Abstract
Motivation: Although genome-wide association studies (GWAS) have identified thousands of variants associated with common diseases and complex traits, only a handful of these variants are validated to be causal. We consider ‘causal variants’ as variants which are responsible for the association signal at a locus. As opposed to association studies that benefit from linkage disequilibrium (LD), the main challenge in identifying causal variants at associated loci lies in distinguishing among the many closely correlated variants due to LD. This is particularly important for model organisms such as inbred mice, where LD extends much further than in human populations, resulting in large stretches of the genome with significantly associated variants. Furthermore, these model organisms are highly structured and require correction for population structure to remove potential spurious associations.
Results: In this work, we propose CAVIAR-Gene (CAusal Variants Identification in Associated Regions), a novel method that is able to operate across large LD regions of the genome while also correcting for population structure. A key feature of our approach is that it provides as output a minimally sized set of genes that captures the genes which harbor causal variants with probability ρ. Through extensive simulations, we demonstrate that our method not only speeds up computation, but also have an average of 10% higher recall rate compared with the existing approaches. We validate our method using a real mouse high-density lipoprotein data (HDL) and show that CAVIAR-Gene is able to identify Apoa2 (a gene known to harbor causal variants for HDL), while reducing the number of genes that need to be tested for functionality by a factor of 2.
Availability and implementation: Software is freely available for download at genetics.cs.ucla.edu/caviar.
Contact: eeskin@cs.ucla.edu
1 Introduction
Genome-wide association studies (GWAS) have been extremely successful in reproducibly identifying variants associated with various complex traits and diseases (Altshuler et al., 2008; Hakonarson et al., 2007; International Multiple Sclerosis Genetics Consortium et al., 2013; Kottgen et al., 2013; Ripke et al., 2013). The most common type of genetic variants comes in the form of single nucleotide polymorphisms (SNPs), which we make the focus of this study. Because of the correlation structure in the genome, a phenomenon referred to as linkage disequilibrium (LD) (Pritchard and Przeworski, 2001; Reich et al., 2001), each GWAS-associated variant will typically have hundreds to thousands of other variants which are also significantly associated with the trait. Identifying the variants responsible for the observed effect on a trait is referred to as fine mapping (Hormozdiari et al., 2014; Kichaev et al., 2014; Maller et al., 2012; Yang et al., 2012). In the context of association studies, the genetic variants which are responsible for the association signal at a locus are referred to in the genetics literature as the ‘causal variants’. Causal variants have biological effect on the phenotype. Generally, variants can be categorized into three main groups. The first group is the causal variants which have a biological effect on the phenotype and are responsible for the association signal. The second group is the variants which are statistically associated with the phenotype due to LD with a causal variant. Even though association tests for these variants may be statistically significant, under our definition, they are not causal variants. The third group is the variants which are not statistically associated with the phenotype and are not causal. We note that this usage of the term causal has little to do with the concept of causal inference as described in the computer science and statistics literatures (Pearl, 2000; Spirtes et al., 2000).
Fine-mapping methods take as input the full set of association signals in a region and attempt to identify a minimum set of variants that explains the association signals. A common approach is to calculate marginal association statistics for each variant and, depending on the study budget, select the top K ranked variants for follow-up studies. However, the local correlation structure at a fine-mapping locus will induce similar association statistics at neighboring, non-causals variants, thereby making this approach suboptimal in this context. Furthermore, it fails to provide a guarantee that the true causal variant is selected. A recent work (Maller et al., 2012) addressed this issue by estimating the probabilities for variants to be causal under the simplifying assumption that each fine-mapping locus contains a single causal variant. Ranking variants based on association strength (similar to top k) and this probabilistic approach (Maller et al., 2012) assuming a single causal variant give identical relative rankings. However, the probabilistic approach provides the added benefit that we can now select enough variants to guarantee that we have captured the true causal variants with ρ level of confidence. Unfortunately, the key underlying assumption that a fine-mapping locus contains a single causal variant is likely to be invalidated at many risk loci (Hormozdiari et al., 2014; Kichaev et al., 2014). For regions that putatively harbor multiple independent signals, a common strategy is to use iterative conditioning to tease out secondary signals (Yang et al., 2012). This process is analogous to forward stepwise regression, where at each iteration, the variant with the strongest association is selected to enter the model and then marginal statistical scores are re-computed for the remaining variants condition on the ones that have been selected. This process is repeated until there are no remaining variants that are statistically significant. However, it has been shown that this approach is highly sub-optimal (Hormozdiari et al., 2014; Kichaev et al., 2014) due to lack of LD consideration. To address these issues, we recently proposed probabilistic fine-mapping methods (Hormozdiari et al., 2014; Kichaev et al., 2014) that build on the concept of a standard confidence interval by providing a well-calibrated, minimally sized confidence set of variants using principled, LD-aware modeling of multiple causal variants. In these methods, we assign probability to each variant to be causal and subsequently select the smallest number of variants that achieve the desired posterior probability. Many accurate fine-mapping methods have been designed for human studies where there are a relatively small number of associated variants in a region. In model organism studies, however, pervasive LD patterns result in GWAS-associated loci that may span several megabases and contain thousands of variants and dozens of genes. For example, in a widely utilized design for mouse studies, the Hybrid Mouse Diversity Panel (HMDP) (Bennett et al., 2010)—the typical associated region—is approximately 1–2 megabases. Identifying which genes underlie an associated locus in model organism studies is a major, labor-intensive process involving generating gene knockouts. Therefore, it is often the case that identifying the causal genes at an associated locus requires a larger effort than the initial GWAS (Flint and Eskin, 2012). In addition to large LD blocks, fine-mapping studies in model organisms are complicated by population structure (i.e. the complex genetic relationship between different individuals in the study; Flint and Eskin, 2012; Kang et al., 2008; Price et al., 2006) that invalidate commonly used association statistics that assume the individuals in the study are independent. Model organisms such as mice have a high level of population structure, typically larger than what is observed in human populations; therefore, correcting for the population structure for mouse GWAS is imperative to mitigate the chance of false positive signals of association (Flint and Eskin, 2012; Kang et al., 2008; Price et al., 2006).
In this article, we propose CAVIAR-Gene (CAusal Variants Identification in Associated Regions), a statistical method for fine mapping that addresses two main limitations of existing methods. First, as opposed to existing approaches that focus on individual variants, we propose to search only over the space of gene combinations that explain the statistical association signal, and thus drastically reduce runtime. Second, CAVIAR-Gene extends existing framework for fine mapping to account for population structure. The output of our approach is a minimal set of genes that will contain the true casual gene at a pre-specified significance level. This gene set together with its individual gene probability of causality provides a natural way of prioritizing genes for functional testing (e.g. knockout strategies) in model organisms. Through extensive simulations, we demonstrate that CAVIAR-Gene is superior to existing methodologies, requiring the smallest set of genes to follow-up in order to capture the true causal gene(s). To validate our approach, we applied CAVIAR-Gene to real mouse data and found that we can successfully recover Apoa2, a known causal gene for high-density lipoprotein (HDL) (Flint and Eskin, 2012; van Nas et al., 2009), for the HDL phenotype in the HMDP.
2 Methods
2.1 Overview of CAVIAR-Gene
CAVIAR-Gene takes as input the marginal statistics for each variant at a locus, an LD matrix consisting of pairwise Pearson correlations computed between the genotypes of a pair of genetic variants, a partitioning of the set of variants in a locus into genes, and the kinship matrix which indicates the genetic similarity between each pair of individuals. Marginal statistics are computed using methods that correct for population structure (Kang et al., 2008; Lippert et al., 2011; Listgarten et al., 2012; Zhou and Stephens, 2012). We consider a variant to be causal when the variant is responsible for the association signal at a locus and aim to discriminate these variants from ones that are correlated due to LD. Our previous proposed method CAVIAR, is a statistical framework that provides a ‘ρ causal set’ that is defined as the set of variants that contain all the causal variants with probability of at least ρ. The intuition is that due to LD structure, it is impossible to identify exactly the causal variants, but it is possible to identify a set which contains these causal variants. CAVIAR was designed to work on human GWAS where we deal with regions that have at most 100 variants in a locus and we consider all possible causal combinations of at most 6 causal variants to detect the ρ causal set. However, in model organisms, the large stretches of LD regions result in a large number of variants associated in each region, thus making CAVIAR computationally infeasible.
CAVIAR-Gene mitigates this problem by associating each variant to a proximal gene, and instead, operating on the gene level, thus reducing the computational burden by an order of magnitude while facilitating interpreting of GWAS results. Similarly, CAVIAR-Gene detects a ‘ρ causal gene set’ which is a set of genes in the locus that will contain the actual causal genes with probability of at least ρ. Note that not all the genes selected in the ρ causal gene set will be causal. A trivial solution to this problem would be to output all the genes as the ρ causal gene set. However, because this provides no additional information, we are interested in detecting the ρ causal gene set which has the minimum number of genes. We demonstrate that CAVIAR-Gene is well-calibrated as it fails to detect the actual causal gene 1−ρ fraction of the time.
2.2 Standard GWAS
Consider a GWAS on a quantitative trait where we collect phenotypic values for n individuals and genotype all the individuals on m variants. Let yi indicate the phenotypic value of the ith individual and indicate the minor allele count of the ith individual for the kth variant. We use Y to denote the vector of phenotypic values and Xk to denote the vector of normalized genotype values for the kth variant for all the n individuals in the study. Without loss of generality, we assume that genotype values for each variant have been standardized to have mean 0 and variance 1 yielding the following relationships: 1T Xk = 0 and , where 1 denotes the vector of ones. We assume that the data generating model follows a linear additive model, and for simplicity the variant c is the only variant associated (causal) with the phenotype. Each variant is categorized into one of the three groups. The first group is variants which are associated with the phenotype and are considered causal. The second group is variants which are statistically associated with the phenotype due to LD with a causal variant—these variants are considered not causal. The third group is variants which are not associated with the phenotype and are considered not causal. Standard GWAS analysis for the cth variant is performed utilizing the following model equation:
| (1) |
where μ is the mean of the phenotypic values, βc is the effect size of the cth variant, and e is the residual noise. In this model, the residual error is the vector of i.i.d and normally distributed error. Let , where I is the (n × n) identity matrix and σe is a covariance scalar. The estimates of βc, which are indicated by , are obtained by maximizing the likelihood,
and the statistics is computed as follows:
where λc is the non-centrality parameter (NCP) and is equal to . We obtain the estimated value for μ, e, and σe as follows: , and .
2.3 The effect of LD in GWAS
In the previous section, we consider that there is only one variant (variant c), and this variant is causal. Now, we extend the previous case and for simplicity we assume there are two variants, c and k. Similar to the previous section, the variant c is causal and variant k is correlated to c through LD but has no phenotypic effect. The correlation between the two variants is r which is approximated by . Thus, the estimate for the effect size for the variant k is as follows:
and the statistics is computed as follows:
We compute the covariance between the estimated effect size of the two variants as follows:
Thus, the joint distribution of the marginal association statistics for the two variants given their NCPs follows a multivariate normal distribution (MVN),
where rij is the genotype correlation between the ith and jth variants. In the case that both variants are not causal, we have . In the case that the jth variant is causal and the ith variant is not causal, we have . In the case that jth variant is not causal and the ith variant is causal, we have . This result is known from previous studies (Han et al., 2009; Hormozdiari et al., 2014; Kichaev et al., 2014; Zaitlen et al., 2010).
2.4 Computing the likelihood of causal SNP status from GWAS data
Given a set of m variants, the pair-wise correlations denoted by Σ, we use the vector to denote the marginal association statistics. We extend the joint distribution mentioned above for m variants. The joint distribution follows an MVN distribution,
| (2) |
where Λ is the vector of normalized true effect sizes and Σ is a matrix of pair-wise genotype correlations between different SNPs. Let be a n × m matrix of genotype. We can approximate Σ using genotype data as follows: .
In CAVIAR (Hormozdiari et al., 2014), we introduce a new parameter C, which is a binary indicator vector used to represent causal status of m SNPs in a region (i.e. is 1 if the ith SNP is causal and 0 otherwise). We define a prior probability on the vector of Λ for a given causal status using an MVN distribution,
| (3) |
where Σc is a diagonal (m × m) matrix. The diagonal elements of Σc are set to or ϵ where ϵ is a very small constant to make sure the matrix Σc is full rank. The ith element on the diagonal is set to if the ith variant is causal and set to ϵ if the ith variant is non-causal. We know that the LD between two variants is symmetric (). We combine Equations (2) and (3) to compute the joint marginal association statistics of all the variants. The joint distribution follows an MVN distribution,
| (4) |
2.5 Computing the posterior probability of causal SNP status from GWAS data
Given the observed marginal association statistics, , we can compute the posterior probability of the causal SNP status as,
| (5) |
where is the set of all possible causal SNPs. Thus, the size of is . Furthermore, is the prior probability for a particular causal SNP status, . We use Z to indicate the normalization factor.
In CAVIAR, we use a simple prior for a causal SNP status. We assume that the probability of an SNP to be causal is independent from other SNPs and the probability of an SNP to be causal is γ. Thus, we compute the prior probability as . In our work, we set γ to 0.01 (Darnell et al., 2012; Eskin, 2008; Jul and Eskin, 2011). It is worth mentioning that although we use a simple prior for our model, CAVIAR can incorporate external information such as functional data or knowledge from previous studies. As a result, we can have SNP-specific prior where γi indicates the prior probability for the ith SNP to be causal. Thus, we can extend the prior probability to a more general case, .
To compute the posterior probability for each causal SNP status, we need to consider all the possible causal SNP status which is the denominator of Equation (5). To ease the computational burden, we assume we have at most six causal SNP in each region. Assuming we have an upper bound on the number of causal variants is a common procedure in fine-mapping methods (Hormozdiari et al., 2014; Kichaev et al., 2014). We show the upper bound of six causal variants have small effect on the results (Hormozdiari et al., 2014). This assumption reduces the size of from to m6 which is computationally feasible.
2.6 ρ causal SNP set
Give a set of SNPs , we define a causal SNP configuration as all the possible causal SNP status which excludes any SNP as causal outside the set . Note, our definition of causal SNP configuration includes the causal SNP status where no SNP is considered as causal. We use to denote the causal SNP configuration for the . We compute the posterior probability of set capturing all the true causal genes,
Let ρ denote the value of the posterior probability, where , and we refer to it as the confidence level of capturing the actual causal SNPs. We refer to as the ‘ρ confidence set’.
Given a confidence threshold , there may exist many confidence sets that have a confidence level greater than the threshold. However, among all the possible confidence sets, the sets which have the minimum number of SNPs are more informative or have higher resolution to detect the actual causal SNPs. Thus, we are interested in finding the confident set with the minimum size (with minimum number of selected SNPs), , where has the minimum size.
2.7 ρ causal gene set
Unfortunately, the ρ causal SNP sets for mice can select many variants due to the high LD. Instead, we would like to find a set of genes that harbors causal variants. We define a ρ causal gene set as a set of genes which captures all the genes which harbor the causal variants with probability at least ρ. One of the benefits of detecting the ρ causal gene set requires less computation than detecting the ρ causal SNP set.
For simplicity, we use genes as a way to group the SNP to detect the causal SNPs. Thus, SNPs are partition to sets and this partition of the SNPs is done based on the genes. As a result, when a gene is selected in the ρ causal gene set, we can consider all the SNPs which are assigned to that gene which are selected in the ρ causal SNP set in the CAVIAR model. We use a simple way to assign SNPs to a gene—we assign an SNP to the closest gene. We would like to emphasize that CAVAIR-Gene can incorporate more complicated SNP to gene assignment.
Let be a set of genes and indicate all the SNPs assigned to the genes in the set . Then, we formally define the ρ causal gene set as a set where the total posterior probability of all the SNPs in that captures all the causal SNPs is ρ. Among all the ρ causal gene set, we are interested in the set which has the minimum number of genes selected.
Thus, to detect the ρ causal gene set, we need to search over all the possible sets of genes. Given genes in loci, we have possible causal gene set which is much smaller than all the possible sets of SNP, which are .
2.8 Greedy algorithm to detect the ρ causal gene set
We would like to emphasize that ρ causal gene set should capture all the causal genes; however, not all the genes selected in the ρ causal gene set are causal. Thus, even if we set an upper bound of six on the number of causal genes, the size of the ρ causal gene set can be larger than six genes. For example, if we have one causal variant and all the variants in that region have perfect LD, just utilizing the marginal statistics is impossible to distinguish which gene is the actual causal gene. Thus, in order to have 95% causal gene set, we have to select all the genes in the region. This is similar to what we observe in the variant level from previous studies (Hormozdiari et al., 2014; Kichaev et al., 2014).
Instead of considering all the possible causal gene set to find the ρ causal gene set, we propose the following greedy algorithm to ease the computational burden. For each gene, we define a weight that indicates the amount that each gene contributes toward the posterior probability of the ρ causal gene set. Genes which have higher weights will have higher probability of being selected in the ρ causal gene set. Thus, we pick the top set of genes for which the summation of their weights is at least ρ fraction of total weights of all genes in the region.
We use as a vector for the weights of all the genes, where wi is the weight of the ith gene and we compute the weight for the ith gene as follow:
| (6) |
We compute the weight for the ith gene by summing over all the causal gene statuses where the ith gene is selected as causal. We show in Section 3 that the proposed greedy and the brute force algorithm which consider all possible causal gene status tend to have similar results.
2.9 Handling marginal statistics corrected for population structure
The linear model which is used in the standard GWAS assumes only one causal SNP as shown in Equation (1). Moreover, in this linear model, we assume that the phenotypic value of each individual is independent from the phenotypic value of another individual. This assumption is not true in general for GWAS, especially in model organisms such as inbred mice. The model that accounts for this dependency is as follows:
| (7) |
Unfortunately, in a typical GWAS, the number of individuals in a study is much smaller than the number of SNPs (). Thus, estimating the effect size of all the SNPs is not possible. We test each SNP one at a time, , where models the random effects. In this model, we assume that each SNP has an effect and the effect of each SNP is distributed normally as . The total genetic variance is defined as and we use as the estimated genetic variance. We compute the variance of the random effect as , where is referred to as the kinship matrix. The kinship matrix defines pair-wise genetic relatedness which is computed from the genotype data. Let V be the total variance of phenotype Y, which is computed as . Let be the estimated environment and measurement error variance. Thus, the total estimated variance is .
We assume that the collected phenotype has an MVN distribution as follows: . Similar to linear regression, we compute the estimate of the effect size of the causal SNP by maximizing the likelihood. Moreover, we can estimate the effect size of the SNP which is indirectly associated to the causal SNP,
and the statistics is computed as follows:
We would like to emphasize all the existing methods (Kang et al., 2008; Lippert et al., 2011; Listgarten et al., 2012; Zhou and Stephens, 2012) which correct for population structure computes the marginal statics for each variant. However, corrected marginal statistics cannot be used by existing fine-mapping methods (Hormozdiari et al., 2014; Kichaev et al., 2014). As in these methods, we assume that the correlation between the computed marginal statistics is equal to the correlation between the two corresponding variants. As shown in our experiment below, the correlation between the marginal statistics which are corrected for population structure is not equal to the correlation of genotypes corresponding to the two variants.
We compute the covariance between the observed statistics for a causal SNP (variant) and an SNP (variant) which is indirectly associated with the causal SNP as follows:
Let matrix L be the Cholesky decomposition of matrix . Let and . We assume that , and are normalized to mean 0 and variance 1. Thus, we can re-write the covariance between the computed statistics for two SNPs as follow:
This indicates that the covariance between the two marginal statistics corrected for population structure follows an MVN where the correlation between the two statistics is the correlation between the transformed genotype for both SNPs. Thus, we re-write Equation (2) for the case the marginal statistics is corrected for population structure as follows: , where is the pair-wise correlation matrix which is computed by transforming the genotyped data and then computing the pair-wise correlation of transformed genotypes. In principle, this result could also be applied to other problems such as imputing the missing variants that utilize the summary statistics (Lee et al., 2013; Pasaniuc et al., 2014).
3 Results
3.1 CAVIAR-Gene is computationally efficient
CAVIAR and CAVIAR-Gene at high level can consider all possible causal combinations for variants and genes, respectively. However, considering all possible causal combinations is intractable. In CAVIAR, we make an assumption that in each locus we have at most six causal variants. However, in CAVIAR, in order to detect the ρ causal variants, we consider all possible causal sets which can be very slow depending on the number of variants selected in the ρ causal variant set. In the worst case, the running time of CAVIAR can be , where m is the total number of variants in a region. In CAVIAR-Gene, we use the proposed greedy method which is mentioned in Section 2.8. This greedy algorithm reduces the complexity of CAVIAR from to . Applying CAVIAR on loci with 100 of variants will take around 30 h. However, it will take 2 h for CAVIAR-Gene to finish on the same loci and 3 h for CAVIAR-Gene to finish on loci with 200 variants. Figure 1 indicates the running time compression between CAVIAR and CAVIAR-Gene for different number of variants in a region.
Fig. 1.
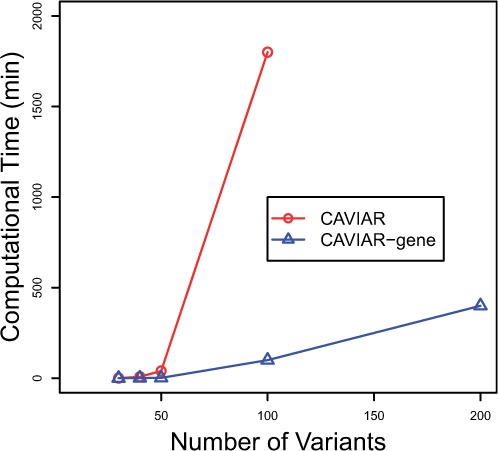
CAVIAR-Gene is computationally more efficient than CAVIAR. Running time comparison between CAVIAR and CAVIAR-Gene. The experiments are run on a 64 bit Intel(R) Xeon(R) 2 G with 5 GB RAM
3.2 CAVIAR-Gene-estimated causal gene sets are well-calibrated
To assess the performance of our method, we conducted a series of simulations. To make our simulations more realistic, we utilize real genotypes from three different datasets: outbred dataset (Zhang et al., 2012), F2 dataset (van Nas et al., 2009), and HMDP dataset (Bennett et al., 2010). After obtaining the real genotype for each dataset, we partition the genome into segments containing 200 genes. For each segment, we implant one, two, or three causal genes in the region where a gene is considered causal if it harbors at least one causal variant. We then generate simulated phenotypes for each segment using a linear mixed model as in the previous studies (Han et al., 2009; Zaitlen et al., 2010).
We extend the existing methods, which are designed to detect the causal variants, to detect the causal genes. For these methods, we consider a gene to be causal if any of the variants in that gene are selected as causal. We run TopK-Gene, conditional method (CM-Gene) (Yang et al., 2012), 1Post-Gene (Maller et al., 2012), and CAVIAR-Gene. Among these methods, CAVIAR-Gene is the only method that is well-calibrated to detect causal genes as shown in Table 1. We consider a method to be well-calibrated if it accurately captures the causal genes in ρ fraction of the time. It is worth mentioning that 1Post-Gene is well-calibrated when we only have one true causal gene; however, 1Post-Gene is mis-calibrated when there are more than one causal gene in the locus as shown in Table1.
Table 1.
CAVIAR-Gene estimated causal gene-sets are well-calibrated
| Causal gene | Recall rate (%) | Causal gene size | ||||
|---|---|---|---|---|---|---|
| 1Post-Gene | CM-Gene | CAVIAR-Gene | 1Post-Gene | CM-Gene | CAVIAR-Gene | |
| 1 | 0.995 | 0.941 | 0.990 | 2.59 | 1.16 | 2.10 |
| 2 | 0.790 | 0.526 | 0.964 | 3.93 | 2.28 | 3.17 |
| 3 | 0.760 | 0.610 | 0.951 | 3.23 | 3.28 | 6.65a |
Note: We implanted one, two, or three causal genes in a region. 1Post-Gene is well-calibrated to detect the causal genes in regions where we have only one true causal gene. CAVIAR-Gene is well-calibrated in all our experiments. We consider a method to be well-calibrated when the recall rate is at least 95%. We compute the recall rate of a method as a percentage of the total simulations where all the true causal variants are detected.
aAlthough we allow for only six causal genes in a region, we can have more than six causal genes in the ρ causal gene set (see Section 2.8).
3.3 CAVIAR-Gene provides better ranking of the causal genes
To compare the performance of each method, we compare the recall rate and the number of causal genes selected by each method. We calculate the recall rate as a percentage of the total simulations where all the true causal variants are detected. Unfortunately, each method selects a different number of genes as causal. Thus, to make the comparison fair, we compute the recall rate for each method as a function of the number of genes each method selects.
The results for all the methods across all three datasets are shown in Figure 2. In this figure, the X-axis is the number of genes selected by each method and the Y-axis is the recall rate for each method. Figure 2c and e indicates the recall rate for Outbred, F2, and HMDP datasets where we have implanted one causal gene. Although the difference between the TopK-Gene and CAVIAR-Gene in the case of one causal gene is negligible, we observe a 10% higher recall rate when there are multiple causal genes in a region (Fig. 2b, d, and f).
Fig. 2.
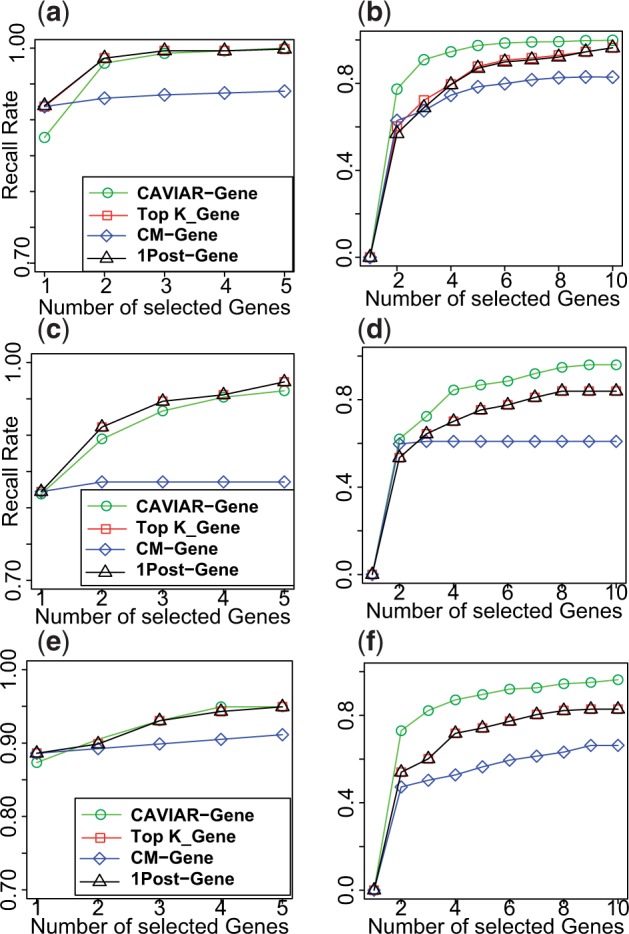
CAVIAR-Gene provides better ranking of the causal genes for Outbred, F2, and HMDP datasets. Panels a and b illustrate the results for Outbred genotypes for case where we have one causal and two causal genes, respectively. Panels c and d illustrate the results for F2 genotypes for case where we have one causal and two causal genes, respectively. Panels e and f illustrate the results for Outbred genotypes for case where we have one causal and two causal genes, respectively
Although in Figure 2 we only compare recall rate of different methods as we vary the number of causal genes selected by each method, these figures are similar to receiver operating characteristic (ROC) curves which are used as a measure to compare results for different methods in statistics and machine learning. In ROC curves, the y-axis is the true positive rate which is equivalent to the recall rate in our result, and the x-axis is the false positive rate which indicates the fraction of simulations where the non-causal genes are selected as causal. Because of the fact that all methods are forced to pick the same number of causal genes, the false positive rate is the same for all the methods. Moreover, similar to ROC curves in our results, as we increase the false positive rate, the recall rate increases and as we reach false positive rate of 1, which means if we select all the genes as causal, we have a recall rate of 1.
3.4 Greedy algorithm and brute force algorithm have similar results
We proposed a greedy algorithm in Section 2.8 to detect the ρ causal gene set in order to speed up the process. In this section, we show that the results obtained from the greedy algorithm and the brute force algorithm are very close. The brute force algorithm considers all the possible different causal gene sets in order to compute the ρ causal gene set. We consider a region with 20 genes and then we simulated data similar to the previous sections. We implant one, two, or three causal genes in the region. We ran both methods and computed the recall rate as well as the size of the ρ causal gene set selected by each method. Table 2 shows the results. We calculate the recall rate as a percentage of the total simulations where all the true causal variants are detected.
Table 2.
Greedy algorithm and brute force algorithm have similar results
| Causal gene | Recall rate (%) | Causal gene size | ||
|---|---|---|---|---|
| Greedy | Brute force | Greedy | Brute force | |
| 1 | 0.999 | 0.999 | 1.72 | 1.67 |
| 2 | 0.983 | 0.990 | 3.84 | 3.30 |
| 3 | 0.956 | 0.976 | 4.82 | 4.73 |
Note: We implanted one, two, or three causal genes in a region. We run both the greedy and brute force algorithm on the simulated data sets. This result indicates that the differences between these two methods are negligible.
3.5 CAVIAR-Gene adjusts for population structure
It is known that in the case where there exists no population structure, the correlation between the marginal statistics of two variants is the same as the correlation between the genotypes from which the statistics were computed. CAVIAR utilizes this fact to compute the likelihood for each possible causal combination. However, when population structure is present and corrected for, this may not hold. We demonstrate in our experiments that the correlation between the marginal statistics for any two variants which are corrected for population structure is the same as the correlation of a transformed version of genotype for the same two variants. We provide the description of this transformation in Section 2. CAVIAR-Gene utilizes this transformation to adjust for the population structure to compute the correct likelihood.
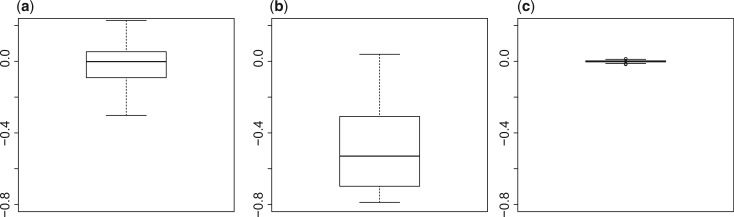
We use an HMDP dataset (Bennett et al., 2010) which we determine to have population structure. We generate phenotypes with population structure and compute the marginal statistics for each variant both corrected and not corrected for population structure. We then compute the correlation between each pair of marginal statistics and the correlation between each pair of variants for the original genotype and the transformed genotype. We calculate the difference between the correlation computed from the marginal statistics for each pair of variants and the correlation of the genotype of the same variants. The boxplot of these differences are shown in Figure 3.
Fig. 3.
CAVIAR-Gene adjusts for population structure. Panel a illustrates the case where the data have population structure and the statistics is not corrected for the population structure. Panels b and c illustrate the cases where we have corrected the statistics for the population structure. However, in Panel b, we compute the correlation between the original genotypes and in Panel c the correlation is computed from the transformed genotypes. Then, we calculate the difference between the correlation computed from the marginal statistics for each pair of variants and the correlation of the genotype of the same variants. The difference between the correlation of the marginal statistics and the correlation of the transformed genotype shown in Panel c is close to zero and their variance is much smaller than other cases as shown in Panels a and b. To compare the results, we plot the residual difference between −0.4 and 0.4, as a result some points for Panel b are not shown
As expected, the difference between the correlation of the marginal statistics and the correlation of the transformed genotype is close to zero and their variance is much smaller than other cases. Thus, the correlation between the marginal statistics when population structure is corrected is closer to the correlation between the genotype which is transformed using the right transformation matrix.
3.6 CAVIAR-Gene identifies Apoa2 as causal gene in HDL
To illustrate an application of our method in real data, we use an HDL dataset which was collected for three different mouse strains: outbred dataset (Zhang et al., 2012), F2 dataset (van Nas et al., 2009), and HMDP dataset (Bennett et al., 2010). We ran CAVIAR-Gene on a region ∼80 megabases in length containing 595 genes (chr1: 120,000,000–197,195,432). This region harbors Apoa2, a gene previously established to influence HDL levels (Flint and Eskin, 2012; van Nas et al., 2009). We applied CAVIAR-Gene on the HMDP dataset considering all the genes in this region which yielded a 95% ρ causal set of 130 genes. Next, we conducted a more refined experiment, using domain-specific knowledge of the phenotype, to create a list of 53 potential candidate genes. CAVIAR-Gene selected a 23 gene subset of this list as the ρ causal gene set. Running CAVIAR-Gene on the Outbred dataset for all 595 genes resulted in a 95% gene set of only 13 genes. Because of the fact that the Outbred mice have a smaller degree of population structure than the HDMP, it is expected that the gene set resolution should be greater in this data. Most importantly, across all the datasets, CAVIAR-Gene includes Apoa2 in the gene set. Figure 4 illustrates the genes which are selected by CAVIAR-Gene for each datasets. The five genes which are common between all the datasets are Nr1i3, Tomm40l, Apoa2, Fcer1g, andNdufs2. All these genes are known to be highly associated with the HDL. This suggests that CAVIAR-Gene not only recovers the actual causal gene, but simultaneously reduced the number of genes that need to undergo functional validation.
Fig. 4.
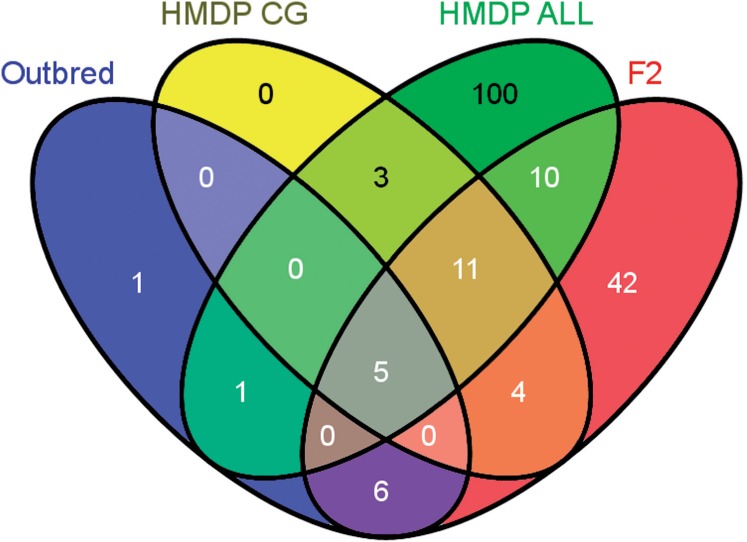
Venn diagram of the genes selected by CAVIAR-Gene on each of the dataset. HMPD ALL is the results of CAVIAR-Gene on HMDP when we utilize all the genes. HMDP CG is the result of CAVIAR-Gene on HMDP when we utilize candidate genes
4 Discussion
In this article, we propose a novel method, CAVIAR-Gene, for performing fine mapping on the gene level. CAVIAR-Gene computes the probability of each set of genes capturing the true causal genes. Then, CAVIAR-Gene selects the set which has the minimum number of genes selected as causal and the probability of the set capturing the true causal gene is higher than a user-defined threshold (e.g. typically 95% or higher). We note that the usage of the term causal has little to do with the concept of causal inference as described in the computer science and statistics literature (Pearl, 2000; Spirtes et al., 2000). In the context of association studies, we consider a variant to be causal if the variant is responsible for the association signal in the locus. CAVIAR-Gene can incorporate marginal statistics which is corrected for population structure. This property makes CAVIAR-Gene suitable for performing fine mapping on the model organism such as inbred mice. We show using simulated data that CAVIAR-Gene has higher recall rate compared with the existing methods for fine mapping on the variants level, while the size of the causal set selected by CAVIAR-Gene is smaller than these methods. CAVIAR-Gene incorporates external information such as functional data as a prior to improve the results.
Funding
This work was supported by the National Science Foundation (0513612, 0731455, 0729049, 0916676, 1065276,1302448, and 1320589 to F.H., W.Y., and E.E.) and the National Institutes of Health (K25-HL080079, U01-DA024417, P01-HL30568, P01-HL28481, R01-GM083198, R01-MH101782, and R01-ES022282 to F.H., W.Y., and E.E.). E.E. is supported in part by the NIH BD2K award, U54EB020403. We acknowledge the support of the National Institute of Neurological Disorders and Stroke Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691 and T32 NS048004-09). G.K. and B.P. are supported in part by the National Institutes of Health (R01 GM053275). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest: none declared.
References
- Altshuler D., et al. (2008) Genetic mapping in human disease. Science, 322, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B.J., et al. (2010) A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res., 20, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium et al. (2013) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet., 45, 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell G., et al. (2012) Incorporating prior information into association studies. Bioinformatics, 28, i147–i153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin E. (2008) Increasing power in association studies by using linkage disequilibrium structure and molecular function as prior information. Genome Res., 18, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Eskin E. (2012) Genome-wide association studies in mice. Nat. Rev. Genet., 13, 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H., et al. (2007) A genome-wide association study identifies kiaa0350 as a type 1 diabetes gene. Nature, 448, 591–594. [DOI] [PubMed] [Google Scholar]
- Han B., et al. (2009) Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet., 5, e1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., et al. (2014) Identifying causal variants at loci with multiple signals of association. Genetics, 198, 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jul J.H., Eskin E. (2011) Increasing power of groupwise association test with likelihood ratio test. J. Comput. Biol., 18, 1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., et al. (2008) Efficient control of population structure in model organism association mapping. Genetics, 5, e1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., et al. (2014) Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet., 10, e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottgen A., et al. (2013) Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet., 45, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., et al. (2013) DIST: direct imputation of summary statistics for unmeasured SNPs. Bioinformatics, 29, 2925–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., et al. (2011) FaST linear mixed models for genome-wide association studies. Nat. Methods, 8, 833–835. [DOI] [PubMed] [Google Scholar]
- Listgarten J., et al. (2012) Improved linear mixed models for genome-wide association studies. Nat. Methods, 9, 525–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller J.B., et al. (2012) Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet., 44, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., et al. (2014) Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics, 30, 2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. (2000) Causality: Models, Reasoning and Inference. Vol. 29 Cambridge University Press, New York, NY. [Google Scholar]
- Price A.L., et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Przeworski M. (2001) Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet., 69, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D.E., et al. (2001) Linkage disequilibrium in the human genome. Nature, 411, 199–204. [DOI] [PubMed] [Google Scholar]
- Ripke S., et al. (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet., 45, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirtes P., et al. (2000). Causation, Prediction, and Search. Vol. 81 MIT press, Cambridge, MA, USA. [Google Scholar]
- van Nas A., et al. (2009) Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology, 150, 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N., et al. (2010) Leveraging genetic variability across populations for the identification of causal variants. Am. J. Hum. Genet., 86, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., et al. (2012) Genome-wide association mapping of quantitative traits in outbred mice. G3 (Bethesda), 2, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. (2012) Genome-wide efficient mixed model analysis for association studies. Nat. Genet., 44, 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]