σ54-dependent transcription have been discovered in the promoters of the sRNAs (Urban et al. 2007; Reichenbach, Gopel and Gorke 2009). Importantly, the promoter architecture allows that transcription initiation by σ70 and σ54 occurs at the same position producing identical GlmY and GlmZ 5′ ends. Expression by σ54 is further dependent on the two-component system GlrK/GlrR (a.k.a. QseF/QseE), which are encoded by genes immediately downstream of glmY (Reichenbach, Gopel and Gorke 2009; Gopel et al. 2011). The QseE/F two-component system responds to quorum-sensing molecule AI-3, as well as the host signals epinephrine and norepinephrine and is involved in virulence of enterohemorrhagic E. coli (EHEC) (Reading et al. 2007). Together with a second two-component system (QseB/C), QseE/F controls the expression of espFU, encoding a translocated virulence factor (Hernandez-Doria and Sperandio 2013). Both, QseB/C and QseE/F also modulate virulence through GlmY and GlmZ; however, this function is independent of glmS activation. Instead, both sRNAs affect the expression of EspFu and other genes from the EHEC pathogenicity islands LEE4 and LEE5 (Gruber and Sperandio 2014).
Changing RNA structure through Hfq
Hfq is a ubiquitous RNA-binding protein that accumulates to high levels. Estimates of the number of Hfq hexamers per cell vary from ∼400 (Carmichael et al. 1975) to ∼5000–10 000 (Kajitani et al. 1994; Ali Azam et al. 1999). Nevertheless, Hfq is a limiting factor for sRNA-mediated regulation in the cell (Wagner 2013). Besides the ∼100 Hfq-binding sRNAs found in a typical enterobacterial genome, Hfq also binds ∼1,250 different mRNA species in Salmonella (Chao et al. 2012). Certain sRNAs, such as the OxyS from E. coli (Altuvia et al. 1997) or VqmR from V. cholerae (Papenfort et al. 2015) can accumulate to >500 copies per cell which could have an indirect effect on Hfq binding of other sRNAs or mRNAs. In fact, overexpression of sRNAs has been reported to disrupt sRNA-mediated gene expression through Hfq titration (Papenfort et al. 2009; Hussein and Lim 2011; Moon and Gottesman 2011).
Binding of Hfq to 5′ UTRs of mRNAs often has a repressive effect on translation (Desnoyers and Masse 2012). This function of Hfq has important implications for genome stability as it also controls the expression of the transposable elements Tn5 and Tn10 (Ross, Wardle and Haniford 2010; Ross et al. 2014). The cirA mRNA of E. coli is repressed by Hfq in the absence of its sRNA regulator, RyhB. Similar to ompA mRNA (Vytvytska et al. 2000), Hfq binding to cirA represses translation through RBS sequestration and facilitates RNase E-mediated decay. RyhB was previously reported to activate cirA expression but the underlying mechanism remained unclear (Masse, Vanderpool and Gottesman 2005). Recently, Salvail et al. (2013) revealed that base pairing of RyhB to the 5′ UTR of cirA promotes structural rearrangements in the mRNA that displace Hfq from the RBS and indirectly facilitate translation initiation. Base pairing of RyhB to cirA occurs far upstream (–58 to –41 relative to translation start) of canonical translational control elements, which is comparable to the RydC-mediated activation of cfa (Fröhlich et al. 2013 and see above). However, base pairing far upstream of the RBS is not automatically an indicator of a translational activation mechanism: the GcvB sRNA inhibits gltI translation by base pairing with a C/A-rich translational enhancer element located at positions –57 to –45 relative to the start codon (Sharma et al. 2007). Hence, the details of the translational control mechanism of the mRNA itself, rather than the location of the binding site for a base pairing sRNA, seem to determine if a target mRNA is activated or repressed. Given that many sRNAs repress some genes and activate others, this feature could be key to the versatile functions of sRNAs.
RNA ‘sponges’ for repressor sRNAs: ChiX and SroC
Repression of a repressor results in activation of gene expression. This concept, which is common in transcription factor networks, has also been recognized for bacterial sRNAs (Beisel and Storz 2010). The ChiX sRNA is part of the chitin utilization network of many enterobacterial species. Uptake of chitosugars requires transport across the outer membrane by the chitoporin, ChiP followed by translocation into the cytoplasm by the ChbBCA transporter (Figueroa-Bossi et al. 2009). The transcriptional regulator NagC represses transcription of the chbBCARFG and chiPQ operons when chitosugars are scarce (Fig. 5C). In addition, the chiP mRNA is repressed by base pairing with the ChiX sRNA; a dual process which involves a translation repression (Figueroa-Bossi et al. 2009; Rasmussen et al. 2009) and Rho-dependent premature transcription termination (Bossi et al. 2012). Expression of ChiX is constitutive and base pairing with the chiP mRNA does not cause degradation of ChiX (Fig. 5C). This mechanism is contrary to the coupled degradation mechanism of many other sRNAs (Masse, Escorcia and Gottesman 2003) and allows efficient repression of chiP under conditions of low chitosugars (Figueroa-Bossi et al. 2009; Overgaard et al. 2009). The ChiX sRNA also base pairs with the intergenic sequence between the chbB–chbC cistrons on the chbBCARFG polycistronic mRNA, which can have two different outcomes. In the absence of chitosugars, ChiX inhibits translation of chbCARFG and reduces chitosugar transport (Plumbridge et al. 2014). This regulatory pattern reinforces repression of chitosugar utilization genes. Relief of NagC repression at the chbB promoter through increasing intracellular concentrations of N-acetylglucosamine-6-phosphate provides the basis for a switch in the system. Induction of the transcription of chbBCARFG orchestrates a feed-forward loop in which ChbR activates the expression of its own operon. At high levels, the chbB–chbC intergenic region no longer functions as a target of ChiX but acts as a sponge that sequesters ChiX and therefore indirectly activates ChiP production (Fig. 5C) (Figueroa-Bossi et al. 2009; Overgaard et al. 2009). Combined expression of ChiP and Chb proteins will further trigger the system loop and facilitate chitosugar utilization. Although ChiX generally employs the same base pairing region to bind the chiP and chbB-C mRNAs, the ChiX-chbB-C interaction engages two additional nucleotides of the 3′ terminal stem of ChiX, which could lead to partial opening of the stem loop and promote 3′ exonucleolytic decay (Figueroa-Bossi et al. 2009). Dependent on the availability of chitosugars, the chbB-C intergenic region can either function as a repressed target or act like a sponge for ChiX. This mechanism is similar to target regulation by the Qrr sRNAs from Vibrio species. Here, base pairing of target mRNAs to a 5′ terminal stem loop in the Qrrs results in destabilization and degradation of the sRNAs; however, different from ChiX, this process seems to involve the activity of RNase E (Shao et al. 2013; Feng et al. 2015).
Another sRNA sponge of E. coli and Salmonella is produced from the intergenic region of the gltI and gltJ genes (Fig. 5D). Genome-wide searches for regulatory RNAs in E. coli initially identified this stable, ∼160-nt mRNA breakdown product which was named SroC (Vogel et al. 2003). Different from most RNA degradation intermediates, SroC tightly associates with Hfq (Sittka et al. 2008; Chao et al. 2012; Tree et al. 2014), spurring the prediction that it may act as a trans-acting sRNA. Indeed, microarray-based searches for SroC target genes revealed an unusually high number of 26 activated mRNAs (Miyakoshi, Chao and Vogel 2015a). How can one sRNA activate so many genes? Besides the activated genes, SroC also repressed 14 targets, the strongest of which was the GcvB sRNA. GcvB is a well-known repressor of mRNAs-encoding proteins involved in amino-acid metabolism and regulates ∼1% of all genes in Salmonella (Urbanowski, Stauffer and Stauffer 2000; Sharma et al. 2007, 2011). Hence, SroC might indirectly activate target genes by modulating the activity of GcvB. In a series of elegant genetic and biochemical experiments, Miyakoshi, Chao and Vogel (2015a) showed that SroC employs two exposed base pairing regions to interact with GcvB, a process which results in degradation of GcvB and upregulation of target mRNAs (Fig. 5D). Indeed, 14 of the activated SroC targets genes were previously reported to be repressed by GcvB (Sharma et al. 2011; Miyakoshi, Chao and Vogel 2015a). The recent discovery of the bacteriophage encoded AgvB sRNA from EHEC could add another layer of complexity to the system. Similar to SroC, AgvB also targets the GcvB sRNA, however, the base pairing regions differ from the GcvB–SroC interaction and AgvB overexpression does not facilitate GcvB degradation (Tree et al. 2014). It is currently unclear how AgvB is controlled and if physiological levels of AgvB can affect GcvB activity.
Both sponge sRNAs, i.e. chbB/C and SroC, share certain commonalities: both reside in the intergenic region of mRNAs and both repress sRNAs through direct base pairing followed by RNA decay. In contrast to chbB/C, SroC accumulates as a stable sRNA and associates with Hfq. Expression of SroC depends on additional factors such as RNase E, which is required to separate SroC from the gltI mRNA. It is interesting to note that the gltI mRNA itself is also a target of GcvB (Sharma et al. 2007). It is not understood how regulation of gltI translation by GcvB affects SroC production, but given the complexity of regulation observed in the ChiX-controlled network of chitosugar utilization, one can speculate that SroC together with GcvB orchestrate an elaborate feed-forward loop that optimizes amino acid metabolism in the cell.
Future perspectives
The mRNA targets themselves may provide the best starting point for further study of activation by sRNAs. As the examples discussed in this review illustrate, there is no defining sRNA characteristic that makes it an activator rather than a repressor; the vast majority of sRNAs that have been characterized thus far act as both depending on the target. Even the position of the sRNA-binding site on the mRNA does not dictate whether the regulation is positive or negative. Instead, the molecular details governing mRNA translation and stability determine whether and how the mRNA is regulated by sRNAs. Thus, future studies would do well to focus on mRNAs of interest for which there are hints of uncharacterized post-transcriptional regulation. RNA-seq analyses are increasingly being used to globally define transcriptional start sites (TSS), and these may reveal transcripts with unusually long 5′ or 3′ UTRs that could well be sites for regulation by sRNAs. For example, recent work defining TSS in V. cholerae revealed that the rpoS transcript has a long 5′ UTR (Papenfort et al. 2015), analogous to the E. coli and Salmonella rpoS 5′ UTR, yet no sRNAs have thus far been identified as activators of rpoS in Vibrio. These observations yield the obvious and easily testable hypothesis that new examples of sRNA-mediated activation of rpoS await discovery in Vibrio species.
Many aspects of activation by sRNAs will necessarily differ from the well-characterized mechanisms of repression. Among the cases highlighted here, there are different requirements for ribonucleases, e.g. RNase E and RNase III, as well as involvement of specific adaptors. The involvement of RapZ in modulating the function of GlmZ and GlmY and of CsrD in controlling activity of CsrB may represent a more common scenario than is currently appreciated. There is clear evidence for involvement of nucleases in addition to RNase III in modulating GadY-dependent activity on the gadX–gadW mRNA (Opdyke et al. 2011). None of the currently known ribonucleases were responsible for the observed RNase III-independent regulation, indicating the involvement of new uncharacterized nucleases. Additional protein cofactors involved in sRNA-mediated activation will likely be identified by detailed studies of individual regulatory examples.
Although examples of sRNA-mediated repression currently far outnumber the confirmed cases of positive regulation, the growing list of activated targets (including some described in this review) along with evidence from global target searches suggests that activation by sRNAs is a vastly undercharacterized phenomenon. Dozens of studies have approached sRNA target identification using transcriptome approaches, e.g. microarrays or RNA-seq, to globally examine transcript abundance after induction of a small RNA of interest. For RyhB (Masse, Vanderpool and Gottesman 2005), SgrS (Papenfort et al. 2012) and RydC (Fröhlich et al. 2013), these experiments lead to the identification of activated mRNA targets (see above), while other sRNAs such as GcvB (Sharma et al. 2011), MicF (Corcoran et al. 2012), MicL (Guo et al. 2014) and MicC (Pfeiffer et al. 2009) seem to act exclusively through repression. In other cases, such experiments revealed activated target mRNAs; however, these have not been validated through additional analyses. For example, microarray analysis of RybB pulse expression indicated activation of ipbAB, adiY and yhbT (Papenfort et al. 2006), the transcripts of which all contain long 5′ UTRs (93, 92 and 102 nt, respectively; Kroger et al. 2013). Interestingly, the ibpA 5′ UTR harbors a temperature-sensitive RNA structure that blocks the RBS at low temperatures but opens when temperature increases (Waldminghaus et al. 2009). Interaction of RybB with the ibpA 5′ UTR could interfere with formation of the inhibitory structure and allow IbpA production under stress conditions other than high temperature. Similarly, target searches for the Spot 42 (Beisel and Storz 2011), FnrS (Boysen et al. 2010), CyaR (De Lay and Gottesman 2009) and OmrA/B (Guillier and Gottesman 2006) sRNAs revealed potential activated target mRNAs which remain to be tested for their underlying mechanisms and their physiological role.
As mentioned above, most of these studies employed pulse expression of the sRNA combined with microarrays or RNA-seq to score global changes in gene expression patterns. This approach comes with the limitation that it can only identify targets with altered mRNA abundance, whereas sequestered transcripts (Moller et al. 2002; Feng et al. 2015), changing only at the level of protein production, remain elusive. In addition, fold changes obtained for repressed targets usually display a larger dynamic range when compared to activated mRNAs and therefore are harder to detect by transcriptomic techniques. The reason for this discrepancy might be found in the different kinetic properties underlying target repression and activation. Translational repression through sRNAs often involves the recruitment of ribonucleases like RNase E, which actively degrade the target mRNA (Masse, Escorcia and Gottesman 2003; Morita, Maki and Aiba 2005). In contrast, target activation is a rather passive process relying on the intrinsic half-live of the mRNA and the transcript stabilizing effect of translating ribosomes. Thus, constitutive sRNA overexpression might allow a more efficient detection of activated targets (Sledjeski, Gupta and Gottesman 1996; Majdalani et al. 2001; Opdyke, Kang and Storz 2004), but comes at the risk of scoring indirect effects (Urban et al. 2007).
Finally, RNA-based regulators are gathering momentum as control devices in synthetic biology and biotechnology (Kang et al. 2014). For example, simultaneous overexpression of DsrA, RprA and ArcZ strongly increased acid tolerance and protection against oxidative stress in E. coli (Gaida et al. 2013). Engineered riboswitches have long been known to allow precisely tuned gene expression through post-transcriptional control (Groher and Suess 2014). Likewise, synthetic sRNAs that activate gene expression could function as analogous, maybe even more versatile regulators. In the recently developed so-called toehold switches (Callura, Cantor and Collins 2012; Green et al. 2014), gene expression is controlled through an inhibitory stem-loop RNA structure covering the RBS and a corresponding trigger-RNA that antagonizes stem-loop formation and allows mRNA translation. Therefore, toehold switches use the same molecular principles that are also employed in naturally occurring anti-antisense regulation of bacterial sRNA (Fig. 1A). Toehold switches offer a large dynamic range, a high degree of orthogo-nality and a high potential for ‘field diagnostics’. Freeze dried cell extracts of E. coli containing a toehold switch and an enzymatic reporter protein reliably detected Ebola virus RNA as well as antibiotic resistance genes (Pardee et al. 2014). RNA regulators are modular, versatile, highly programmable and therefore ideal candidates for synthetic biology approaches. The concept of anti-antisense regulation has recently been applied in toehold switches but other forms of activation, e.g. through inhibition of ribonucleolytic decay or protein sequestration, could provide equally efficient technologies. Understanding the molecular details of these processes will be key to exploiting the full potential of RNA-based gene control for synthetic regulatory purposes.
Acknowledgments
We thank Kathrin Fröhlich for comments on the manuscript and help with figures.
FUNDING
KP is supported by a long-term post-doctoral fellowship from the Human Frontiers in Science (HFSP) organization. CK acknowledges funding through grant R01 GM092830 (National Institutes of Health).
Conflict of interest. None declared.
REFERENCES
- Ali Azam T, Iwata A, Nishimura A, et al. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–70. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, et al. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Ansong C, Yoon H, Porwollik S, et al. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal M, Bordeau V, Douchin V, et al. A small bacterial RNA regulates a putative ABC transporter. J Biol Chem. 2005;280:7901–8. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–63. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Bak G, Han K, Kim D, et al. Roles of rpoS-activating small RNAs in pathways leading to acid resistance of Escherichia coli. Microbiologyopen. 2014;3:15–28. doi: 10.1002/mbo3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS, Eory LA, Yakhnin H, et al. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine–Dalgarno sequence. J Bacteriol. 2007;189:5472–81. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian D, Vanderpool CK. Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J Bacteriol. 2013;195:4620–30. doi: 10.1128/JB.00586-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–82. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41:286–97. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito Y, Lina G, Greenland T, et al. trans-complementation Complementation of a Staphylococcus aureus agr mutant by Staphylococcus lugdunensis agr RNAIII. J Bacteriol. 1998;180:5780–3. doi: 10.1128/jb.180.21.5780-5783.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Gene Dev. 2007;21:1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeau V, Felden B. Curli synthesis and biofilm formation in enteric bacteria are controlled by a dynamic small RNA module made up of a pseudoknot assisted by an RNA chaperone. Nucleic Acids Res. 2014;42:4682–96. doi: 10.1093/nar/gku098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Schwartz A, Guillemardet B, et al. A role for Rho-dependent polarity in gene regulation by a non-coding small RNA. Gene Dev. 2012;26:1864–73. doi: 10.1101/gad.195412.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, et al. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–37. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Boysen A, Moller-Jensen J, Kallipolitis B, et al. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J Biol Chem. 2010;285:10690–702. doi: 10.1074/jbc.M109.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. P Natl Acad Sci USA. 2012;109:5850–5. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael GG, Weber K, Niveleau A, et al. The host factor required for RNA phage Qbeta RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J Biol Chem. 1975;250:3607–12. [PubMed] [Google Scholar]
- Caswell CC, Oglesby-Sherrouse AG, Murphy ER. Sibling rivalry: related bacterial small RNAs and their redundant and non-redundant roles. Front Cell Infect Microbiol. 2014;4:151. doi: 10.3389/fcimb.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol B, Fortas E, Martret C, et al. Riboregulation of the bacterial actin-homolog MreB by DsrA small non-coding RNA. Integr Biol. 2015;7:128–41. doi: 10.1039/c4ib00102h. [DOI] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, et al. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Chavez RG, Alvarez AF, Romeo T, et al. The physiological stimulus for the BarA sensor kinase. J Bacteriol. 2010;192:2009–12. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010;7:316–21. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- Corcoran CP, Podkaminski D, Papenfort K, et al. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol. 2012;84:428–45. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- Cronan JE., Jr Phospholipid modifications in bacteria. Curr Opin Microbiol. 2002;5:202–5. doi: 10.1016/s1369-5274(02)00297-7. [DOI] [PubMed] [Google Scholar]
- Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65:373–85. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. The Crp-activated small non-coding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191:461–76. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol. 2012;86:524–38. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Masse E. Non-canonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Gene Dev. 2012;26:726–39. doi: 10.1101/gad.182493.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Cornelis P, Williams P, et al. 4-quinolone signalling in Pseudomonas aeruginosa: old molecules, new perspectives. Int J Med Microbiol. 2006;296:83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Dimastrogiovanni D, Fröhlich KS, Bandyra KJ, et al. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife. 2014;3:e05375. doi: 10.7554/eLife.05375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duss O, Michel E, Yulikov M, et al. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature. 2014;509:588–92. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- Feng L, Rutherford ST, Papenfort K, et al. A qrr non-coding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160:228–40. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, et al. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Gene Dev. 2009;23:2004–15. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Papenfort K, Fekete A, et al. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32:2963–79. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida SM, Al-Hinai MA, Indurthi DC, et al. Synthetic tolerance: three non-coding small RNAs, DsrA, ArcZ and RprA, acting supra-additively against acid stress. Nucleic Acids Res. 2013;41:8726–37. doi: 10.1093/nar/gkt651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopel Y, Luttmann D, Heroven AK, et al. Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in Enterobacteriaceae. Nucleic Acids Res. 2011;39:1294–309. doi: 10.1093/nar/gkq986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopel Y, Papenfort K, Reichenbach B, et al. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Gene Dev. 2013;27:552–64. doi: 10.1101/gad.210112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AA, Silver PA, Collins JJ, et al. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–39. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groher F, Suess B. Synthetic riboswitches—a tool comes of age. Biochim Biophys Acta. 2014;1839:964–73. doi: 10.1016/j.bbagrm.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Gruber CC, Sperandio V. Post-transcriptional control of microbe-induced rearrangement of host cell actin. MBio. 2014;5:e01025–13. doi: 10.1128/mBio.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–47. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Rhodius VA, Ahuja N, et al. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol. 2007;189:1963–73. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MS, Updegrove TB, Gogol EB, et al. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Gene Dev. 2014;28:1620–34. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Bio. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. P Natl Acad Sci USA. 2007;104:11145–9. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle H, Vecerek B, Resch A, et al. Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature. RNA Biol. 2013;10:1834–41. doi: 10.4161/rna.27100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Doria JD, Sperandio V. Nutrient and chemical sensing by intestinal pathogens. Microbes Infect. 2013;15:759–64. doi: 10.1016/j.micinf.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37:5465–76. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Boisset S, Saveanu C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–35. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. P Natl Acad Sci USA. 2011;108:1110–5. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idone V, Brendtro S, Gillespie R, et al. Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect Immun. 2003;71:4351–60. doi: 10.1128/IAI.71.8.4351-4360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–80. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, et al. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–57. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen MG, Nielsen JS, Boysen A, et al. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol Microbiol. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen MG, Thomason MK, Havelund J, et al. Dual function of the McaS small RNA in controlling biofilm formation. Gene Dev. 2013;27:1132–45. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani M, Kato A, Wada A, et al. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol. 1994;176:531–4. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamorz F, Reichenbach B, Marz W, et al. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol. 2007;65:1518–33. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- Kang Z, Zhang C, Zhang J, et al. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biot. 2014;98:3413–24. doi: 10.1007/s00253-014-5569-y. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Koide Y, Morita T, et al. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol. 2006;61:1013–22. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Kawsar HI, Ohtani K, Okumura K, et al. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol Lett. 2004;235:289–95. doi: 10.1016/j.femsle.2004.04.047. [DOI] [PubMed] [Google Scholar]
- Kim BH, Kim S, Kim HG, et al. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology. 2005;151:209–18. doi: 10.1099/mic.0.27265-0. [DOI] [PubMed] [Google Scholar]
- Kimata K, Tanaka Y, Inada T, et al. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 2001;20:3587–95. doi: 10.1093/emboj/20.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach AR, Hoff KE, Canty JT, et al. Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA. 2014;20:1548–59. doi: 10.1261/rna.044032.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, Boyle MD, Buttaro BA, et al. Group A Streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Kroger C, Colgan A, Srikumar S, et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar typhimurium. Cell Host Microbe. 2013;14:683–95. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, et al. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–53. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA: RNA interactions at multiple loci. P Natl Acad Sci USA. 1998;95:12456–61. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Smith D, McDonough K, et al. The small non-coding DsrA RNA is an acid resistance regulator in Escherichia coli. J Bacteriol. 2004;186:6179–85. doi: 10.1128/JB.186.18.6179-6185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Lewis RJ, Mader U, et al. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol. 2012;84:1005–17. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–19. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Coleman ML, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–6. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. P Natl Acad Sci USA. 2009;106:19292–7. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Niu G, Xie Z, et al. The Streptococcus mutans irvA gene encodes a trans-acting riboregulatory mRNA. Mol Cell. 2015;57:179–90. doi: 10.1016/j.molcel.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mu C, Ying X, et al. RNAIII activates map expression by forming an RNA–RNA complex in Staphylococcus aureus. FEBS Lett. 2011;585:899–905. doi: 10.1016/j.febslet.2011.02.021. [DOI] [PubMed] [Google Scholar]
- Liu Z, Trevino J, Ramirez-Pena E, et al. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol Microbiol. 2012;86:140–54. doi: 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Abel CA, Feig AL, et al. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2010;78:622–35. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–89. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- Ma Z, Richard H, Tucker DL, et al. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW) J Bacteriol. 2002;184:7001–12. doi: 10.1128/JB.184.24.7001-7012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JD, McKay FC, Ramachandran V, et al. Allelic variants of streptokinase from Streptococcus pyogenes display functional differences in plasminogen activation. FASEB J. 2008;22:3146–53. doi: 10.1096/fj.08-109348. [DOI] [PubMed] [Google Scholar]
- McCullen CA, Benhammou JN, Majdalani N, et al. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol. 2010;192:5559–71. doi: 10.1128/JB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, et al. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol. 2001;39:1382–94. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, et al. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. P Natl Acad Sci USA. 1998;95:12462–7. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–26. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, et al. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. P Natl Acad Sci USA. 2004;101:13318–23. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LC, Yakhnin H, Camacho MI, et al. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol. 2011;80:1637–56. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Gene Dev. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–71. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Busse S, Possling A, et al. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Gene Dev. 2005;19:2770–81. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014;11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi M, Chao Y, Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015a doi: 10.15252/embj.201490546. doi: 10.15252/embj.201490546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi M, Chao Y, Vogel J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol. 2015b;24C:132–9. doi: 10.1016/j.mib.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O, et al. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–14. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T, Franch T, Udesen C, et al. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Gene Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro C, Papenfort K, Hentrich K, et al. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–62. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Six DA, Lee HJ, et al. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol Microbiol. 2013;89:52–64. doi: 10.1111/mmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Taylor D, von Gabain A, et al. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–77. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, El-Kazzaz W, Tanaka Y, et al. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem. 2003;278:15608–14. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Gene Dev. 2005;19:2176–86. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Gene Dev. 1996;10:1143–51. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- Mustelin T. A brief introduction to the protein phosphatase families. Method Mol Biol. 2007;365:9–22. doi: 10.1385/1-59745-267-X:9. [DOI] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Obana N, Nomura N, Nakamura K. Structural requirement in Clostridium perfringens collagenase mRNA 5′ leader sequence for translational induction through small RNA–mRNA base pairing. J Bacteriol. 2013;195:2937–46. doi: 10.1128/JB.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana N, Shirahama Y, Abe K, et al. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol Microbiol. 2010;77:1416–28. doi: 10.1111/j.1365-2958.2010.07258.x. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Hirakawa H, Tashiro K, et al. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe. 2010;16:258–64. doi: 10.1016/j.anaerobe.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Okumura K, Ohtani K, Hayashi H, et al. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J Bacteriol. 2008;190:7719–27. doi: 10.1128/JB.01573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Fozo EM, Hemm MR, et al. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol. 2011;406:29–43. doi: 10.1016/j.jmb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, et al. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. P Natl Acad Sci USA. 2011;108:13059–64. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, et al. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Forstner KU, Cong JP, et al. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. P Natl Acad Sci USA. 2015;112:E766–75. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, et al. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–88. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Podkaminski D, Hinton JC, et al. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. P Natl Acad Sci USA. 2012;109:E757–64. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–58. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, et al. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–37. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116–27. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front Cell Infect Microbiol. 2014;4:91. doi: 10.3389/fcimb.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K, Green AA, Ferrante T, et al. Paper-based synthetic gene networks. Cell. 2014;159:940–54. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson-Fortin LM, Vakulskas CA, Yakhnin H, et al. Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J Mol Biol. 2013;425:3662–77. doi: 10.1016/j.jmb.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Curtis JE, Fang X, et al. Structural model of an mRNA in complex with the bacterial chaperone Hfq. P Natl Acad Sci USA. 2014;111:17134–9. doi: 10.1073/pnas.1410114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, et al. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–6. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14:3958–65. doi: 10.1002/j.1460-2075.1995.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J, Bossi L, Oberto J, et al. Interplay of transcriptional and small RNA-dependent control mechanisms regulates chitosugar uptake in Escherichia coli and Salmonella. Mol Microbiol. 2014;92:648–58. doi: 10.1111/mmi.12573. [DOI] [PubMed] [Google Scholar]
- Porcheron G, Habib R, Houle S, et al. The small RNA RyhB contributes to siderophore production and virulence of uropathogenic Escherichia coli. Infect Immun. 2014;82:5056–68. doi: 10.1128/IAI.02287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost K, Salvail H, Desnoyers G, et al. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–73. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- Quereda JJ, Ortega AD, Pucciarelli MG, et al. The Listeria small RNA Rli27 regulates a cell wall protein inside eukaryotic cells by targeting a long 5′-UTR variant. PLoS Genet. 2014;10:e1004765. doi: 10.1371/journal.pgen.1004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Pena E, Trevino J, Liu Z, et al. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–47. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Johansen J, Nielsen JS, et al. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol. 2009;72:566–77. doi: 10.1111/j.1365-2958.2009.06688.x. [DOI] [PubMed] [Google Scholar]
- Reading NC, Torres AG, Kendall MM, et al. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol. 2007;189:2468–76. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach B, Gopel Y, Gorke B. Dual control by perfectly overlapping sigma 54- and sigma 70- promoters adjusts small RNA GlmY expression to different environmental signals. Mol Microbiol. 2009;74:1054–70. doi: 10.1111/j.1365-2958.2009.06918.x. [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Maes A, Kalamorz F, et al. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–80. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Gottesman S. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J Bacteriol. 2001;183:4012–23. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Gottesman S. Temperature sensing by the dsrA promoter. J Bacteriol. 2003;185:6609–14. doi: 10.1128/JB.185.22.6609-6614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A, Afonyushkin T, Lombo TB, et al. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA. 2008;14:454–9. doi: 10.1261/rna.603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A, Vecerek B, Palavra K, et al. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol. 2010;7:796–802. doi: 10.4161/rna.7.6.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, Balasubramanian D, Vanderpool CK. Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. P Natl Acad Sci USA. 2012;109:E2691–8. doi: 10.1073/pnas.1207927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, Vanderpool CK. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 2011;39:3806–19. doi: 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KE, Orans J, Kovach AR, et al. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014;42:2736–49. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol. 2013;15:313–24. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romilly C, Caldelari I, Parmentier D, et al. Current knowledge on regulatory RNAs and their machineries in Staphylococcus aureus. RNA Biol. 2012;9:402–13. doi: 10.4161/rna.20103. [DOI] [PubMed] [Google Scholar]
- Ross JA, Trussler RS, Black MD, et al. Tn5 transposition in Escherichia coli is repressed by Hfq and activated by over-expression of the small non-coding RNA SgrS. Mob DNA. 2014;5:27. doi: 10.1186/s13100-014-0027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Wardle SJ, Haniford DB. Tn10/IS10 transposition is downregulated at the level of transposase expression by the RNA-binding protein Hfq. Mol Microbiol. 2010;78:607–21. doi: 10.1111/j.1365-2958.2010.07359.x. [DOI] [PubMed] [Google Scholar]
- Rutherford ST, van Kessel JC, Shao Y, et al. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Gene Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Caron MP, Belanger J, et al. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 2013;32:2764–78. doi: 10.1038/emboj.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Masse E. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA. 2012;3:26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. P Natl Acad Sci USA. 2011;108:13065–70. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyll E, Van Melderen L. The ribonucleoprotein csr network. Int J Mol Sci. 2013;14:22117–31. doi: 10.3390/ijms141122117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol. 2012;83:599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Feng L, Rutherford ST, et al. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 2013;32:2158–71. doi: 10.1038/emboj.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, et al. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Gene Dev. 2007;21:2804–17. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, et al. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 2011;81:1144–65. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Ba-Thein W, Tamaki M, et al. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J Bacteriol. 1994;176:1616–23. doi: 10.1128/jb.176.6.1616-1623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Yaguchi H, Ohtani K, et al. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol Microbiol. 2002;43:257–65. doi: 10.1046/j.1365-2958.2002.02743.x. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, et al. Deep sequencing analysis of small non-coding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. P Natl Acad Sci USA. 1995;92:2003–7. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Abdou L, Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. P Natl Acad Sci USA. 2009;106:21866–71. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Gonzalez N, Sorger-Domenigg T, et al. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol. 2011;80:868–85. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, et al. Positive regulation by small RNAs and the role of Hfq. P Natl Acad Sci USA. 2010;107:9602–7. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–50. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–17. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA–RNA duplex formation during phage infection. Nucleic Acids Res. 2011;39:4890–9. doi: 10.1093/nar/gkr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Futschik ME, Lindell D, et al. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ringdahl U, Homeister JW, et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–6. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, et al. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Gene Dev. 2006;20:2605–17. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson MD, Sjobring U, Luo F, et al. Roles of the plasminogen activator streptokinase and the plasminogen-associated M protein in an experimental model for streptococcal impetigo. Microbiology. 2002;148:3933–45. doi: 10.1099/00221287-148-12-3933. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Kiefer P, Reimmann C, et al. Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J Biol Chem. 2009;284:34976–85. doi: 10.1074/jbc.M109.052571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason MK, Fontaine F, De Lay N, et al. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JJ, Granneman S, McAteer SP, et al. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Papenfort K, Thomsen J, et al. A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J Mol Biol. 2007;373:521–8. doi: 10.1016/j.jmb.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–68. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–33. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–9. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–89. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–43. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt SL, Evans AD, Guest RL, et al. The Cpx envelope stress response regulates and is regulated by small non-coding RNAs. J Bacteriol. 2014;196:4229–38. doi: 10.1128/JB.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Sharma CM, Mitschke J, et al. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 2014;8:2056–68. doi: 10.1038/ismej.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O, Moll I, Kaberdin VR, et al. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Gene Dev. 2000;14:1109–18. [PMC free article] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. P Natl Acad Sci USA. 2007;104:20454–9. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG. Cycling of RNAs on Hfq. RNA Biol. 2013;10:619–26. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Gaubig LC, Klinkert B, et al. The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol. 2009;6:455–63. doi: 10.4161/rna.6.4.9014. [DOI] [PubMed] [Google Scholar]
- Wang AY, Cronan JE., Jr The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol Microbiol. 1994;11:1009–17. doi: 10.1111/j.1365-2958.1994.tb00379.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, et al. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–63. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, et al. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Baker CS, Vakulskas CA, et al. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol. 2013;87:851–66. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Ohtani K, Yoshizawa S, et al. Complex transcriptional regulation of citrate metabolism in Clostridium perfringens. Anaerobe. 2012;18:48–54. doi: 10.1016/j.anaerobe.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–24. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Zhao X, Koestler BJ, Waters CM, et al. Post-transcriptional activation of a diguanylate cyclase by quorum sensing small RNAs promotes biofilm formation in Vibrio cholerae. Mol Microbiol. 2013;89:989–1002. doi: 10.1111/mmi.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]