Abstract
Objectives
Ribavirin concentrations may impact hepatitis C virus (HCV) treatment outcome. We modelled ribavirin serum and intracellular ribavirin monophosphate (RBV-MP) and ribavirin triphosphate (RBV-TP) pharmacokinetics in red blood cells (RBC) using samples collected during the NIAID SPARE trial to explore associations with treatment outcome and the development of anaemia.
Patients and methods
Individuals infected with HCV genotype 1 (GT1) received 400 mg of sofosbuvir and either low-dose or weight-based ribavirin as part of the NIAID SPARE trial. Concentrations were modelled using NONMEM and associated with treatment outcomes using unpaired t-tests or Pearson's rho correlations.
Results
Average day 14 RBV-MP concentrations were higher in subjects with haemoglobin nadir <10 g/dL relative to patients with haemoglobin nadir ≥10 g/dL (6.54 versus 4.48 pmol/106 cells; P = 0.02). Additionally, day 14 RBV-MP average concentrations trended towards being higher in subjects that achieved sustained virological response (SVR) as compared with patients who relapsed (4.97 versus 4.09 pmol/106 cells; P = 0.07). Receiver operating characteristic curves suggested day 14 RBV-MP concentration thresholds of 4.4 pmol/106 cells for SVR (P = 0.06) and 6.1 pmol/106 cells for haemoglobin nadir <10 versus ≥10 g/dL (P = 0.02), with sensitivity and specificity ≥60%. Dosing simulations showed that 800 mg of ribavirin once daily produced day 14 RBV-MP concentrations within the 4.4–6.1 pmol/106 cells range.
Conclusions
RBV-MP concentrations in RBC at day 14 were related to anaemia and SVR. A therapeutic range was identified for RBV-MP in persons with HCV GT1 disease receiving 24 weeks of sofosbuvir plus ribavirin, suggesting a potential pharmacological basis for individualized ribavirin dosing in IFN-free regimens.
Keywords: ribavirin monophosphate, anaemia, SVR, interferon-free therapy, red blood cells
Introduction
Ribavirin has been a key component of hepatitis C virus (HCV) infection treatment for decades. Although its exact mechanism of antiviral activity in vivo is unknown, several hypotheses have been proposed.1 Ribavirin is administered as a prodrug, which undergoes successive intracellular phosphorylation to monophosphate (RBV-MP), diphosphate and triphosphate (RBV-TP) anabolites.2 Phosphorylated forms of ribavirin have been associated with antiviral effects observed in vitro. RBV-TP has been shown to act as a nucleoside analogue, causing chain termination upon integration by the HCV RNA-dependent RNA polymerase, as well as a direct RNA mutagen.3 RBV-MP has been shown to inhibit inosine monophosphate dehydrogenase (IMPDH), resulting in decreased GTP production, leading to suppressed viral replication.3 The phosphorylated anabolites are also thought to cause haemolytic anaemia, the major dose-limiting toxicity of ribavirin. Phosphorylated forms of the drug accumulate in red blood cells (RBC), which lack dephosphorylation enzymes.4,5 Despite in vitro data suggesting the intracellular phosphorylated forms are associated with antiviral response and cellular toxicity,3 there are few data on the intracellular kinetics of RBV-MP and RBV-TP in vivo6 and no data in the context of IFN-free treatment (i.e. treatment combinations that do not use pegylated IFN).
Several direct-acting antiviral (DAA) agents have recently received regulatory approval for the treatment of HCV. These agents include NS3 protease inhibitors, NS5A inhibitors and nucleotide and non-nucleoside NS5B polymerase inhibitors. While several antiviral combinations have shown efficacy without ribavirin in clinical trials,7–10 ribavirin remains a critical component to optimize sustained virological response (SVR) rates in many treatment regimens. For example, the combination of ribavirin and the NS5B nucleotide polymerase inhibitor sofosbuvir is the preferred regimen for the treatment of genotype 2 and 3 disease.11 Ribavirin is also an essential component with ritonavir-boosted paritaprevir, ombitasvir and dasabuvir for the treatment of HCV genotype 1a, as well as in patients with cirrhosis or who are treatment experienced.12–14 Defining the intracellular pharmacology of ribavirin and determining concentration–effect associations can inform and improve the use of ribavirin within IFN-free regimens.
The NIAID SPARE trial investigated the use of sofosbuvir with either low-dose (600 mg daily) or weight-based (1000 or 1200 mg daily) ribavirin in genotype 1 patients for 24 weeks.15 In an ITT analysis, SVR was achieved in 48% (12/25) and 68% (17/25) of subjects randomized to either low-dose or weight-based ribavirin, respectively,15 which suggests that higher ribavirin exposures may increase the odds of SVR. However, 32% (8/25) of subjects in the weight-based dosing arm became anaemic (haemoglobin nadir ≤10.9 g/dL) and 6 subjects required dose reductions,15 indicating that ribavirin-induced haemolytic anaemia still occurs in the context of IFN-free therapy. There is a clear need to define the pharmacokinetic (PK) parameter for ribavirin that best predicts outcomes. To this end, establishing ribavirin doses and dosing strategies that result in concentrations that maximize the likelihood of SVR while minimizing the risk of haemolytic anaemia is imperative.
The objectives of this work were to: (i) characterize the serum and intracellular PK of ribavirin; (ii) define concentration–effect associations for serum and intracellular ribavirin; (iii) identify target concentration ranges that effectively balance the likelihood of SVR with the risk of anaemia; and (iv) determine ribavirin dosing strategies that achieve desired PK exposures in an IFN-free HCV treatment regimen of sofosbuvir and ribavirin.
Patients and methods
Subjects and study design
Complete study design and subject details have been published previously.15 Briefly, participants provided written informed consent for a single-centre, randomized, open-label Phase 2 clinical trial at the Clinical Research Center of the NIH in Bethesda, MD, USA (NCT01441180). The study protocol conformed to the ethics guidelines of the 1975 Declaration of Helsinki as reflected by NIH/NIAID Institutional Review Board approval. HCV treatment-naive participants were chronically infected with HCV genotype 1 and had baseline platelet counts of ≥50 000 cells/μL and haemoglobin ≥11 g/dL (women) or 12 g/dL (men). All fibrosis stages (including compensated cirrhosis) were allowed.
The study was conducted in two phases. The first phase was an open-label study in which 10 patients were treated with sofosbuvir (400 mg/day) and weight-based ribavirin (400 mg in the morning and 600 mg in the evening if <75 kg or 600 mg twice a day if ≥75 kg) for 24 weeks. In the second phase, 50 patients were randomized (n = 25 in each arm) to receive sofosbuvir with either low-dose (600 mg once daily) or weight-based ribavirin. For each participant, ribavirin, RBV-MP and RBV-TP were quantified using single serum and RBC samples that were collected at multiple study visits (average of 6.5 serum and 3.6 RBC samples per patient). This sparse sampling approach using varied treatment timepoints can be analysed using population PK, as this approach is powered for model building without the need for intensive sampling of each individual.
Bioanalytical methods
Ribavirin serum levels were quantified using a validated HPLC-MS/MS method linear between 0.05 and 10 mg/L. Whole blood was drawn into PAXgene RNA isolation tubes (Qiagen, Valencia, CA, USA), stored at −80°C and then diluted 1 : 50 with 1 mL of 70% methanol solution prior to extraction. These samples were treated as isolated RBC samples during data analysis, as a strong correlation was established to concentration results from purified RBC samples (r2 = 0.9984; Figure S1, available as Supplementary data at JAC Online). Samples were separated into RBV-MP and RBV-TP fractions using a Waters QMA strong anion-exchange solid-phase extraction (SPE) cartridge, dephosphorylated and then prepared for a validated HPLC-MS/MS analysis using a Varian BondElut Phenylboronic Acid SPE cartridge.16 The assay was linear between 0.5 and 200 pmol/sample and reported concentrations are normalized to a per million cell count (pmol/106 cells).
The inosine triphosphatase (ITPA) genotype was determined from DNA extracted from whole blood using the PAXgene Blood DNA Kit (PreAnalytiX, a Qiagen/BD Company). PCR was performed using TaqMan SNP genotyping assays (Life Technologies, Grand Island, NY, USA) for the ITPA variants rs7270101 and rs1127354 using a 7500 Real Time PCR System (Applied Biosystems, Grand Island, NY, USA) as previously described.15,17 IFN-λ3 (IFNL3, also known as IL28B) and IFN-λ4 (IFNL4) genotypes were determined as previously described.15,18
Model development
Population PK models were developed to characterize ribavirin PK in serum and RBC RBV-MP and RBV-TP PK and to explore the clinical factors that might affect serum and intracellular PK. Non-linear mixed-effects modelling (NONMEM version 7.2, New Hanover, MD, USA) was used to construct the population PK models. First-order conditional estimation and proportional plus additive error were used for all models. Serum concentrations (n = 339) from 52 subjects were modelled using a two-compartment model (Figure S2A) parameterized on clearance (CL), following previous descriptions in the literature.6,19–21 The absorption rate (ka) was fixed at 2.0 h−1, as insufficient data during the absorption period were available to fit this parameter reliably.19 Concentrations (n = 171) from 47 subjects were used to develop one-compartment models parameterized on the elimination rate (ke) (Figure S2B) for intracellular RBV-MP and RBV-TP. In this model, Kin represents an aggregate rate of intracellular formation and volume (V) acts as a scalar; as such, ke was the only intracellular parameter tested for association with covariates. The effect of sex, race, weight, age, serum creatinine, glomerular filtration rate [GFR; calculated using the Modification of Diet in Renal Disease (MDRD) equation],22 fibrosis stage [histology activity index (HAI) fibrosis 0–4] and ITPA phenotype on serum ribavirin and RBC RBV-MP and RBV-TP concentrations were first explored using Pearson's rho correlations for continuous covariates and unpaired t-tests for categorical covariates in GraphPad Prism (GraphPad Software, San Diego, CA, USA) using a cut-off of P < 0.10. Clinical factors associated with PK were added to the model in a nested fashion and the significance of the parameter–covariate relationship was assessed in a variety of ways, including the reduction of model variability, improvement of diagnostic plots and a drop in the objective function value of >3.84 (P ≤ 0.05). Covariates were added in a forward inclusion process until no significant covariates remained, at which point backward elimination, also at a significance level of P < 0.05, was performed.
PK/pharmacodynamic (PD) associations
Final population PK models for ribavirin serum, RBV-MP and RBV-TP were used to determine average concentrations (Cave) at first dose and on days 3, 7, 14, 28, 56 and 84 of treatment. Cave were calculated using the equation Cave,t = Css × (1 − e−ket), where Css was determined using Css = AUC/τ (where AUC = dose × F/CL and τ is the dosing interval) and ke was utilized from the population PK model. These PK parameters were tested for association with PD outcomes, including treatment outcome (SVR or relapse), baseline haemoglobin (g/dL), haemoglobin nadir (g/dL), change in haemoglobin (g/dL) and whether the subject required a dose reduction for anaemia (yes or no). PK/PD associations were evaluated using Pearson's rho correlations (continuous) or unpaired t-tests (dichotomous). Logistic regression (SAS version 9.4) was used to evaluate the associations between clinical covariates and SVR and to control for the relevant covariates in evaluating the relationship between PK and SVR. Finally, receiver operating characteristic (ROC) curves were constructed to determine potential exposure thresholds corresponding to efficacy and toxicity outcomes.
Dosing simulations
Once threshold concentrations were identified, we sought to determine ribavirin doses that achieved these exposure targets. The SIM program in ADAPT V (Biomedical Simulations Resource, Los Angeles, CA, USA) was used to simulate concentrations expected to result from a variety of ribavirin dosing schemes, including those used in the SPARE trial (600 mg once daily, 600 mg twice daily and 400 mg in the morning/600 mg in the evening), 800–1400 mg once daily and a 600 mg in the morning/800 mg in the evening regimen. Final population PK parameter estimates were utilized for simulations using the ‘population with output error simulation’ option. Data were simulated at every hour through 12 weeks of dosing and the simulated concentration data were plotted in order to visually compare and contrast the dosing regimens and to determine which doses provided exposures that balanced SVR and anaemia as determined by thresholds from the ROC analyses.
Results
Subject demographics
Serum and/or whole blood samples were available for analysis from 52 subjects enrolled in the SPARE trial. The demographics of these 52 subjects are described in Table 1. Participants were mostly black [79% (41/52)], infected with HCV genotype 1a [73% (38/52)] and had unfavourable IFNL3/IFNL4 genotypes [rs12979860: 83% CT or TT (43/52) and rs368234815: 85% ΔG/TT or ΔG/ΔG (44/52)]. Most subjects [73% (38/52)] had minimal fibrosis (HAI fibrosis ≤2) based on liver histology obtained by biopsy. Twelve patients (23%) had bridging fibrosis (F3) and two (4%) had cirrhosis (F4) based on liver histology. Twenty-two subjects (42%) received low-dose ribavirin, while 30 (58%) received weight-based dosing [21 (70%) on 600 mg twice daily and 9 (30%) on 600 mg/400 mg]. Of those receiving weight-based dosing, six (20%) had ribavirin dose reductions as a result of anaemia (10 g/dL, except in those with a history of coronary artery disease, which had a cut-off of 12 g/dL). Of the 52 subjects included in this analysis, 34 (65%) achieved SVR, 17 (33%) relapsed and 1 dropped out of the study.
Table 1.
Subject demographics (n = 52)
| Gender | 18 (35%) women |
| 34 (65%) men | |
| Race | 41 (79%) African-American |
| 9 (17%) non-African-American | |
| 2 (4%) Hispanic | |
| Weight (kg), median (range) | 85.6 (45–153.4) |
| Age (years), median (range) | 54 (25–78) |
| MDRD (mL/min/1.73 m2), median (range) | 105 (63.1–161) |
| Baseline HCV RNA | 18 (35%) ≤800 000 IU/mL |
| 34 (65%) >800 000 IU/mL | |
| ITPA activity phenotype | 38 (73%) = 100% |
| 7 (13%) = 60% | |
| 5 (10%) = 30% | |
| 1 (2%) = 5% | |
| 1 (2%) = missing | |
| IFNL3/IL28B (rs12979860) | 9 (17%) CC |
| 26 (50%) CT | |
| 17 (33%) TT | |
| IFNL4 (rs368234815) | 8 (15%) TT/TT |
| 25 (48%) ΔG/TT | |
| 19 (37%) ΔG/ΔG | |
| HCV genotype | 38 (73%) HCV genotype 1a |
| 14 (27%) HCV genotype 1b | |
| Fibrosis stage | 12 (23%) = 0 |
| 25 (48%) = 1 | |
| 1 (2%) = 2 | |
| 12 (23%) = 3 | |
| 2 (4%) = 4 |
Population PK model
Median (range) steady-state ribavirin serum concentrations were 1.95 (0.70–5.26) mg/L and the half-life was 7.1 (1.7–25.8) days. The clinical covariates that significantly explained ribavirin serum PK were sex on the central compartment volume (Vc) and estimated GFR on the CL parameter. Male sex was associated with a 73% higher Vc (5130 versus 2960 L; P = 0.005) while higher GFR was positively correlated with ribavirin CL, with a 25% increase in GFR predicting a 10% increase in ribavirin CL (P = 0.02). Model parameter estimates and calculated PK parameters are shown in Table 2 and Table 3, respectively. Diagnostic plots support the appropriateness of the final model (Figure S3).
Table 2.
Ribavirin serum model parameter estimates
| Parameter | Base model | Final model |
|---|---|---|
| CL (L/h) | 18.8 (16.8–20.8), 37.4% | 12.2 + 7.11 (MDRD/104.7), 35.1% |
| V2 (L) | 4230 (3280–5180), 66% | women: 2960 (1920–4000) men: 5130 (4030–6230) 60.3% |
| Q (L/h) | 7.87 (1.01–14.7), 35.4% | 6.97 (0.678–13.3), 43.8% |
| V3 (L) | 2900 (1810–3990), 80.2% | 2680 (1710–3650), 81.9% |
| ka (h−1) (fixed) | 2.0 | 2.0 |
Data are mean (95% CI), with percentage interindividual variability.
Table 3.
Steady-state PK parameter estimates
| Low dosea | Weight baseda | |
|---|---|---|
| Serum ribavirin | ||
| AUCτ (mg · h/L) | 37.2 (39%) | 33.9 (39%) |
| Css (mg/L) | 1.55 (38%) | 2.07 (39%) |
| t1/2 (days) | 8.60 (59%) | 6.84 (64%) |
| RBV-MP | ||
| AUCτ (pmol · h/106 cells) | 175 (49%) | 123 (38%) |
| Css (pmol/106 cells) | 7.30 (49%) | 8.82 (55%) |
| t1/2 (days) | 13.7 (24%) | 12.5 (21%) |
| RBV-TP | ||
| AUCτ (pmol · h/106 cells) | 2538 (53%) | 1823 (46%) |
| Css (pmol/106 cells) | 106 (53%) | 130 (60%) |
| t1/2 (days) | 12.3 (64%) | 12.7 (43%) |
Data are mean (CV).
aAUCτ is 0–24 h for the low-dose group (who were dosed every 24 h) and 0–12 h for the weight-based group (who were dosed every 12 h).
Median (range) steady-state concentrations were 8.34 (2.05–20.5) pmol/106 cells for RBV-MP and 122 (33.4–320) pmol/106 cells for RBV-TP. Median (range) half-lives were 12.8 (7.8–24.0) and 11.3 (4.4–29.5) days for RBV-MP and RBV-TP, respectively. Only the impact of ITPA phenotype on RBV-TP elimination was found to be significant, as reduced ITPA functionality corresponded to a 37% slower ke (i.e. higher RBV-TP concentration) compared with those with 100% ITPA functionality (P = 0.002; Figure S4). No covariates were retained in the RBV-MP model. Final PK parameters for the intracellular model are shown in Table 3. Diagnostic plots support the RBV-MP and RBV-TP models and are shown in Figure S5 and Figure S6, respectively.
PK/PD associations
The population mean (%CV) Cave values used to evaluate PK/PD relationships are provided in Table S1. Cave ribavirin serum (P < 0.002 through day 7) and RBV-MP [P < 0.05 on days 14 (Figure 1a) through 84], but not RBV-TP (P > 0.10), were inversely correlated with haemoglobin nadir. Mean (SD) day 14 RBV-MP was higher [6.54 (1.70) pmol/106 cells] in those with a haemoglobin nadir <10 g/dL relative to those with a haemoglobin nadir ≥10 g/dL [4.48 (1.49) pmol/106 cells; P = 0.02]. Change in haemoglobin over the duration of the study was inversely correlated with both RBV-MP (P < 0.02 at all studied times) and RBV-TP (P = 0.04; day 3) Cave. Serum ribavirin on day 14 (P = 0.02), but not RBV-MP or RBV-TP (P ≥ 0.11), exhibited a significant direct correlation with the need for ribavirin dose reduction. Neither serum nor intracellular ribavirin were significantly associated with SVR (P ≥ 0.07); however, mean (SD) day 14 RBV-MP concentrations trended towards being higher [4.97 (1.66) pmol/106 cells] in those that achieved SVR compared with those that relapsed [4.09 (1.46) pmol/106 cells; P = 0.07; Figure 1b]. Besides RBV-MP concentration, baseline HCV RNA ≤800 000 IU/mL and female sex were also univariately associated with SVR. The odds of SVR were 5.9 times greater (P = 0.03) in females, as well as in individuals with baseline HCV RNA ≤800 000 IU/mL, as compared with males and those with HCV RNA >800 000 IU/mL, respectively. In a multivariable logistic regression model, after controlling for the effect of baseline HCV RNA ≤800 000 IU/mL, the trend between day 14 RBV-MP and SVR reached statistical significance (P = 0.01). Sex was not significant in the multivariable model (P = 0.14).
Figure 1.
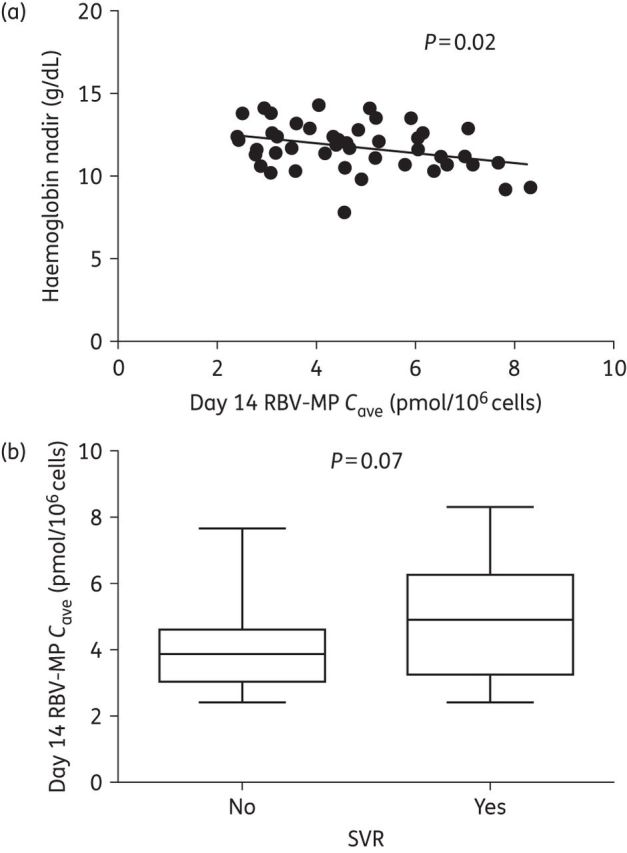
Day 14 RBV-MP Cave were associated with both haemoglobin nadir (a) and SVR (b). Box and whisker plots show medians, quartiles and ranges of data.
ROC analysis
The associations between SVR and haemoglobin nadir with day 14 RBV-MP Cave led us to attempt to identify exposure thresholds using ROC curves (Figure 2). These ROC curves suggested a day 14 RBV-MP Cave threshold of 4.4 pmol/106 cells for SVR and 6.1 pmol/106 cells for a haemoglobin nadir <10 g/dL. The SVR ROC curve had an AUC of 0.67 (P = 0.06) while the curve describing the haemoglobin nadir had an AUC of 0.82 (P = 0.02). The sensitivities (true positive rate) of the thresholds were 69% and 60% for SVR and anaemia, respectively, while the specificities (true negative rate) were 76% and 83%. The negative predictive value of the RBV-MP threshold for SVR was 59%, suggesting that treatment failure would be predicted in 59% of individuals with RBV-MP concentrations <4.4 pmol/106 cells. This suggests that a day 14 RBV-MP Cave target window of between 4.4 and 6.1 pmol/106 cells may reduce the risk of anaemia while still providing high rates of SVR. Once controlled for baseline HCV RNA ≤800 000 IU/mL, the SVR ROC curve AUC increased to 0.81 (P < 0.0001), suggesting that the likelihood of SVR at this threshold is moderated by baseline HCV RNA.
Figure 2.
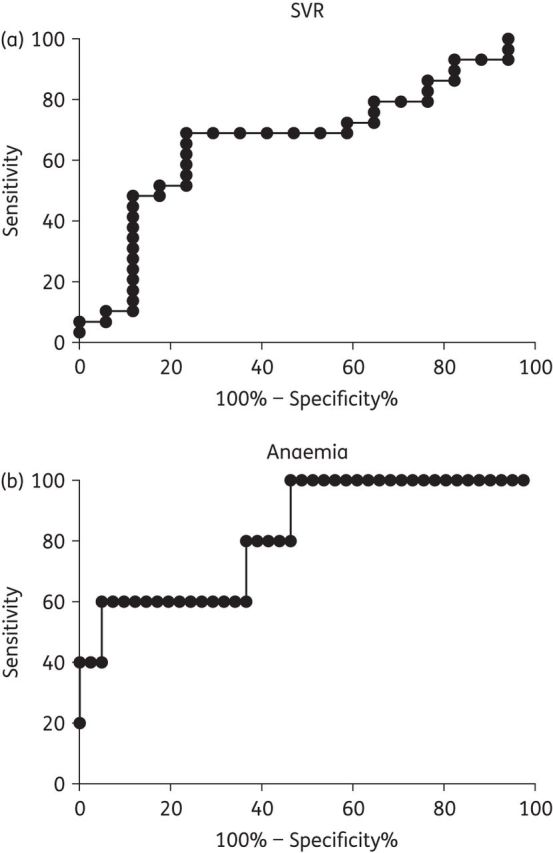
ROC curve analysis of (a) SVR and (b) anaemia (haemoglobin nadir <10 g/dL) with day 14 RBV-MP Cave levels.
Dosing simulations
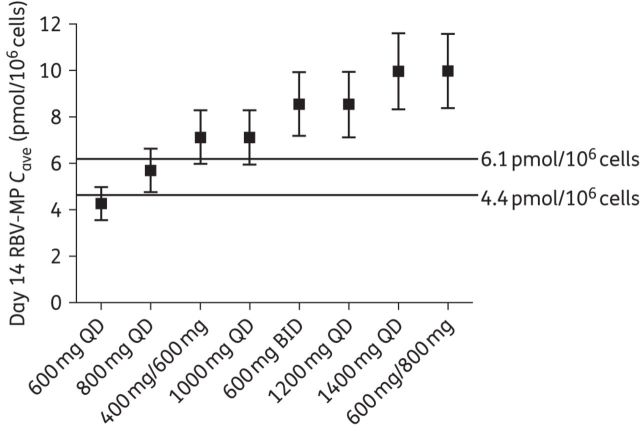
We next attempted to identify ribavirin dosing regimens that could generate day 14 concentrations within the target therapeutic window of 4.4–6.1 pmol/106 cells. Simulated average (SD) concentrations resulting from the various regimens are shown in Figure 3, with lines delineating the identified thresholds. The results indicate that the 800 mg daily dose provided Cave at day 14 within the desired range. Only the simulated Cave resulting from the 600 and 800 mg daily doses remained below the established threshold for anaemia. However, only 44% of the simulated subjects at the 600 mg dose reached the SVR threshold of 4.4 pmol/106 cells.
Figure 3.
Population mean (SD) RBV-MP Cave on day 14 resulting from simulated dosing schemes. Lines are drawn at the threshold levels determined by ROC analysis to be associated with a high likelihood of SVR (4.4 pmol/106 cells) and anaemia (6.1 pmol/106 cells). QD, once daily dosing; BID, twice daily dosing.
Discussion
This study utilized clinical samples collected during the NIAID SPARE trial to understand the serum and cellular disposition of ribavirin and to explore and define ribavirin PK/PD associations and dosing requirements in the context of an IFN-free regimen consisting of sofosbuvir and either low-dose or weight-based ribavirin. We demonstrated that ribavirin exhibits a long half-life in both serum (7.1 days) and RBC (12.8 and 11.3 days for the RBV-MP and RBV-TP moieties, respectively). Analysis of the relationship between drug PK and PD suggested day 14 RBC RBV-MP concentrations were significantly associated with anaemia and trended towards an association with SVR. Finally, we utilized these PK/PD relationships to identify a target therapeutic range of 4.4–6.1 pmol/106 cells and, by simulating dosing regimens, found that an 800 mg daily dose would provide exposures within this therapeutic range.
Model estimates of ribavirin serum CL (CL/F = 19.3 L/h) for an individual with median renal function were similar to previously published reports, which ranged from 9.06 to 25.0 L/h.19–21,23,24 Ribavirin CL was also significantly correlated with renal function, consistent with prior studies.25 Men were associated with higher Vc, which could help explain why relapse was more common in men during SPARE.15 Model-estimated serum half-lives and steady-state concentrations agree well with previous literature on ribavirin serum PK, which have reported half-lives between 187 and 303 h and steady-state concentrations near 2 mg/L.6,20,21,24,26
This is the first description of in vivo RBC RBV-MP and RBV-TP concentrations in an IFN-free regimen. While no covariates were significantly associated with RBV-MP PK, we observed a significant reduction in RBV-TP elimination (and thus increased RBV-TP concentrations) in subjects with decreased ITPA functionality, similar to our previous observations in subjects receiving pegylated IFN, ribavirin and telaprevir.6 ITPA is present in RBC and responsible for the breakdown of inosine triphosphate (ITP), which has been shown to stand in for GTP in the biosynthesis of ATP.27 Due to the chemical similarities between ITP, GTP and RBV-TP (a guanosine analogue), ITPA may have some affinity for RBV-TP and thus may contribute to the elimination of RBV-TP. ITPA genotypes have also been shown to impact ribavirin plasma PK and PD, with lower-functionality phenotypes resulting in decreased ribavirin plasma concentrations as well as being protective against anaemia.28,29 Our models suggest that both moieties were long-lived in RBC, similar to both in vitro and in vivo reports of RBC PK.6,30 The long half-lives suggest that ribavirin does not reach intracellular steady-state concentrations until approximately week 12 on therapy [corresponding to approximately seven half-lives (>99% of steady-state) for each moiety].
Ribavirin PK was evaluated for relationships with treatment outcomes, including SVR, anaemia (haemoglobin nadir and change in haemoglobin) and the need for ribavirin dose reduction. We found that RBV-MP concentrations at day 14 trended towards an association with SVR and reached significance when the association was controlled for baseline HCV RNA. Previous reports in the literature have described several mechanisms by which ribavirin might exert its antiviral effect in HCV, including the activation of endogenous clearance mechanisms, RNA mutagenicity and the potential for RBV-TP to inhibit HCV replication.3 Our findings that the RBV-MP trended towards an association with SVR support the proposed mechanism via which RBV-MP decreases the production of GTP by inhibiting IMPDH.3,31
Average day 14 RBV-MP concentrations were utilized to establish a threshold concentration anticipated to correlate with a high likelihood of SVR and a cut-off associated with high risk of anaemia (haemoglobin nadir <10 g/dL), using ROC curves. Day 14 was found to be the earliest timepoint correlated with haemoglobin nadir and associated with SVR. Though not a perfect test, the sensitivity and specificity were ≥60% for both thresholds and the area under the ROC curves suggested good predictive power of ≥67%, supporting threshold validity. Additionally, these thresholds are likely robust for a variety of patient populations, since there were no clinical factors associated with modelled RBV-MP PK. These results suggest that day 14 average RBV-MP concentrations may provide a pharmacological basis behind the efficacy and toxicity of ribavirin in IFN-free regimens and, as such, could be used to guide initial dosing strategies in future studies of IFN-free regimens that contain ribavirin. Additionally, the use of day 14 concentrations has the potential benefit of being early enough in therapy to allow dose modification to improve patient outcomes.
A major question in the IFN-free era is the amount of ribavirin needed to generate optimal outcomes. While higher ribavirin doses may increase the likelihood of SVR, ribavirin-induced anaemia remains a challenge even with IFN-free DAA regimens. In the HCV Target trial, an ongoing study characterizing DAA use and outcomes in ‘real world’ populations, anaemia occurred in 18% of the 612 patients on sofosbuvir/ribavirin and 24% of the 183 patients on sofosbuvir/ribavirin plus simeprevir.32 We used our RBV-MP thresholds as a target therapeutic range in order to determine a ribavirin dose capable of generating RBV-MP concentrations predictive of a high likelihood of SVR while maintaining minimal risk of anaemia. Table 4 shows the percentage of patients at each simulated ribavirin dosing scheme exceeding the 4.4 and 6.1 pmol/106 cell thresholds. The percentage of the population above the determined threshold for elevated anaemia risk was 37% at the 800 mg dose, but jumped to 75% for the 1000 mg daily regimen. At the 600 mg daily dose, only 44% of individuals were above the 4.4 pmol/106 cells SVR threshold, which compares well with the findings of the SPARE trial (48% SVR at 600 mg daily in the ITT analysis).15 This percentage increased to 86% at the 800 mg daily dose. These results showed an 800 mg daily dose best achieved exposures within the target therapeutic range. One small study of 10 genotype 3a subjects found that 800 mg of ribavirin plus sofosbuvir for 12 weeks achieved a 60% SVR rate, which compares favourably with the 56% SVR rate observed in the FISSION trial of 183 genotype 3 patients receiving sofosbuvir plus weight-based dosing for 12 weeks.33,34 Though these thresholds need to be confirmed in larger subject populations, and clinical data are needed to support the success of an 800 mg daily dose, our findings suggest that 800 mg of ribavirin daily may warrant evaluation in clinical studies of IFN-free regimens.
Table 4.
Percentage of a simulated population (n = 1000) with average day 14 RBV-MP concentrations above the determined thresholds for SVR (4.4 pmol/106 cells) and anaemia (6.1 pmol/106 cells)
| Dose (mg) | Cave (pmol/106 cells) | Percentage above SVR threshold (4.4 pmol/106 cells) | Percentage above anaemia threshold (6.1 pmol/106 cells) |
|---|---|---|---|
| 600 | 4.27 | 44.4 | 2.3 |
| 800 | 5.69 | 85.6 | 37.0 |
| 1000 | 7.12 | 96.3 | 74.8 |
| 400/600 | 7.13 | 96.7 | 75.6 |
| 1200 | 8.54 | 98.8 | 90.9 |
| 600/600 | 8.55 | 99.0 | 91.5 |
| 1400 | 9.96 | 99.5 | 96.5 |
| 600/800 | 9.98 | 99.6 | 96.9 |
Though these results help fill gaps in the knowledge of ribavirin pharmacology, a few limitations exist. The first limitation is that post-dose times were not recorded during the study. However, given the long half-life of ribavirin both in serum and RBC compared with the dosing interval, concentrations are relatively constant within the dosing interval. Additionally, not all study doses were observed, meaning that PK results might be impacted by non-adherence. However, this increases the real-world applicability of our study, as non-adherence is likely to occur in the clinical setting. While this analysis yielded a strong estimate for the anaemia threshold (ROC AUC = 0.82), the ROC curve was only moderately predictive of the concentration threshold for SVR (ROC AUC = 0.67). It is likely that the SVR threshold was limited by the inverse association between baseline HCV RNA and SVR, suggesting the need to confirm the utility of this SVR threshold in a larger patient population. Finally, these thresholds were established using RBC levels, which is highly relevant for predicting anaemia, but may or may not be a valid surrogate for efficacy, as the primary site of antiviral effect is the hepatocyte. However, the collection of hepatocytes is more invasive. Furthermore, the relationships found in RBC can be extended to dried blood spot samples, which primarily consist of RBC and offer several benefits, including cheaper and easier sample processing, elimination of biological sample shipping requirements and costs and a minimal blood requirement, which is advantageous for anaemic patients or other special populations with blood volume limitations.35
In summary, treatment outcomes for HCV-infected patients treated with sofosbuvir and ribavirin appeared to be best associated with average day 14 RBV-MP concentrations. These results led to the determination of day 14 RBV-MP concentration thresholds that were predictive of SVR and anaemia. These thresholds provide guidance for clinical studies of ribavirin dosing in the context of IFN-free DAA regimens.
Funding
This research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (grant number R03 DK 096121 to J. J. K.) and the intramural programme of the National Institute of Allergy and Infectious Diseases, NIH, Bethesda.
Transparency declarations
A. O. and J. G. M. are employees and stock-holders of Gilead Sciences. All other authors: none to declare.
Supplementary data
Acknowledgements
These data have previously been presented at The Liver Meeting 2014 of the American Association of the Study of Liver Diseases (Poster 967).
References
- 1.Brillanti S, Mazzella G, Roda E. Ribavirin for chronic hepatitis C: and the mystery goes on. Dig Liver Dis 2011; 43: 425–30. [DOI] [PubMed] [Google Scholar]
- 2.Page T, Connor JD. The metabolism of ribavirin in erythrocytes and nucleated cells. Int J Biochem 1990; 22: 379–83. [DOI] [PubMed] [Google Scholar]
- 3.Lau JY, Tam RC, Liang TJ, et al. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 2002; 35: 1002–9. [DOI] [PubMed] [Google Scholar]
- 4.Homma M, Matsuzaki Y, Inoue Y, et al. Marked elevation of erythrocyte ribavirin levels in interferon and ribavirin-induced anemia. Clin Gastroenterol Hepatol 2004; 2: 337–9. [DOI] [PubMed] [Google Scholar]
- 5.D'Avolio A, De Nicolo A, Simiele M, et al. Development and validation of a useful HPLC-UV method for quantification of total and phosphorylated-ribavirin in blood and erythrocytes of HCV+ patients. J Pharm Biomed Anal 2012; 66: 376–80. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, Rower J, Burton J, et al. Population pharmacokinetic modeling of plasma and intracellular ribavirin concentrations in patients with chronic hepatitis C virus. Antimicrob Agents Chemother 2015; 59: 2179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afdhal N, Reddy KR, Nelson DR, et al. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. New Engl J Med 2014; 370: 1483–93. [DOI] [PubMed] [Google Scholar]
- 8.Everson GT, Sims KD, Rodriguez-Torres M, et al. Efficacy of an interferon- and ribavirin-free regimen of daclatasvir, asunaprevir, and BMS-791325 in treatment-naive patients with HCV genotype 1 infection. Gastroenterology 2014; 146: 420–9. [DOI] [PubMed] [Google Scholar]
- 9.Lawitz E, Sulkowski MS, Ghalib R, et al. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: the COSMOS randomised study. Lancet 2014; 384: 1756–65. [DOI] [PubMed] [Google Scholar]
- 10.Hezode C, Serfaty L, Vierling J, et al. Safety and efficacy of the all-oral regimen of MK-5172/MK-8742 plus/minus ribavirin in treatment-naive, non-cirrhotic patients with hepatitis C virus genotype 1 infection: the C-WORTHy study. In: Abstracts of The International Liver Congress 2014, London, UK, 2014. Abstract O10 European Association for the Study of the Liver, Geneva, Switzerland. [Google Scholar]
- 11. AASLD/IDSA/IAS-USA. Recommendations for Testing, Managing, and Treating Hepatitis C. http://www.hcvguidelines.org.
- 12.Poordad F, Hezode C, Trinh R, et al. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. New Engl J Med 2014; 370: 1973–82. [DOI] [PubMed] [Google Scholar]
- 13.Zeuzem S, Jacobson IM, Baykal T, et al. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. New Engl J Med 2014; 370: 1604–14. [DOI] [PubMed] [Google Scholar]
- 14.Curry MP, Forns X, Chung RT, et al. Sofosbuvir and ribavirin prevent recurrence of HCV infection after liver transplantation: an open-label study. Gastroenterology 2015; 148: 100–7.e1. [DOI] [PubMed] [Google Scholar]
- 15.Osinusi A, Meissner EG, Lee YJ, et al. Sofosbuvir and ribavirin for hepatitis C genotype 1 in patients with unfavorable treatment characteristics: a randomized clinical trial. JAMA 2013; 310: 804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimmerson LC, Ray ML, Bushman LR, et al. Measurement of intracellular ribavirin mono-, di- and triphosphate using solid phase extraction and LC-MS/MS detection. J Chromatogr B Analyt Technol Biomed Life Sci 2015; 978–979: 163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson AJ, Fellay J, Patel K, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology 2010; 139: 1181–9.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner EG, Bon D, Prokunina-Olsson L, et al. IFNL4-ΔG genotype is associated with slower viral clearance in hepatitis C, genotype-1 patients treated with sofosbuvir and ribavirin. J Infect Dis 2014; 209: 1700–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruchfeld A, Lindahl K, Schvarcz R, et al. Dosage of ribavirin in patients with hepatitis C should be based on renal function: a population pharmacokinetic analysis. Ther Drug Monit 2002; 24: 701–8. [DOI] [PubMed] [Google Scholar]
- 20.Kamar N, Chatelut E, Manolis E, et al. Ribavirin pharmacokinetics in renal and liver transplant patients: evidence that it depends on renal function. Am J Kidney Dis 2004; 43: 140–6. [DOI] [PubMed] [Google Scholar]
- 21.Wade JR, Snoeck E, Duff F, et al. Pharmacokinetics of ribavirin in patients with hepatitis C virus. Br J Clin Pharmacol 2006; 62: 710–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levey A, Bosch J, Lewis J, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med 1999; 130: 461–70. [DOI] [PubMed] [Google Scholar]
- 23.Preston SL, Drusano GL, Glue P, et al. Pharmacokinetics and absolute bioavailability of ribavirin in healthy volunteers as determined by stable-isotope methodology. Antimicrob Agents Chemother 1999; 43: 2451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin R, Fossler MJ, McHutchison JG, et al. Population pharmacokinetics and pharmacodynamics of ribavirin in patients with chronic hepatitis C genotype 1 infection. AAPS J 2012; 14: 571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta SK, Kantesaria B, Glue P. Pharmacokinetics, safety, and tolerability of ribavirin in hemodialysis-dependent patients. Eur J Clin Pharmacol 2012; 68: 415–8. [DOI] [PubMed] [Google Scholar]
- 26.Jin R, Cai L, Tan M, et al. Optimum ribavirin exposure overcomes racial disparity in efficacy of peginterferon and ribavirin treatment for hepatitis C genotype 1. Am J Gastroenterol 2012; 107: 1675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hitomi Y, Cirulli ET, Fellay J, et al. Inosine triphosphate protects against ribavirin-induced adenosine triphosphate loss by adenylosuccinate synthase function. Gastroenterology 2011; 140: 1314–21. [DOI] [PubMed] [Google Scholar]
- 28.Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature 2010; 464: 405–8. [DOI] [PubMed] [Google Scholar]
- 29.D'Avolio A, Ciancio A, Siccardi M, et al. Inosine triphosphatase polymorphisms and ribavirin pharmacokinetics as determinants of ribavirin-associate anemia in patients receiving standard anti-HCV treatment. Ther Drug Monit 2012; 34: 165–70. [DOI] [PubMed] [Google Scholar]
- 30.Homma M, Inoue Y, Hasegawa Y, et al. Blood ribavirin concentration in high-dose ribavirin for adenovirus-induced haemorrhagic cystitis—a case report. J Clin Pharm Ther 2008; 33: 75–8. [DOI] [PubMed] [Google Scholar]
- 31.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 2005; 436: 967–72. [DOI] [PubMed] [Google Scholar]
- 32.Jensen D, O'Leary J, Pockros P, et al. Safety and efficacy of sofosbuvir-containing regimens for hepatitis C: real-world experience in a diverse, longitudinal observational cohort. In: Abstracts of the Liver Meeting, Boston, MA, 2012. Abstract 45 American Association for the Study of Liver Diseases, Alexandria, VA, USA. [Google Scholar]
- 33.Gane EJ, Stedman CA, Hyland RH, et al. Once daily sofosbuvir (GS-7977) regimens in HCV genotype 1-3: the ELECTRON trial. In: Abstracts of the Liver Meeting, Boston, MA, 2014. Abstract 229 American Association for the Study of Liver Diseases, Alexandria, VA, USA. [Google Scholar]
- 34.Lawitz E, Mangia A, Wyles D, et al. Sofosbuvir for previously untreated chronic hepatitis C infection. New Engl J Med 2013; 368: 1878–87. [DOI] [PubMed] [Google Scholar]
- 35.Jimmerson LC, Zheng JH, Bushman LR, et al. Development and validation of a dried blood spot assay for the quantification of ribavirin using liquid chromatography coupled to mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2014; 944: 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.