Abstract
The mitochondrion plays wide-ranging roles in eukaryotic cell physiology. In pathogenic fungi, this central metabolic organelle mediates a range of functions related to disease, from fitness of the pathogen to developmental and morphogenetic transitions to antifungal drug susceptibility. In this review, we present the latest findings in this area. We focus on likely mechanisms of mitochondrial impact on fungal virulence pathways through metabolism and stress responses, but also potentially via control over signaling pathways. We highlight fungal mitochondrial proteins that lack human homologs, and which could be inhibited as a novel approach to antifungal drug strategy.
Keywords: fungal pathogenesis, mitochondria, Candida albicans
Pathogenic fungi cause a range of human infections, including invasive life-threatening disease. Here we discuss mitochondrial activity in fungal disease and therapeutic approaches.
Graphical Abstract Figure.
Pathogenic fungi cause a range of human infections, including invasive life-threatening disease. Here we discuss mitochondrial activity in fungal disease and therapeutic approaches.
INTRODUCTION
All eukaryotic species contain in their cells a specialized organelle, the mitochondrion, which functions as a master regulator of metabolism. The mitochondrion has originated from the engulfment of an alpha-proteobacterium by the ancestor of eukaryotes. It is this event, and the capacity for energy production and sophisticated metabolic control that came from it, that enabled the evolution of complex life (Lane and Martin 2010). In modern eukaryotic cells, mitochondria are not only a key energy generator through the process of oxidative phosphorylation, but also a central ‘hub’ that coordinates multiple aspects of cell physiology. Several metabolic pathways converge on mitochondria, and the mitochondrial respiratory chain is a major site of reactive oxygen species (ROS) production (Drose and Brandt 2012). Stress responses that are critical for cell survival under adverse conditions, and process such as cellular aging, apoptotic programmed cell death and tumorigenesis, all involve mitochondrial functions (Guarente 2008; Hsu and Sabatini 2008; Green, Galluzzi and Kroemer 2014).
Multiple mitochondria exist in every cell, forming a dynamic tubular network that constantly fuses and divides. These processes known as ‘mitochondrial dynamics’ are required to maintain mitochondrial health and enable organelle inheritance during cell division, ultimately equipping the cell to meet metabolic demand (Labbe, Murley and Nunnari 2014). In addition, mitochondrial function is intimately linked to other eukaryotic organelles for global control over cell physiology. Physical tethers bridge mitochondria to the endoplasmic reticulum (ER) and to the vacuole (Kornmann et al. 2009; Elbaz-Alon et al. 2014; Honscher et al. 2014). These inter-organellar communications mediate lipid trafficking and possibly also calcium homeostasis (Kornmann and Walter 2010). Moreover, mitochondria communicate with the nucleus by multiple mechanisms to regulate the expression of genes by metabolic and ROS-dependent signaling pathways (Jazwinski 2013; Schroeder, Raimundo and Shadel 2013).
Given these central physiological roles, it is not surprising that mitochondrial activity has been implicated in multiple aspects of fungal cell biology. These encompass pathways that are essential for virulence of critical human pathogens such as Candida sp., Cryptococcus sp. and Aspergillus sp. (Brown et al. 2012). In fungal pathogens, mitochondria play roles in developmental and morphogenetic switches such as hyphal differentiation and biofilm formation, adaptation to stress, cell wall biosynthesis and structure, innate immune cell interaction and susceptibility to antifungal drugs (Shingu-Vazquez and Traven 2011; Morales et al. 2013; She et al. 2013). Importantly, fungi have their own repertoire of mitochondrial-specific proteins that are critical to the activities mentioned above. New advances in our understanding of how mitochondria are made and regulated predominantly in the major human pathogen Candida albicans are to be covered in this review. Much of the published data are based upon knockout strains in mitochondrial genes. As these strains have various types of dysfunctions mentioned above, especially growth and virulence, we suggest translational insights based upon the development of new antifungal therapies that target key fungal-specific mitochondrial proteins.
MITOCHONDRIAL BIOGENESIS AND DYNAMICS IN FUNGAL PATHOGENESIS
Several factors that regulate mitochondrial morphology and biogenesis are distinct in fungi compared to metazoans, and therefore represent potential therapeutic targets. Recent studies have begun to shed light on the mechanisms of mitochondrial biogenesis and dynamics in fungal virulence, revealing shared and divergent mechanisms between pathogenic fungi and the model yeast Saccharomyces cerevisiae.
Mitochondrial protein import—lessons from C. albicans
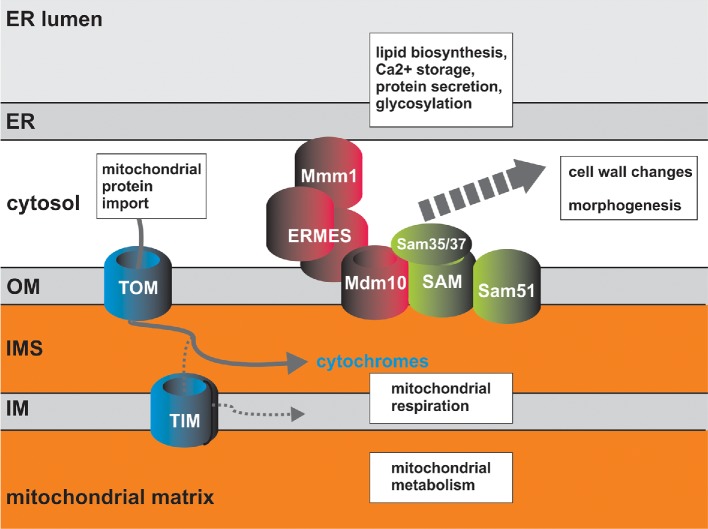
The vast majority of the mitochondrial proteome is encoded in the nucleus, and proteins are imported into the organelle from their place of translation in the cytoplasm. Mitochondrial protein import is thus essential for organelle biogenesis. Most of what is known about factors and mechanisms of mitochondrial protein import comes from the S. cerevisiae model, and the mechanisms are largely conserved across eukaryotes (Chacinska et al. 2009). In a first assessment of the mitochondrial protein import machinery in a human fungal pathogen, Hewitt et al. (2012) demonstrated the almost all of the components as known in S. cerevisiae can be identified in C. albicans (Fig. 1). An exception is the genes TOM70 and TOM71 encoding subunits of the TOM (translocase of the outer membrane) complex, which arose through genome duplication in S. cerevisiae. Only one TOM70-like gene is found in the C. albicans genome (Hewitt et al. 2012). Several new discoveries of mitochondrial protein import mechanisms were made in the C. albicans system. For example, it appears that C. albicans uses a different pathway than S. cerevisiae to import cytochromes c1 and b2 into the mitochondrial intermembrane space (Hewitt et al. 2012), reviewed in Hewitt, Gabriel and Traven (2014). These substrates are imported via the so-called stop transfer pathway in S. cerevisiae, but the C. albicans proteins lack N-terminal sequences needed for this import mechanism (Hewitt et al. 2012). What import pathway is used in C. albicans for these substrates remains to be determined. This difference between C. albicans and S. cerevisiae is interesting from the perspective of the evolution of mitochondrial import mechanisms in eukaryotes, and we have suggested that metabolic features in host environments, such as access to oxygen, might have influenced the selection of pathways for importing these substrates into mitochondria (Hewitt et al. 2012), reviewed in Hewitt, Gabriel and Traven (2014). Does the ‘evolutionary rewiring’ of the import routes for cytochromes c1 and b2 have relevance for C. albicans biology? We do not know yet, but cytochrome b2 is essential for growth on lactate, an important carbon source found in human host niches (Ueno et al. 2011; Hewitt, Gabriel and Traven 2014).
Figure 1.
The mitochondrial biogenesis apparatus in C. albicans. Proteins required for mitochondrial protein import are shown. All components of the mitochondrial import apparatus as described in the model baker's yeast can be found by bioinformatics searches in C. albicans. TOM is the ‘gateway’ to mitochondria through which all imported proteins pass. TIM enables translocation into the matrix and targeting to the inner membrane. In C. albicans, some cytochromes (b2 and c1) follow a distinct route to the intermembrane space to what is known in S. cerevisiae. The pathways required for their mitochondrial import in C. albicans remain to be discovered. Candida albicans contains an additional Omp85 family member, Sam51, which is part of the SAM complex, and data suggest that SAM and ERMES are bridged together via Mdm10 (Victoria Hewitt, A.T. and Trevor Lithgow, in preparation). The Sam37 and Sam35 subunits of the SAM complex and entire ERMES complex are not conserved in humans, and Sam37 and Mmm1 have been found to be required for virulence of C. albicans in the systemic mouse model (Becker et al. 2010; Qu et al. 2012). Links between SAM and ERMES function and cell wall integrity and hyphal morphogenesis have been made (Dagley et al. 2011; Qu et al. 2012), reenforcing the concept that mitochondria have broad impact on pathways required for fungal pathogenesis.
So far, our studies in C. albicans have mostly focused on two complexes that are located in the mitochondrial outer membrane: the SAM (sorting and assembly machinery) and the ERMES (ER-mitochondria encounter structure) (Fig. 1). Based on studies in S. cerevisiae, SAM and ERMES play central roles in mitochondrial biogenesis and function by mediating insertion of proteins into the outer membrane, membrane lipid homeostasis, maintenance of the tubular structure of the mitochondrial network, inheritance of mitochondria by the daughter cell mediated by ERMES and quality control through the roles of ERMES in mitophagy (Burgess, Delannoy and Jensen 1994; Gratzer et al. 1995; Dimmer et al. 2002; Boldogh et al. 2003; Gentle et al. 2004; Ishikawa et al. 2004; Meisinger et al. 2004, 2007; Altmann and Westermann 2005; Kornmann et al. 2009; Yamano, Tanaka-Yamano and Endo 2010; Dagley et al. 2011; Bockler and Westermann 2014), reviewed in (Chacinska et al. 2009; Labbe, Murley and Nunnari 2014). These two complexes exemplify the concept of mitochondria as a ‘hub’ that coordinates multiple aspects of cell physiology (Kornmann and Walter 2010). For example, in C. albicans the function of SAM is linked to cell wall biogenesis, echinocandin susceptibility and hyphal morphogenesis (Dagley et al. 2011; Qu et al. 2012). The bioinformatics analysis performed by Hewitt et al. (2012) revealed that in C. albicans, in addition to the ‘standard’ Omp85-like protein Sam50 that is the core, essential component of the SAM, a second Omp85-family protein is found that we named Sam51(Fig. 1). Sam51 can also be found in CTG clade Candida species (members of this clade translate CTG as serine instead of leucine), and some other fungal species such as Kluyveromyces lactis, but was lost in S. cerevisiae following the whole genome duplication event (Hewitt et al. 2012). In contrast to SAM50, loss of SAM51 in C. albicans does not lead to lethality, and it does not even obviously impair growth under standard laboratory conditions, although biochemical analysis suggests a function in mitochondrial outer membrane biogenesis (Hewitt et al. 2012). Moreover, loss of SAM51 did not impair virulence of C. albicans in the mouse systemic tail vein injection model of candidiasis (Yue Qu and A.T. unpublished). Unlike Sam51, the Sam37 subunit of the SAM complex is required for fitness and virulence of C. albicans (Qu et al. 2012). Bioinformatic analysis suggested that fungal Sam37 proteins are very different from their functional homologs in metazoans, the metaxins (Qu et al. 2012). Therefore, the activity of the SAM complex could be considered for antifungal therapy through targeting Sam37. Another possible target is fungal Sam35, a second metaxin-like essential SAM subunit (Qu et al. 2012). Similarly, ERMES, that is involved in ER–mitochondria contacts in yeast and has essential roles in mitochondrial homeostasis as described above, is widely conserved in fungi but homologs are not found in metazoans (Wideman et al. 2013). Of note, ER–mitochondria contacts occur in higher eukaryotes including humans through the so-called mitochondria-associated membranes, but factors and mechanisms other than ERMES are likely to mediate these contacts (Vance 2014). Studies in S. cerevisiae suggest that there is crosstalk between the SAM and ERMES complexes via the Mdm10 subunit, which is a subunit of both (Meisinger et al. 2004; Wideman et al. 2013) (Fig. 1). The biochemical details of how these two complexes might be cooperating are presently not clear, but using C. albicans as a model, our data suggest that the two complexes are bridged by Mdm10 into a ‘super-complex’ (Victoria Hewitt, A.T. and Trevor Lithgow, in preparation, see Fig. 1). ERMES mutants of S. cerevisiae grow slower than wild type (WT) but can be easily recovered on fermentative carbon sources, whereas in C. albicans the entire complex is practically essential for growth: only very, very slow growing colonies can be recovered following homozygous deletion of any of the ERMES genes at 30°C on glucose-containing media, and growth is arrested at human body temperature of 37°C (Timothy Tucey and A.T. in preparation). Consistent with a requirement for virulence of C. albicans, a tet-off conditional mutant in the ERMES subunit MMM1 assessed by Backer et al. (2010) as part of a large mutant collection screen was found to be essential for pathogenesis in the mouse systemic model. Therefore, ERMES is another fungal-specific mitochondrial complex that should be considered in the context of antifungal therapy.
Mitochondrial dynamics in fungal pathogenesis
An important function of ERMES is in assuring that the tubular structure of the mitochondrial network is preserved (Labbe, Murley and Nunnari 2014). Based on studies in S. cerevisiae and mammalian systems, it is clear that mutations that cause changes to mitochondrial network structure by disrupting fusion, fission or impacting on tubulation are detrimental to mitochondrial health and function (Labbe, Murley and Nunnari 2014). However, little is known about mitochondrial dynamics and morphology in fungal virulence mechanisms. The C. albicans mitochondrial fusion mutant in the mitofusin homolog, the GTPase FZO1, displayed drastic fitness impairment, perturbed mitochondrial phospholipid levels and increased susceptibility to the azole drugs that inhibit ergosterol biosynthesis (Thomas et al. 2013). Organelle fragmentation that results from mitochondrial fusion defects is known to cause mitochondrial DNA loss in S. cerevisiae (Rapaport et al. 1998). This was also the case in the C. albicans fzo1 mutant (Thomas et al. 2013), providing an explanation for the much more pronounced fitness defect of the C. albicans mutant compared to S. cerevisiae (mitochondrial genome loss is significantly less tolerated in the petite-negative C. albicans than in the petite-positive S. cerevisiae). It follows from here that, more generally, dysfunction of mitochondrial fusion will be detrimental for fitness in C. albicans, and other petite-negative pathogenic species. A protein that cooperates with Fzo1 in mitochondrial fusion, Ugo1, has no homologs in humans (Okamoto and Shaw 2005), and therefore represents another fungal-specific mitochondrial protein that could be of interest for drug development. To our knowledge, the functions of Ugo1 have not been addressed directly in fungi outside of S. cerevisiae as yet.
To maintain mitochondrial network structure, organelle fusion is balanced by fission. Fission of mitochondria has been widely studied in both S. cerevisiae and mammals (Okamoto and Shaw 2005; Labbe, Murley and Nunnari 2014). A link between mitochondrial fission and fungal virulence comes from studies of the hypervirulent Cryptococcus gattii strain responsible for the outbreak on Vancouver Island (Ma et al. 2009; Voelz et al. 2014). The hypervirulent Cr. gattii is able to switch to tubularized mitochondrial morphology upon engulfment by macrophages, suggesting that the environment in innate immune phagocytes promotes a shift in the mitochondrial fusion–fission balance towards fusion. Mitochondrial hypertubularization promotes survival of the fungus in the face of macrophage attack, and results in overall increased fitness of the pathogen (Voelz et al. 2014). The precise mechanism of mitochondrial hypertubularization in Cr. gattii remains to be identified, but it was linked to ROS generated by the macrophage (Voelz et al. 2014). Oxidative stress and other stimuli, such as acetic acid, can induce cell death in yeast, and this can under certain conditions result in tipping of the mitochondrial fusion–fission balance towards mitochondrial fragmentation (Fannjiang et al. 2004; Tar et al. 2014). ROS produced by macrophages might be promoting the opposite in Cr. gattii, i.e. hyperfusion of mitochondria. It should be noted that, as was demonstrated by studies in C. albicans, the levels of oxidative stress sensed by fungal pathogens in macrophages are likely to be significantly different than the high doses used in in vitro experiments to cause cell death (Enjalbert et al. 2007). It would be worth exploring if mechanisms converging on the regulation of the mitochondrial fission apparatus are responsible for promoting tubularization of the mitochondrial network in Cr. gattii strains that is observed inside macrophages.
MITOCHONDRIAL RESPIRATORY ACTIVITY IN FUNGAL PATHOGENESIS: THE LATEST FROM C. ALBICANS
Cellular respiration is a key function of mitochondria. Mitochondrial respiratory activity has been implicated in several virulence-related functions in human fungal pathogens including transitions between cell types and growth modes (yeast, hyphae, biofilms), susceptibility to antifungal drugs, cell wall biogenesis and immune interactions (Watanabe et al. 2006; Bambach et al. 2009; Dagley et al. 2011; Shingu-Vazquez and Traven 2011; Qu et al. 2012; Morales et al. 2013; She et al. 2013; Sun et al. 2013; Lindsay et al. 2014). Below we review recent work on fungal-specific mitochondrial proteins that impact on mitochondrial respiration, and which could be promising for antifungal drug development.
Goa1p is required for mitochondrial complex I (CI) activity but is not a subunit of CI.
Our focus on mitochondria led to studies of the CI of C. albicans. CI is one of five complexes of the electron transport chain (ETC) that is responsible mitochondrial generation of cellular ATP. We initially screened a C. albicans null mutant library to identify proteins that were required for oxidant adaptation (Bambach et al. 2009). Of those primary ‘hit’ mutants, several were of interest, including an unstudied gene that we named GOA1 because of its critical roles in cell growth and oxidant adaptation. Two lines of evidence suggest that Goa1p localizes to mitochondria during stress conditions in cells. First, a Goa1p-GFP was detected in mitochondria following stress, and second, more recently, the protein was found enriched in mitochondrial fractions (Bambach et al. 2009 and unpublished data).
GOA1 is found only among members of the CTG clade of the Ascomycetous subphylum Saccharomycotina that includes all Candida pathogens except C. glabrata (Fitzpatrick et al. 2006). CTG clade specificity suggested that any anti-Goa1p therapeutic would not broadly target pathogens but nevertheless, could be potentially useful against most Candida species. Oppositely, the CTG specificity was of interest. Why does this specificity occur, and, even more important, are there other mitochondrial proteins that are more broadly fungal specific or even others that are CTG specific?
GOA1 functions in C. albicans
Subsequently, an isogenic set of strains was constructed that included a null mutant and a single gene-reconstituted strain were compared phenotypically to parental WT cells (Bambach et al. 2009; Li et al. 2011; Chen et al. 2012; She et al. 2013, 2015; Sun et al. 2013; Khamooshi et al. 2014). The inability of the goa1Δ to grow on mitochondrial respiratory-specific substrates such as glycerol, along with its loss of mitochondrial membrane potential, reduced oxidative respiration, and reduced ATP synthesis suggested that Goa1p was required for the activity of the mitochondrial ETC. But how?
Since energy production was profoundly reduced in the goa1Δ, CI–CIV enzyme activities of the ETC were measured. Only CI complex was reduced in goa1Δ (Li et al. 2011). Other mutant phenotypes included an inability to maintain a chronological life span or aging (Chen et al. 2012) and elevated ROS, which often is associated with intrinsic cell apoptosis as O2 acquires free electrons to generate superoxide in CI dysfunctions (Li et al. 2011).
The goa1Δ was avirulent in a blood-borne, murine model of invasive candidiasis and displayed reduced recognition by a human macrophage cell line and human neutrophils (Bambach et al. 2009; She et al. 2013). Surprisingly, the latter studies suggested an escape of host cell recognition by the mutant, but in spite of its reduced phagocytosis and recognition by innate immune cells, extracellular killing by neutrophils was enhanced compared to control strains (She et al. 2013).
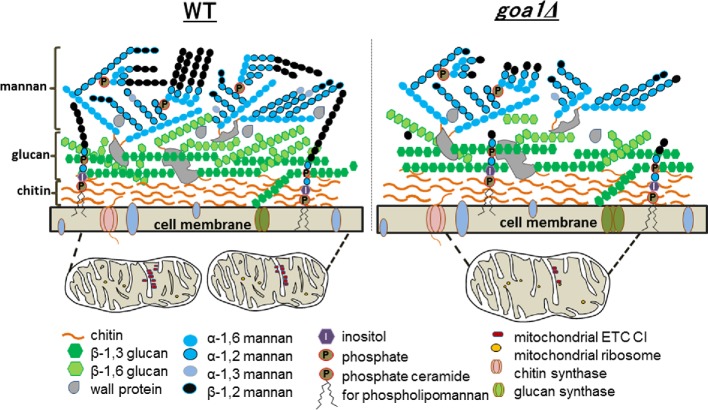
A reduction in the recognition of the null mutant by innate immune cells suggested changes in the organization and/or composition of the goa1Δ cell wall. Transcriptional profiling of goa1Δ showed significant downregulation of genes associated with cell wall biosynthesis including several types of adhesins such as those of the ALS family (agglutinin-like sequences) (Chen et al. 2012). These data were supported by gel permeation chromatography, which indicated that the goa1Δ mannan was smaller and less complex than that of WT and the GOA1-reconstituted cells (manuscript in preparation). Also, increased sensitivity of the mutant to cell wall inhibitors such as alcian blue, caspofungin and Congo Red indicates other wall changes. The cell wall phenotypes and mitochondria of WT and the goa1Δ strains are depicted in Fig. 2 and explained below (Chen et al. 2012). The cell wall defects of the goa1Δ mutant strain are consistent with a key function of mitochondria in ensuring proper fungal cell wall functions in C. albicans (Dagley et al. 2011; Qu et al. 2012), reviewed in Shingu-Vazquez and Traven (2011).
Figure 2.
Cell wall changes upon respiratory dysfunction in the C. albicans goa1Δ mutant. The cell wall structures of WT (left) and the null goa1Δ (right) are shown. Cell wall polysaccharides and their linkages are color coded. In addition, phosphate, inositol and phosphate ceramide are shown as a GPI-anchored cell wall glycoprotein. Below the cell membrane, mitochondria are depicted with fewer mitochondrial ETC CI in the goa1Δ. The reduction of β-1,2 mannan and β-1,6 glucan (unpublished) is shown in the mutant cell wall.
Downregulation of genes associated with β-1,2 mannosylation (BMT 3,6–9) was observed in the goa1Δ which may indicate a reduction in β-1,2 oligomannan (Fig. 2) (Chen et al. 2012). β-1,2 Mannosylation of cell wall mannan is not found in S. cerevisiae, but is in C. albicans where it is a component of both phosphopeptidomannan and phospholipomannan (Mille et al. 2004, 2008, 2012; Fradin et al. 2008). There is an association of length of β-linked mannosyl residues with serotype, which is higher in serotype A than in serotype B C. albicans. β-Mannosylation of both N- and O-linked mannan has been observed (Fradin et al. 2008). In C. albicans, β-mannosylation is a component of adhesins and induces innate immune cells to produce cytokines. Surprisingly, loss of BMT1–4 genes had a minor impact on antigenic expression in these mutants when analyzed by western blot analysis (Mille et al. 2008). Functional activities of some members of the BMT gene family, based upon studies with null mutants, include virulence, the induction of biofilm formation, proinflammatory response by macrophages and regulation of multidrug transporters (Karababa et al. 2004; Mille et al. 2004; Fradin et al. 2008; Bonhomme et al. 2011; Devillers et al. 2013; Ueno et al. 2013). As several of the ALS family of adhesins has been associated with virulence, their transcriptional downregulation, as well as that of β-1,2 oligomannosylation BMT genes, may account for reduced survival in mice infected with goa1Δ (Bambach et al. 2009; Chen et al. 2012). While it is clear that mitochondrial function is necessary for proper cell wall structuring in C. albicans, it is not understood what the biochemical links are. It also appears that different mitochondrial mutants might have distinct perturbations in cell wall topography. For example, the SAM complex mutant sam37Δ does not display altered glucan/mannan levels in the wall, although the cell wall is thicker than in WT cells (Qu et al. 2012). Therefore, while we can predict that disruption of mitochondrial function in C. albicans, and likely other fungal pathogens, will cause cell wall defects, the precise nature of those defects and therefore the impact on virulence pathways might be distinct depending on the affected mitochondrial activities.
TRANSCRIPTIONAL REGULATORS OF MITOCHONDRIAL RESPIRATION IN C. ALBICANS
If Goa1p regulates CI activity in some fashion, what regulates GOA1? To answer this question, we screened a library of transcriptional regulatory knockout mutants (TRKO) for their growth on glycerol, a mitochondrial, respiratory-specific substrates (Homann et al. 2009; Khamooshi et al. 2014). We identified 5 of 143 mutants that had reduced growth on glycerol, but grew on glucose. Three of those mutants, dpb4Δ, hfl1Δ and rbf1Δ displayed a transcriptional downregulation of GOA1, indicating that each is probably a positive regulator of GOA1 (directly or indirectly). Rbf1p is a repressor of the yeast → hyphal morphogenesis (Ishii et al. 1997). We showed that both the dpb4Δ and the hfl1Δ were similarly constitutively filamentous (Khamooshi et al. 2014). As the goa1Δ is not constitutively filamentous, we suggest that Rbf1p, Dpb4p and Hfl1p have other gene targets unrelated to GOA1.
The sensitivity of the TRKO strains and goa1Δ to antifungal drugs was compared using standardized MIC assays. The goa1Δ, rbf1Δ and hfl1Δ, but not dpb4Δ were hypersensitive to caspofungin or fluconazole (Khamooshi et al. 2014). Each of these TRKO strains had reduced oxygen consumption, reduced CI and CIV enzyme activity, and were hypersensitive to the CI inhibitor, rotenone. Transcriptional profiling indicated more similarities for rbf1Δ and hfl1Δ compared to dpb4Δ, while DPB4 appeared to regulate mtDNA maintenance and mtRNA processing. These data indicate a common role for these transcriptional regulators on GOA1 transcription and mitochondrial activities in general, but also distinct functions suggesting broad direct or indirect regulatory activity in cells.
Fungal-specific CI subunit proteins
Most of the CI subunit proteins of fungi are broadly conserved among eukaryotic species (Azevedo et al. 1992; McDonough et al. 2002; Videira and Duarte 2002; Abdrakhmanova et al. 2004; Gabaldon, Rainey and Huynen 2005; Bridges, Fearnley and Hirst 2010; Pereira, Videira and Duarte 2013). In C. albicans, CI is composed of ∼39 subunit proteins, including at least two fungal-specific subunits (see below). Most of these subunits have not been functionally studied in human fungal pathogens aside from their presumed enzyme activity as NADH:Ubiquinone Oxidoreductases (Gabaldon, Rainey and Huynen 2005; Noble et al. 2010). The fungal-specific subunits were first identified as the NUXM and NUZM in Neurospora crassa (Azevedo et al. 1992). NUXM is broadly conserved in C. albicans, N. crassa, Yarrowia lipolytica, Arabidopsis thaliana and Chlamydomonas reinhardtii. NUXM is orf19.6607 in the Candida Genome Database, while NUZM is orf19.287. These two genes have been named NUO1 and NUO2 as they are both NADH:ubiquione oxidoreductases (She et al. 2015). The null mutants of NUO1 and NUO2 had normal profiles of respiratory dysfunction, including reduced oxygen consumption and membrane potential, elevated ROS and reduction of CI enzyme activity (She et al. 2015). These phenotypes were similar to the phenotypes for a mutant in NDH51, which is broadly conserved among eukaryotic CI subunit proteins (Gabaldon, Rainey and Huynen 2005). The fungal specificity of Nuo1p and Nuo2p as well as important activities, such as virulence (see below), suggests that both may be attractive targets for therapeutic intervention.
Previous to our current studies of NUO1 and NUO2 (She et al. 2015), we had screened for fluconazole susceptibility a library of CI mutants (Sun et al. 2013). Of the 12 suspected CI mutants tested, nine were hypersusceptible to fluconazole. Each of the nine hypersusceptible mutants was unable to grow on glycerol-yeast-peptone agar medium (YP-glycerol) but could in glucose-yeast-peptone (YP-glucose). The fluconazole hypersusceptible mutants included orf19.6607 (NUO1) and orf19.287 (NUO2). The three mutants that grew as well as WT cells in the presence of fluconazole were also able to grow in both YP-glycerol and YP-glucose, indicating they were very likely not required for mitochondrial respiration and perhaps unlikely to be CI subunit proteins. These data indicate a strong fluconazole hypersusceptibility with the loss of CI and point out another interesting feature of CI subunit proteins such as Nuo1p and Nuo2p.
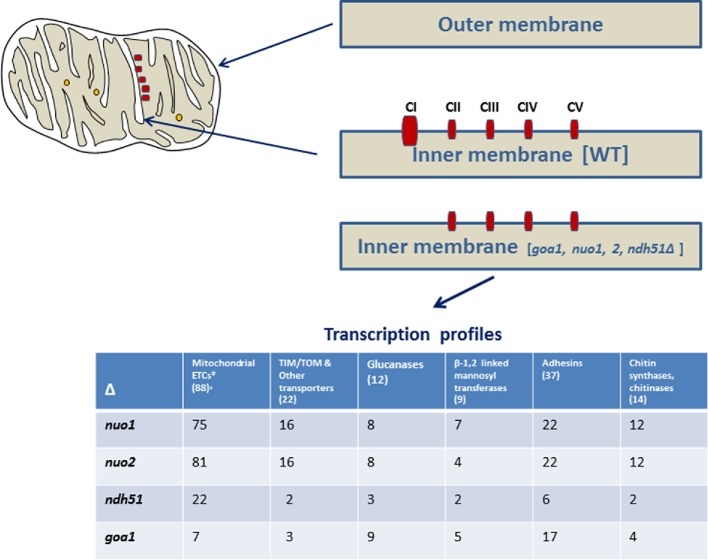
Both nuo1Δ and nuo2Δ are avirulent in a murine model of hematogenously disseminated candidiasis (She et al. 2015). Relevant to the avirulence phenotype, we expected many similarities among the CI mutants as a consequence of gene loss. Relatively unexpected was, however, that the transcription profiles of nuo1Δ and nuo2Δ were quite different than that of ndh51Δ. Especially noted were the changes in genes relevant to cell wall and mitochondrial functions (Fig. 3). These data may imply additional functions (besides CI enzyme activity) associated with NUO1, NUO2 and NDH51. Below, we present data on differences among cell wall encoding genes in Nuo1p, Nuo2pand Ndh51p (CI subunit proteins) as well as Goa1p, which is not a subunit protein of CI.
Figure 3.
Left. A WT mitochondrion is shown with an intact CI–CV ETC chain (red squares). The five subunits of the ETC are shown as rectangles. As for the individual subunit proteins of CI mentioned below, Ndh51p is critically located at the ubiquinone reduction site within the CI where other NADH oxidoreductases are located. The location of Nuo1p and Nuo2p is not known. Right. The outer/inner membranes are shown. In the CI mutants as well as goa1Δ, a loss (disassembly) of the CI complex from the inner membrane is depicted. The CI in the WT is represented as larger in molecular mass versus the other complexes. For each mutant, the disassembly of the CI is represented. Table below. Upregulated and downregulated genes in each of the CI mutants and in the putative CI regulator (goa1Δ) are listed for specific gene categories. Nuo1p and Nuo2p are nearly functionally redundant. All of the mutants except goa1Δ have major upregulation in genes of the ETC complexes, except CI (left column). These data may suggest that mutants have ETC compensatory activities that include one of the two alternate oxidases AOX2 and the NDE1.
The three mitochondrial CI subunit proteins Nuo1p, Nuo2p and Ndh51p were not created equally
We were interested in explaining the loss of CI enzyme activity of nuo1Δ and nuo2Δ mutants. For these experiments, we also included the ndh51Δ, which as mentioned above is a broadly conserved CI subunit protein (Gabaldon, Rainey and Huynen 2005). Are the reason(s) for reduced CI activity the same for each mutant? We do not know for certain, but a significant 70–80% loss of the CI complex was observed (She et al. 2015) (Fig. 3). We refer to this reduction in the amount of CI as disassembly, also reported in N. crassa for the loss of CI subunit proteins, summarized in Mimaki et al. (2012). Thus, our hypothesis is that a reduction in CI enzyme activity occurs because the CI is not assembled properly. A similar loss of CI is observed with the goa1Δ (Li et al. 2011). In comparison to the CI subunit proteins, we believe that Goa1p is either responsible for the assembly of the C. albicans CI subunit proteins individually or more likely is needed for the assembly of one or more subcomplexes of CI. Future experiments need to be done to resolve this hypothesis. In any case, similar defects in each of the mutants suggest that a CI assembly defect causes reduced respiratory activity.
Are other activities of each subunit protein uniform, or are they different? Transcriptional profiling revealed general similarities among Nuo1p and Nuo2p that are quite different from the Ndh51p in families of β-glucanases, chitinases and mannosyl transferases, and compensatory upregulation of a larger number of genes encoding the mitochondrial biogenesis apparatus in nuo1Δ and nuo2Δ compared to ndh51Δ. These observations suggest subunit-specific activities particularly in regard to effects on cell wall integrity and potentially mitochondrial biogenesis (Fig. 3).
MITOCHONDRIAL ACTIVITY IN C. ALBICANS DEVELOPMENTAL TRANSITIONS
Widely studied virulence pathways of C. albicans are the developmental transitions between yeast and hyphal morphology, as well as the ability to adhere to substrates for biofilm growth (Finkel and Mitchell 2011; Sudbery 2011). Signaling mechanisms and gene expression regulators that mediate the morphological and physiological changes in response to environmental cues are well characterized (Finkel and Mitchell 2011; Sudbery 2011). Metabolic reprograming accompanies the yeast to hyphae morphogenesis and biofilm formation (Bonhomme et al. 2011; Nobile et al. 2012; Fox et al. 2014, 2015; Guedouari et al. 2014; Holland et al. 2014). It is easy to imagine that metabolic reprograming could signal directly to the pathways that regulate hyphal and biofilm growth, and/or could be involved in quorum-sensing mechanisms in the multicellular biofilms. While there is evidence of their involvement, how precisely mitochondrial functions are wired into the complex cellular pathways that control hyphal and biofilm growth remains to be fully defined.
Generally, mitochondrial dysfunction cripples hyphal morphogenesis in C. albicans, reviewed in Shingu-Vazquez and Traven (2011). Some studies however suggest the opposite, for example treatment with the mitochondrial inhibitor antimycin promoted hyphal morphogenesis in one study (Mulhern, Logue and Butler 2006), while in another study it had no effect (Watanabe et al. 2006). A recent report using biochemical assays to measure outputs such as TCA cycle and oxygen consumption suggests that robust mitochondrial activity is necessary to support growth of hyphae (Guedouari et al. 2014). It has been suggested that mitochondrial respiration might be signaling to RAS/PKA (Watanabe et al. 2006), a critical signal transduction pathway for the morphogenetic switch in C. albicans (Hogan and Sundstrom 2009). Direct proof of this remains to be established. Interestingly, several reports in S. cerevisiae link mitochondrial activity with RAS/PKA pathway status. A recent report using the filamentous Sigma1278b strain of S. cerevisiae indicated that mitochondrial dysfunction reduced Ras/PKA pathway activity, thereby inhibiting the expression of the adhesin FLO11 and impairing the ability to grow as pseudohyphal filaments (Aun, Tamm and Sedman 2013). A previous study by one of us supports the idea that a S. cerevisiae mitochondrial petite mutant displays reduced Ras/PKA pathway activity (Traven et al. 2001), but another report from the Nunnari lab found the opposite—hyperactivation of Ras/PKA upon mitochondrial inhibition (Graef and Nunnari 2011). Future studies should directly test the effects of mitochondrial dysfunction on hyphal signaling pathways in C. albicans. In any case, if future antifungal drugs are developed that target mitochondrial activity, these compounds are likely to block hyphal formation—this could be beneficial for reducing virulence of C. albicans, and also for modulating the inflammatory response that is triggered by the hyphal form and which likely contributes to pathology (Traven and Naderer 2014).
Robust hyphal formation is needed for C. albicans biofilm maturation, and thus the prediction is that mitochondrial respiration is necessary for biofilm formation. In support for this idea, a mutant in the kinase domain of the transcriptional regulator Mediator displays higher mitochondrial respiration and is able to form biofilms more robustly than the WT strain (Lindsay et al. 2014). Accordingly, Mediator kinase domain mutants are resistant to the inhibitory effects on morphogenesis of the phenazine pyocyanin (Lindsay et al. 2014). Pyocyanin is produced by Pseudomonas aeruginosa and it mediates P. aeruginosa–C. albicans interspecies interactions by targeting fungal succinate dehydrogenase. Pyocyanin inhibits the formation of biofilms by C. albicans (Morales et al. 2013), suggesting that targeting mitochondria could be beneficial for blocking biofilm formation.
While it appears that cellular respiration is necessary for C. albicans hyphal morphogenesis and consequently biofilm maturation, transcriptomic and metabolomics studies suggest that mitochondrial activity is downregulated as the biofilms mature (Nobile et al. 2012; Zhu et al. 2013; Holland et al. 2014; Fox et al. 2015) (Fig. 4). These seemingly contradictory results might be reconciled if one thinks about the temporal context during biofilm formation, and the structural complexity of three-dimensional biofilm growth (Fig. 4). Hyphal morphogenesis is an early event that follows substrate adhesion in the biofilm formation process (Finkel and Mitchell 2011). It is possible that at this early stage, mitochondrial activity is necessary and beneficial (Fig. 4). As the biofilms mature, the changes in nutrient and oxygen availability might drive a decline in mitochondrial function (Fig. 4). In fact, it has recently been shown that biofilms do contain hypoxic pockets (Fox et al. 2014). Could a decline in mitochondrial function be beneficial for biofilm growth? Analysis of S. cerevisiae colonies as a proxy for biofilm structures provides some insight in this regard (Cap et al. 2012; Traven et al. 2012). Fractionation of the inside from the outside colony layers and subsequent transcriptomics showed that the outside cells are active, but their mitochondrial function might be reduced, while the inside cells do not appear to be actively growing, but mitochondrial activity is present, as the capacity of these inner cells to consume oxygen is higher than in the outside cells (Cap et al. 2012). It has been suggested that reduction in mitochondrial activity observed on the outside colony layers might protect the colony from the detrimental effect of oxidative damage by ROS produced via mitochondrial respiration (Cap et al. 2012). How these results with S. cerevisiae colonies correlate to C. albicans biofilms is not clear at the moment, but these studies nevertheless suggest that modulation of mitochondrial activity in biofilms might be triggering some of the biofilm phenotypes, such as stress resistance. In fact, Cap et al. (2012) suggested that it might be promoting prolonged survival of biofilm cells (Fig. 4). Direct studies of mitochondrial activity in C. albicans biofilms, and its effects on stress resistance and antifungal drug susceptibility should provide new insight into the interplay between metabolic reprograming and developmental switches in C. albicans, and inform how targeting mitochondria might be beneficial for biofilm-related Candida infections.
Figure 4.
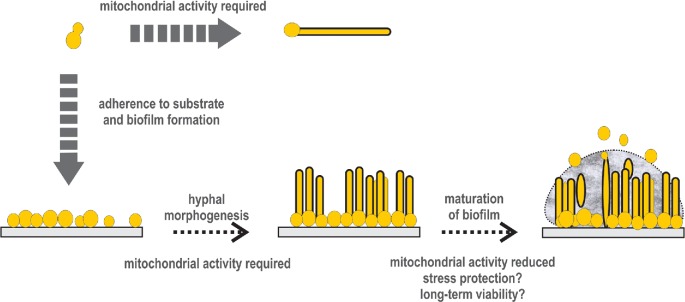
Roles of mitochondrial activity in C. albicans morphogenesis and biofilm formation. Mitochondrial activity is necessary for the transition from yeast to hyphal growth, but in mature biofilms mitochondria might be repressed perhaps due to changes in nutrients or access to oxygen. Reduction of mitochondrial activity might be driving protection from stresses, such as oxidative stress, thereby promoting cellular fitness. Whether and how mitochondrial functions impact on antifungal susceptibility of Candida biofilms remains to be determined.
MITOCHONDRIA AS A SOURCE FOR POTENTIAL ANTIFUNGAL DRUG TARGETS
The most recent additions to the antifungal armature are remodeled triazoles (Calderone et al. 2014). An approach to finding antifungal drugs is to screen repurposed- or off-patent compounds that have anticancer, antidepressant and other established activities. Several are known to have antifungal activities (Roemer and Krysan 2014). Still, new targets and correspondingly, new antifungal compounds to those targets are needed. The new compounds should optimally be fungal specific, not toxic to humans, fungicidal and protective in animal models. Fungal specificity of potential targets is the easiest of prerequisites to establish, given the availability of genome databases. However, what is time consuming, in most cases, is verification that a target is important to growth and virulence of pathogens. In this regard, there are examples such as the hybrid histidine kinases (Defosse et al. 2015). There is also a growing literature that loss of fungal-specific mitochondrial targets begat avirulence in gene-specific mutants (Bambach et al. 2009; Becker et al. 2010; Noble et al. 2010; Qu et al. 2012; Guan et al. 2015). Repurposed compounds are plentiful and available upon request from the NIH libraries Program [The Molecular libraries Small Molecule Repository (MLSMR) http://mli.gov/mil/mlp-overview]. With regard to antifungal drug discovery that targets CI, a dual platform high-throughput screen of compounds could be established. The assay could combine a screen of compounds to inhibit growth in the presence of glycerol and a secondary screen utilizing heterozygous CI or non-CI mitochondrial mutants with a read out of haploinsufficiency or resistance (of null mutants).
As we have discussed previously, mitochondrial dysfunction in some fungi, notably S. cerevisiae and C. glabrata, can lead to drug resistance, including reduced susceptibility to the azoles (Shingu-Vazquez and Traven 2011). However, we observed the exact opposite with CI mutants in C. albicans, i.e. increased azole susceptibility (Sun et al. 2013), and generally mitochondrial dysfunction leads to higher sensitivity to the echinocandins, reviewed in Shingu-Vazquez and Traven (2011). Candida glabrata and S. cerevisiae lack CI NADH-oxidoreductases (Koszul et al. 2003), so that antifungal drug resistance in both of these organisms very likely reflects other mechanisms. However, a better understanding of the molecular basis of mitochondrial function in drug susceptibility is needed to fully appreciate the dangers of drug resistance if targeting mitochondrial function in pathogens. Having said that, as we have previously argued, the severe viability defects upon mitochondrial dysfunction in pathogenic fungi suggest that inhibition of mitochondria would be an effective way to combat fungal disease (Shingu-Vazquez and Traven 2011).
CONCLUSIONS AND OUTLOOK
The central place for mitochondrial activity in eukaryotic cell physiology makes the mitochondrion a prime target for the development of new therapeutics to treat fungal disease. The obvious concern is the conservation of mitochondrial proteins and their functions between fungi and humans, which could lead to host toxicity of mitochondria-targeted drugs. As we have discussed in this review, there are several mitochondrial proteins that play an essential role in organelle biogenesis or function, and yet they lack structural homologs in humans. There is now evidence from studies in C. albicans that some of these pathogen-specific mitochondrial proteins are virulence determinants in the systemic model in mice (Bambach et al. 2009; Becker et al. 2010; Noble et al. 2010; Qu et al. 2012; Guan et al. 2015). In some cases, the pathogen-specific mitochondrial proteins are broadly conserved in fungi and are likely to play important roles in fungal virulence more broadly than only in CTG species of Candida. This means that mitochondrial inhibitors have a chance to act as pan-fungal drugs. Finally, targeting mitochondrial function would be a novel approach for antifungal therapy, which is a significant advantage when considering the limited spectrum of current antifungal therapeutics.
Acknowledgments
The authors wish to thank Trevor Lithgow for his help with Figures 1 and 4.
FUNDING
The authors wish to acknowledge bridge support from the Georgetown University Medical Center Biomedical Graduate Research Organization and an NIH-NIAID grant (AI090290) to RC. Work on fungal mitochondria in the AT laboratory is supported by grants from the Australian National Health and Medical Research Council (APP1023973) and the Australian Research Council (DP130100578).
Conflict of interest. None declared.
REFERENCES
- Abdrakhmanova A, Zickermann V, Bostina M, et al. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim Biophys Acta. 2004;1658:148–56. doi: 10.1016/j.bbabio.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Altmann K, Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5410–7. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aun A, Tamm T, Sedman J. Dysfunctional mitochondria modulate cAMP-PKA signaling and filamentous and invasive growth of Saccharomyces cerevisiae. Genetics. 2013;193:467–81. doi: 10.1534/genetics.112.147389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo JE, Nehls U, Eckerskorn C, et al. Primary structure and mitochondrial import in vitro of the 20.9 kDa subunit of complex I from Neurospora crassa. Biochem J. 1992;288(Pt 1):29–34. doi: 10.1042/bj2880029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambach A, Fernandes MP, Ghosh A, et al. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell. 2009;8:1706–20. doi: 10.1128/EC.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JM, Kauffman SJ, Hauser M, et al. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. P Natl Acad Sci USA. 2010;107:22044–9. doi: 10.1073/pnas.1009845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockler S, Westermann B. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell. 2014;28:450–8. doi: 10.1016/j.devcel.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang HC, et al. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–27. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme J, Chauvel M, Goyard S, et al. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol. 2011;80:995–1013. doi: 10.1111/j.1365-2958.2011.07626.x. [DOI] [PubMed] [Google Scholar]
- Bridges HR, Fearnley IM, Hirst J. The subunit composition of mitochondrial NADH:ubiquinone oxidoreductase (complex I) from Pichia pastoris. Mol Cell Proteomics. 2010;9:2318–26. doi: 10.1074/mcp.M110.001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–91. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone R, Sun N, Gay-Andrieu F, et al. Antifungal drug discovery: the process and outcomes. Future Microbiol. 2014;9:791–805. doi: 10.2217/fmb.14.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cap M, Stepanek L, Harant K, et al. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell. 2012;46:436–48. doi: 10.1016/j.molcel.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, et al. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Calderone R, Sun N, et al. Caloric restriction restores the chronological life span of the Goa1 null mutant of Candida albicans in spite of high cell levels of ROS. Fungal Genet Biol. 2012;49:1023–32. doi: 10.1016/j.fgb.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Dagley MJ, Gentle IE, Beilharz TH, et al. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol Microbiol. 2011;79:968–89. doi: 10.1111/j.1365-2958.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- Defosse TA, Sharma A, Mondal AK, et al. Hybrid histidine kinases in pathogenic fungi. Mol Microbiol. 2015;95:914–24. doi: 10.1111/mmi.12911. [DOI] [PubMed] [Google Scholar]
- Devillers A, Courjol F, Fradin C, et al. Deficient beta-mannosylation of Candida albicans phospholipomannan affects the proinflammatory response in macrophages. PLoS One. 2013;8:e84771. doi: 10.1371/journal.pone.0084771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Fritz S, Fuchs F, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–53. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drose S, Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol. 2012;748:145–69. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Enjalbert B, MacCallum DM, Odds FC, et al. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun. 2007;75:2143–51. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannjiang Y, Cheng WC, Lee SJ, et al. Mitochondrial fission proteins regulate programmed cell death in yeast. Gene Dev. 2004;18:2785–97. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–18. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, et al. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EP, Bui CK, Nett JE, et al. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 2015 doi: 10.1111/mmi.13002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EP, Cowley ES, Nobile CJ, et al. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 2014;24:2411–6. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C, Slomianny MC, Mille C, et al. Beta-1,2 oligomannose adhesin epitopes are widely distributed over the different families of Candida albicans cell wall mannoproteins and are associated through both N- and O-glycosylation processes. Infect Immun. 2008;76:4509–17. doi: 10.1128/IAI.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) J Mol Biol. 2005;348:857–70. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, et al. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 2011;30:2101–14. doi: 10.1038/emboj.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzer S, Lithgow T, Bauer RE, et al. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G, Wang H, Liang W, et al. The mitochondrial protein Mcu1 plays important roles in carbon source utilization, filamentation, and virulence in Candida albicans. Fungal Genet Biol. 2015 doi: 10.1016/j.fgb.2015.01.006. in press. [DOI] [PubMed] [Google Scholar]
- Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–6. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedouari H, Gergondey R, Bourdais A, et al. Changes in glutathione-dependent redox status and mitochondrial energetic strategies are part of the adaptive response during the filamentation process in Candida albicans. Biochim Biophys Acta. 2014;1842:1855–69. doi: 10.1016/j.bbadis.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Hewitt VL, Gabriel K, Traven A. The ins and outs of the intermembrane space: diverse mechanisms and evolutionary rewiring of mitochondrial protein import routes. Biochim Biophys Acta. 2014;1840:1246–53. doi: 10.1016/j.bbagen.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Hewitt VL, Heinz E, Shingu-Vazquez M, et al. A model system for mitochondrial biogenesis reveals evolutionary rewiring of protein import and membrane assembly pathways. P Natl Acad Sci USA. 2012;109:E3358–66. doi: 10.1073/pnas.1206345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 2009;4:1263–70. doi: 10.2217/fmb.09.106. [DOI] [PubMed] [Google Scholar]
- Holland LM, Schroder MS, Turner SA, et al. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014;10:e1004365. doi: 10.1371/journal.ppat.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, et al. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honscher C, Mari M, Auffarth K, et al. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Ishii N, Yamamoto M, Yoshihara F, et al. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology. 1997;143(Pt 2):429–35. doi: 10.1099/00221287-143-2-429. [DOI] [PubMed] [Google Scholar]
- Ishikawa D, Yamamoto H, Tamura Y, et al. Two novel proteins in the mitochondrial outer membrane mediate beta-barrel protein assembly. J Cell Biol. 2004;166:621–7. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM. The retrograde response: when mitochondrial quality control is not enough. Biochim Biophys Acta. 2013;1833:400–9. doi: 10.1016/j.bbamcr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karababa M, Coste AT, Rognon B, et al. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Ch. 2004;48:3064–79. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamooshi K, Sikorski P, Sun N, et al. The Rbf1, Hfl1 and Dbp4 of Candida albicans regulate common as well as transcription factor-specific mitochondrial and other cell activities. BMC Genomics. 2014;15:56. doi: 10.1186/1471-2164-15-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–81. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123:1389–93. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Malpertuy A, Frangeul L, et al. The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 2003;534:39–48. doi: 10.1016/s0014-5793(02)03749-3. [DOI] [PubMed] [Google Scholar]
- Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Bi. 2014;30:357–91. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–34. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- Li D, Chen H, Florentino A, et al. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot Cell. 2011;10:672–82. doi: 10.1128/EC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AK, Morales DK, Liu Z, et al. Analysis of Candida albicans mutants defective in the Cdk8 module of mediator reveal links between metabolism and biofilm formation. PLoS Genet. 2014;10:e1004567. doi: 10.1371/journal.pgen.1004567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Hagen F, Stekel DJ, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. P Natl Acad Sci USA. 2009;106:12980–5. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough JA, Bhattacherjee V, Sadlon T, et al. Involvement of Candida albicans NADH dehydrogenase complex I in filamentation. Fungal Genet Biol. 2002;36:117–27. doi: 10.1016/S1087-1845(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfannschmidt S, Rissler M, et al. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–39. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Rissler M, Chacinska A, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Mille C, Bobrowicz P, Trinel PA, et al. Identification of a new family of genes involved in beta-1,2-mannosylation of glycans in Pichia pastoris and Candida albicans. J Biol Chem. 2008;283:9724–36. doi: 10.1074/jbc.M708825200. [DOI] [PubMed] [Google Scholar]
- Mille C, Fradin C, Delplace F, et al. Members 5 and 6 of the Candida albicans BMT family encode enzymes acting specifically on beta-mannosylation of the phospholipomannan cell-wall glycosphingolipid. Glycobiology. 2012;22:1332–42. doi: 10.1093/glycob/cws097. [DOI] [PubMed] [Google Scholar]
- Mille C, Janbon G, Delplace F, et al. Inactivation of CaMIT1 inhibits Candida albicans phospholipomannan beta-mannosylation, reduces virulence, and alters cell wall protein beta-mannosylation. J Biol Chem. 2004;279:47952–60. doi: 10.1074/jbc.M405534200. [DOI] [PubMed] [Google Scholar]
- Mimaki M, Wang X, McKenzie M, et al. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817:851–62. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Morales DK, Grahl N, Okegbe C, et al. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio. 2013;4:e00526–12. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhern SM, Logue ME, Butler G. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot Cell. 2006;5:2001–13. [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, et al. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–8. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–36. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Pereira B, Videira A, Duarte M. Novel insights into the role of Neurospora crassa NDUFAF2, an evolutionarily conserved mitochondrial complex I assembly factor. Mol Cell Biol. 2013;33:2623–34. doi: 10.1128/MCB.01476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Jelicic B, Pettolino F, et al. Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence. Eukaryot Cell. 2012;11:532–44. doi: 10.1128/EC.05292-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, et al. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–5. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Roemer T, Krysan DJ. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect Med. 2014;4:a019703. doi: 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013;17:954–64. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Zhang L, Chen H, et al. Cell surface changes in the Candida albicans mitochondrial mutant goa1Delta are associated with reduced recognition by innate immune cells. Cell Microbiol. 2013;15:1572–84. doi: 10.1111/cmi.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She XKK, Gao Y, Shen Y, et al. Fungal-specific subunits of the Candida albicans mitochondrial complex I drive diverse functions such as cell wall synthesis. Cell Microbiol. 2015 doi: 10.1111/cmi.12438. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu-Vazquez M, Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell. 2011;10:1376–83. doi: 10.1128/EC.05184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–48. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Sun N, Fonzi W, Chen H, et al. Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob Agents Ch. 2013;57:532–42. doi: 10.1128/AAC.01520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tar K, Dange T, Yang C, et al. Proteasomes associated with the Blm10 activator protein antagonize mitochondrial fission through degradation of the fission protein Dnm1. J Biol Chem. 2014;289:12145–56. doi: 10.1074/jbc.M114.554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E, Roman E, Claypool S, et al. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob Agents Ch. 2013;57:5580–99. doi: 10.1128/AAC.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Janicke A, Harrison P, et al. Transcriptional profiling of a yeast colony provides new insight into the heterogeneity of multicellular fungal communities. PLoS One. 2012;7:e46243. doi: 10.1371/journal.pone.0046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Naderer T. Microbial egress: a hitchhiker's guide to freedom. PLoS Pathog. 2014;10:e1004201. doi: 10.1371/journal.ppat.1004201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Wong JM, Xu D, et al. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J Biol Chem. 2001;276:4020–7. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- Ueno K, Matsumoto Y, Uno J, et al. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS One. 2011;6:e24759. doi: 10.1371/journal.pone.0024759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Okawara A, Yamagoe S, et al. The mannan of Candida albicans lacking beta-1,2-linked oligomannosides increases the production of inflammatory cytokines by dendritic cells. Med Mycol. 2013;51:385–95. doi: 10.3109/13693786.2012.733892. [DOI] [PubMed] [Google Scholar]
- Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta. 2014;1841:595–609. doi: 10.1016/j.bbalip.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Videira A, Duarte M. From NADH to ubiquinone in Neurospora mitochondria. Biochim Biophys Acta. 2002;1555:187–91. doi: 10.1016/s0005-2728(02)00276-1. [DOI] [PubMed] [Google Scholar]
- Voelz K, Johnston SA, Smith LM, et al. ‘Division of labour’ in response to host oxidative burst drives a fatal Cryptococcus gattii outbreak. Nat Commun. 2014;5:5194. doi: 10.1038/ncomms6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Ogasawara A, Mikami T, et al. Hyphal formation of Candida albicans is controlled by electron transfer system. Biochem Bioph Res Co. 2006;348:206–11. doi: 10.1016/j.bbrc.2006.07.066. [DOI] [PubMed] [Google Scholar]
- Wideman JG, Gawryluk RM, Gray MW, et al. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol. 2013;30:2044–49. doi: 10.1093/molbev/mst120. [DOI] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 2010;11:187–93. doi: 10.1038/embor.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Wang H, Shang Q, et al. Time course analysis of Candida albicans metabolites during biofilm development. J Proteome Res. 2013;12:2375–85. doi: 10.1021/pr300447k. [DOI] [PubMed] [Google Scholar]