Abstract
Organisms and cells, in response to environmental influences or during development, undergo considerable changes in DNA methylation on a genome-wide scale, which are linked to a variety of biological processes. Using MethylC-seq to decipher DNA methylome at single-base resolution is prohibitively costly. In this study, we develop a novel approach, named MBRidge, to detect the methylation levels of repertoire CpGs, by innovatively introducing C-hydroxylmethylated adapters and bisulfate treatment into the MeDIP-seq protocol and employing ridge regression in data analysis. A systematic evaluation of DNA methylome in a human ovarian cell line T29 showed that MBRidge achieved high correlation (R > 0.90) with much less cost (∼10%) in comparison with MethylC-seq. We further applied MBRidge to profiling DNA methylome in T29H, an oncogenic counterpart of T29's. By comparing methylomes of T29H and T29, we identified 131790 differential methylation regions (DMRs), which are mainly enriched in carcinogenesis-related pathways. These are substantially different from 7567 DMRs that were obtained by RRBS and related with cell development or differentiation. The integrated analysis of DMRs in the promoter and expression of DMR-corresponding genes revealed that DNA methylation enforced reverse regulation of gene expression, depending on the distance from the proximal DMR to transcription starting sites in both mRNA and lncRNA. Taken together, our results demonstrate that MBRidge is an efficient and cost-effective method that can be widely applied to profiling DNA methylomes.
Keywords: DNA methylome, MB-seq, ridge regression, single-base resolution
Introduction
DNA methylation plays a crucial role in epigenetic regulation and it has been recognized as the ‘fifth base’ in most mammalian genomes. As a covalent modification, it predominantly occurs at the C5 position of cytosine (5mC) within CpG dinucleotides (CpGs), but also presents at non-CpG cytosines (CHG and CHH, where H = A, T, or C) in embryonic stem cells and brains of mammals (Lister et al., 2009, 2013; Xie et al., 2012). It is generally accepted that pattern of DNA methylation can be stably transmitted to daughter cells through cell mitosis in higher eukaryotic organisms and the next generation via sperm as demonstrated in zebrafish (Jones, 2012; Jiang et al., 2013). However, in response to environmental influences and during developmental process, DNA methylation undergoes considerable changes (Meissner et al., 2008; Christensen et al., 2009; Baylin and Jones, 2011). Aberrant DNA methylation is unequivocally associated with pathogenesis and progression of many diseases, including developmental disorders, cancer, and immunological dysfunction (Robertson and Wolffe, 2000; Robertson, 2005). Therefore, detecting and estimating the DNA methylome is of great importance for understanding relevance of DNA methylation in diseases and biological processes.
Since introduction of next-generation sequencing technologies, several methods have been developed aiming at profiling DNA methylation on genome-wide scale. Of these, whole-genome bisulfite sequencing (MethylC-seq or WGBS) has been proved to be the most powerful and complete strategy for quantitative genome-wide detection of 5mC at single-base resolution (Cokus et al., 2008; Beck, 2010; Harris et al., 2010). However, MethylC-seq requires substantial sequencing effort—at least 30-fold coverage of the entire genome, which equates to a minimum of ∼90 Gb aligned data for a human genome (Lister et al., 2009; Satterlee et al., 2010). Within mammals and plants, 5mC accounts for ∼1%–6% of total C nucleotides in a genome, with the vast majority of 5mC occurring at CpGs (Lister and Ecker, 2009). Therefore, despite falling of sequencing costs, under-representation of CpGs in the genome (CpGs account for 4.76% of the total C nucleotides in a human genome) makes MethylC-seq expensive and inefficient in terms of sequencing per CpG site because only 20%–30% of the MethylC-seq data provides relevant information about DNA methylation (Ziller et al., 2013). In addition, mining underlying biological implication from resultant sequencing data is extremely computing-intensive. For example, it typically takes ∼22 days on a single 8-core processor with 24 GB RAM to obtain the DNA methylation levels at single-base resolution from 100 Gb data, even when using a fast tool such as BSMAP (Kunde-Ramamoorthy et al., 2014).
Restriction enzyme-based methods, including RRBS (Meissner et al., 2008) and methylation-sensitive restriction enzyme sequencing (MRE-seq) (Ball et al., 2009), combine digestion of genomic DNA using certain restriction enzymes and high-throughput sequencing of the digested fragments. RRBS reduces the amount of data required, saturating at ∼3 Gb, and generates a single-base methylation profile covering ∼10% of all CpGs, which includes most CpG islands (CGIs) in the human genome (Meissner et al., 2008; Bock et al., 2010; Wang et al., 2012). MRE-seq relies on restriction enzymes that are sensitive to methylated CpG (mCpG) and targets approximately 6% of unmethylated CpGs in the human genome (Maunakea et al., 2010). However, most tissue and cancer-specific differential methylation regions (DMRs) occur in CGI shores rather than CGIs (Irizarry et al., 2009). Moreover, ∼38% of CpGs in the human genome occur in repetitive elements (REs), in which they are usually heavily methylated, especially in Alu elements and long interspersed nucleotide elements (LINEs) (Kochanek et al., 1993; Bestor, 1998; Schmid, 1998). As Nichol and Pearson (2002) demonstrated, aberrant DNA methylation of these repeat sequences can significantly affect their genetic stability; therefore, the spectrum of whole DNA methylome cannot be adequately represented by the one derived from RRBS and MRE-seq.
Affinity enrichment-based methods, such as methylated DNA immunoprecipitation sequencing (MeDIP-seq) and methyl-binding domain sequencing (MBD-seq), capture methylated fraction of genomic DNA with 5-methylcytosine-specific antibody (5mC antibody) and MBD2 protein, respectively (Down et al., 2008; Serre et al., 2010). In contrast to restriction enzyme-based methods that target specific genomic regions, affinity enrichment-based methods enable identification of all potentially methylated genomic regions on genome-wide scale. However, MeDIP-seq and MBD-seq suffer from low resolution (∼100 bp) and bias derived from preferential affinity enrichment of methylated DNA fragments (Laird, 2010). Therefore, they provide only relative quantification of the DNA methylation levels in comparison with the absolute levels obtained using bisulfite-based methods (Takayama et al., 2014). To overcome this limitation, researchers have developed bioinformatic approaches to calibrate the DNA methylation levels from affinity enrichment-based sequencing data, including: BALM (Lan et al., 2011) for MBD-seq; Batman (Down et al., 2008), MEDIPS (Chavez et al., 2010), and MEDME (Pelizzola et al., 2008) for MeDIP-seq; BayMeth (Riebler et al., 2014) for both MBD-seq and MeDIP-seq; and methylCRF for MeDIP-seq/MRE-seq (Stevens et al., 2013). While these bioinformatic tools possess their respective advantages, none of these tools offers a good balance among single-base resolution, computational efficiency, accuracy, and flexibility (Riebler et al., 2014).
To achieve genome-wide coverage at reduced cost without sacrificing single-base resolution, we developed a novel and efficient DNA methylome profiling method, MeDIP-bisulfite sequencing (MB-seq), which was modified from MeDIP-seq protocol to encompass bisulfite treatment. In MB-seq, our innovative introduction of C-hydroxylmethylated Illumina adapters (all the cytosines in the adapters were hydroxylmethylated) rendered it more operation-friendly than MeDIP-Bseq (Takayama et al., 2014). In data process, we developed an accurate correction method to obtain the absolute methylation levels from data derived from MB-seq based on ridge regression. We applied the ridge regression model to correct data generated by MB-seq in an ovarian epithelial cell line (T29) and its oncogenic counterpart (T29H), resulted in respective methylomes covering repertoire CpGs with single-base resolution. Further analysis identified 131790 DMRs with high accuracy between T29 and T29H, which are mainly enriched in carcinogenesis-related pathways, and substantially different from the ones obtained by RRBS. Taken together, we demonstrated that MB-seq combined with ridge regression, namely MBRidge, can accurately detect the whole DNA methylome at dramatically reduced cost compared to MethylC-seq. Thus, our method is a promising tool for large-scale and genome-wide studies on DNA methylation.
Results
Convenient manipulation and high reproducibility of MB-seq
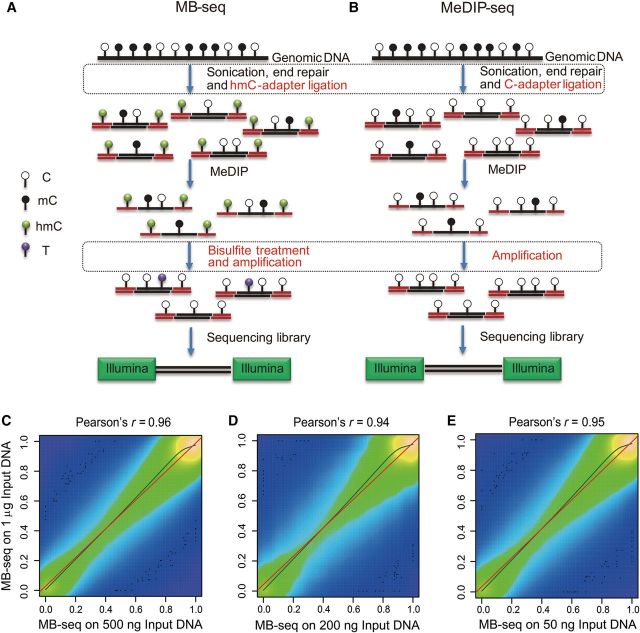
As shown in Figure 1A and B, we developed MB-seq for profiling a DNA methylome at single-base resolution. MB-seq differed from MeDIP-seq as follows: (i) C-hydroxylmethylated Illumina multiplexing adapters were applied in the adapter-ligation step to replace the corresponding Illumina multiplexing adapters in which all Cs are unmethylated; and (ii) bisulfite treatment was performed after MeDIP enrichment step. In addition, we assessed amplification efficiencies of several commercial Taq polymerases under the presence of cytosine 5-methylenesulfonate (CMS) generated by bisulfite conversion of hydroxylmethylated C (5hmC) which could hamper the Taq binding with DNA templates. We found KAPA 2G robust polymerase achieved the best performance (Supplementary Figure S1).
Figure 1.
Schematic diagram and reproducibility of MB-seq. (A) Schematic diagram of the MB-seq approach. Genomic DNA is randomly fragmented to 100–500 bp and ligated to hydroxylmethylated Illumina adapters. The ligated fragments are captured using 5mC antibody. The antibody-enriched DNA fragments are treated with bisulfite and amplified by PCR using Illumina paired-end PCR primers. PCR products of 270–370 bp in length are size-selected on gel and sequenced on the Illumina platform. (B) Schematic diagram of the MeDIP-seq approach. The modified steps are marked with rectangular areas for a comparison with MeDIP-seq. (C–E) Scatter plots of PCC for MB-seq-measured DNA methylation levels of CpG sites between different DNA input libraries, 500 ng vs. 1 µg (C), 200 ng vs. 1 µg (D), and 50 ng vs. 1 µg (E). The color shade describes the relative difference in numeric terms. The more bright color indicates higher CpGs density. Black dots exhibit the 100 most ‘sparse’ points plotted over the smoothed density plot. The olive and red lines are curves of kernel and linear regression, respectively.
To ensure wide application of our DNA methylome profiling technology, it is critical that only the low quantity of genomic DNA starting material is required. We therefore evaluate sensitivity and reproducibility of MB-seq. As shown in Table 1, the same batch of genomic DNA from T29 cell line was performed in our MB-seq analysis by using the various amounts of genomic DNA as input, including 1 µg, 500 ng, 200 ng, and 50 ng, respectively. After removing adapter contaminated, low qualitative, and clonal reads, 8.58, 10.25, 8.85, and 14.57 Gb of clean data were generated from each replicate of the starting amount of genomic DNA, respectively. By counting CpGs with at least 10-fold sequencing depth, we determined the following Pearson correlation coefficient (PCC) values, calculated against MB-seq #1 (1 µg): 0.96 for MB-seq #2 (500 ng), 0.94 for MB-seq #3 (200 ng), and 0.95 for MB-seq #4 (50 ng), respectively (Figure 1C–E). The high correlation between our technical replicates suggested that MB-seq could achieve satisfied reproducibility, even when the starting amount of genomic DNA was used as low as 50 ng.
Table 1.
General information of the sequencing data for MethylC-seq, MB-seq, RRBS, and MeDIP-seq.
| Sample | T29 |
T29H |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Methods and duplicate no. | MethylC-seq | MB-seq #1 | MB-seq #2 | MB-seq #3 | MB-seq #4 | RRBSa | MeDIP-seq | MB-seq | RRBSa |
| Input | 3 µg | 1 µg | 500 ng | 200 ng | 50 ng | 500 ng | 100 ng | 50 ng | 500 ng |
| Raw data (Gbp) | 152.6 | 8.97 | 10.7 | 9.17 | 15.44 | 8.88 | 8.36 | 16.06 | 8.29 |
| Clean data (Gbp) | 147.03 | 8.58 | 10.25 | 8.85 | 14.57 | 8.39 | 7.42 | 15.74 | 7.76 |
| Mapped data (Gbp) | 134.19 | 7.06 | 8.38 | 7.73 | 12.73 | 7.5 | 5.5 | 11.17 | 6.27 |
| Unique mapped data (Gbp) | 128.42 | 6.47 | 7.67 | 7.18 | 11.82 | 6.82 | 4.73 | 10.53 | 5.45 |
| Estimated conversion rate (%) | 99.5 | 99.62 | 99.75 | 99.4 | 99.45 | 99.33 | # | 99.62 | 99.82 |
| Methylation level of C (%) | 4.63 | 7.23 | 7.08 | 7.05 | 6.65 | 6.96 | # | 5.35 | 5.92 |
| Methylation level of CG (%) | 54.48 | 80.21 | 79.93 | 82.1 | 81.06 | 48.64 | # | 72.16 | 42.31 |
| Methylation level of CHG (%) | 0.52 | 0.51 | 0.36 | 0.59 | 0.56 | 0.63 | # | 0.41 | 0.17 |
| Methylation level of CHH (%) | 0.5 | 0.38 | 0.25 | 0.6 | 0.55 | 0.67 | # | 0.38 | 0.18 |
| Total number of mCG identified (Mbp) | 24.43 | 5.38 | 6.75 | 13.79 | 20.15 | 1.87 | # | 17.82 | 2.25 |
| Percent of covered base (%) ≥1× | 91.25 | 41.83 | 46.3 | 65.24 | 73.71 | 11.15 | 33.58 | 77.66 | 12.75 |
| Percent of covered C (%) ≥1× | 97.62 | 45.54 | 50.39 | 72.45 | 81.25 | 13.91 | 32 | 85.48 | 14.52 |
| Percent of covered CpGs (%) ≥1× | 96.09 | 59.05 | 63.65 | 81.8 | 88.11 | 26.76 | 51.12 | 90.02 | 27.06 |
aGeneral information of the sequencing data for RRBS is obtained from the previous study (Wang et al., 2014b).
Cost-effectiveness of genome-wide identification of CpGs and mCpGs by MB-seq
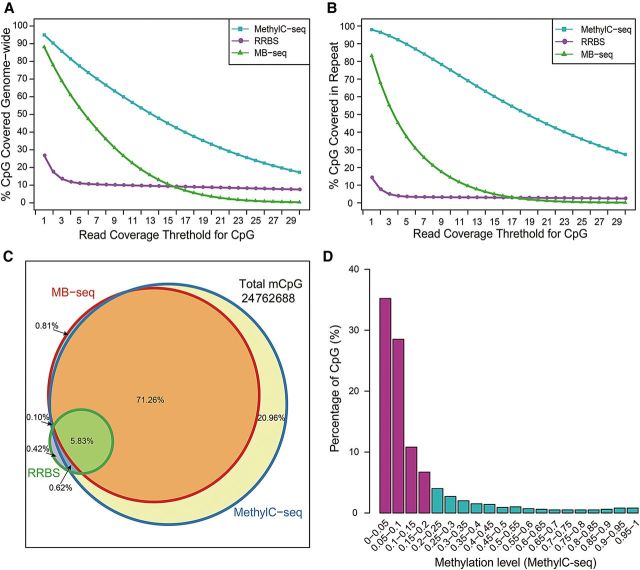
In order to evaluate efficiency of MB-seq, we compared the total number and local context of CpGs and mCpGs in MB-seq with those in MethylC-seq and RRBS using the same batch of genomic DNA from T29. Analyzing 147.03, 14.57, and 8.39 Gb clean data generated by MethylC-seq, MB-seq, and our previously published RRBS (Table 1), respectively, we plotted total coverage of genome-wide CpGs and their respective depth (Figure 2A). MB-seq covered 88.1% of the total CpGs in the human genome, whereas MethylC-seq and RRBS covered 96.1% and 26.8%, respectively. By calculating the CpG recovery rate per Gb clean data in these three methods, MB-seq, MethylC-seq and RRBS were observed as 6.05%, 0.65% and 3.20%, respectively, suggesting that MB-seq is the most cost-efficient method per CpG on genome-wide scale. We also observed 83.2% genome-wide CpGs coverage in REs (Figure 2B) by MB-seq, which was substantially greater than the one observed by RRBS (14.3%) and less than that observed by MethylC-seq (98.0%). By subdividing the data of MB-seq into different genomic features according to hg18 annotation in UCSC database and calculating the proportion of each feature in all data (Supplementary Figure S2A), we found that REs, introns and Alu elements occupy 39.0%, 29.6%, and 18.2% of total MB-seq data, respectively, which is similar to those found in genomic background. From these results, we conclude that MB-seq is also a cost-effective method for profiling DNA methylation in REs and other genomic features (Supplementary Figure S2B).
Figure 2.
The comparison of different DNA methylation profiling methods used at single-CpG resolution. (A and B) CpG coverage as a function of read coverage threshold for MethylC-seq (cyan), RRBS (medium-orchid), and MB-seq (green). X-axis denotes sequencing depth and y-axis denotes the fraction of CpGs that are at or above a given sequencing depth. The percentage of CpGs that were covered genome-wide (A) or in repeat (B) are plotted. (C) Venn diagram shows the overlap of mCpGs from three methylation profiling methods. The total mCpGs measured by all three methods and percentages for each color block are shown. The three circles represent MethylC-seq (blue), RRBS (green), and MB-seq (red), respectively. (D) Barplot represents the fraction of mCpG covered only by MethylC-seq, but not by MB-seq or RRBS.
To further evaluate the coverage of mCpGs by MB-seq, we calculated the total number of mCpGs identified by MB-seq, MethylC-seq, and RRBS (Figure 2C), respectively. Among a total of 24762688 mCpGs identified in all three methods, 77.9%, 98.8%, and 6.9% of the total mCpGs were detected by MB-seq, MethylC-seq, and RRBS, respectively. Of these, 0.81%, 20.96%, and 0.42% of the total mCpGs were uniquely detected by MB-seq, MethylC-seq, and RRBS, respectively. From those detected by MB-seq uniquely, we randomly chose 120 mCpGs derived from 15 loci to perform locus-specific bisulfite sequencing for validation (Supplementary Table S2). Our result showed that all 120 mCpGs exhibited high levels of methylation. We also explored distribution of the methylation level among mCpGs detected by MethylC-seq uniquely (Figure 2D) and found that 63.7% and 81.2% mCpGs presented the methylation levels lower than 10% and 20%, respectively. This result indicated that the unique mCpGs detected by MethylC-seq were almost hypo-methylated, which is consistent with the results in which MB-seq and MeDIP-seq were more preferential to detect highly methylated regions (Supplementary Figure S3A). By calculating the mCpG recovery rate per Gb clean data in these three methods, MB-seq, MethylC-seq and RRBS were observed as 5.35%, 0.67%, and 0.82%, respectively. In summary, we found that with much less data output than MethylC-seq, MB-seq could efficiently detect a greater proportion of mCpGs in the human genome.
Using MB-seq to exclude false positives generated by non-specific enrichment of MeDIP
Currently, MeDIP-seq cannot be used to identify individual 5mC sites in captured reads or distinguish un-methylated reads captured by 5mC antibody due to its non-specific binding (Chavez et al., 2010; Harris et al., 2010); therefore, it significantly increase rate of false positive in detecting mCGs. It is expected that the rate of false positive in MeDIP-seq may be reduced by encompassing of bisulfite treatment in MB-seq. By plotting ROC curve, we investigated the sensitivity and specificity in detection of mCpGs (predefined by MethylC-seq with a binomial test, P < 0.01) among four different methods (MethylC-seq, MB-seq, MeDIP-seq, and MEDIPS (normalized MeDIP-seq)), respectively (Figure 3A). Our data showed that the area under the curve (AUC) value were 99.6%, 97.9%, 78.0%, and 73.0% for MethylC-seq, MB-seq, MeDIP-seq, and MEDIPS, respectively. It clearly indicated that MB-seq, similar to MethylC-seq, exhibited robust the sensitivity and specificity to detect mCpGs in contrast to MeDIP-seq and MEDIPS which is derived from MeDIP-seq by employing normalized algorithm with 50-bp resolution, suggesting that MB-seq is capable of excluding false positives derived from MeDIP, thereby improving its accuracy.
Figure 3.
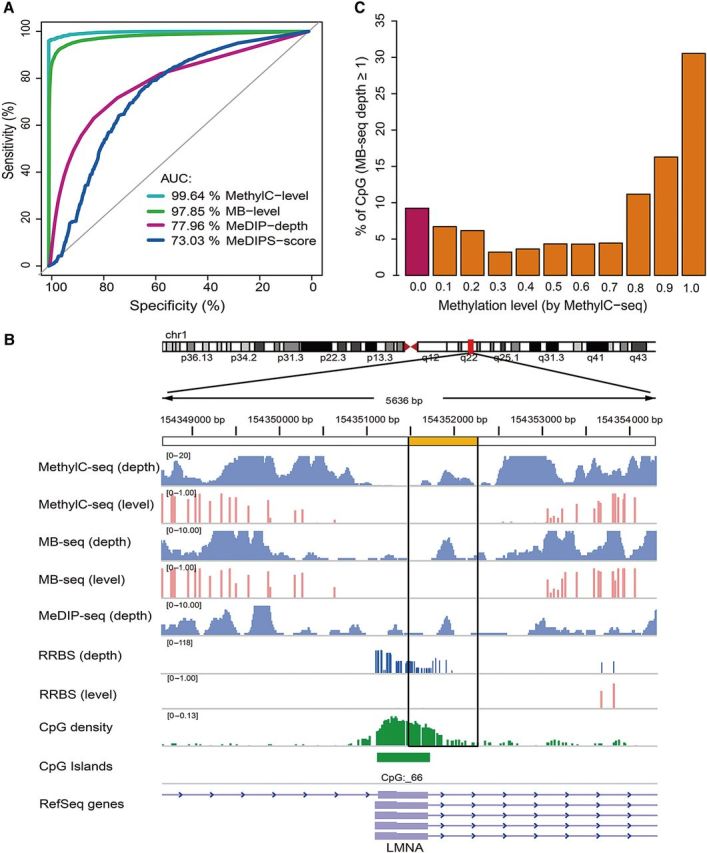
MB-seq can exclude MeDIP-seq false positives. (A) ROC curves are plotted by pROC in R CPAN for MethylC-level (cyan), MB-level (green), MeDIP-depth (violet red), and MEDIPS-score (navy). The AUC value is reported for each measurement. (B) Barplot represents the fraction of CpGs covered by MeDIP-seq. The different methylation levels were measured by MethylC-seq. At least 9% of all CpGs covered by MeDIP-seq are credibly considered as artificial signals, because they present a zero level of methylation (violet bar). (C) IGV screenshot of a region demonstrates that MB-seq can exclude false positives from MeDIP-seq. The black box (yellow-labeled) shows a captured region that has no methylated sites and presents non-specific DNA reads captured by 5mC antibody.
To give a snapshot on excluding the false positives, we profiled and annotated a specific genomic region. As shown in Figure 3B, although the region was covered by reads in three sequencing methods (MethylC-seq, MB-seq, or MeDIP-seq), this region was identified as un-methylated in both MB-seq and MethylC-seq. Thus, the region was a false positive signal which was presumably derived from non-specific binding of 5mC antibody to un-methylated DNA fragments in MeDIP-seq. In addition, we analyzed the methylation level across the entire dataset of MB-seq and found that 9.0% of 17659484 CpGs covered by MB-seq were unmethylated (Figure 3C). Similarly, 6.4% of 11279092 CpGs covered by MeDIP-seq were unmethylated (Supplementary Figure S3B). Therefore, it is reasonable to conclude that 6.4%–9.0% of the CpGs identified by MeDIP-seq are highly likely to be the false positive signals.
Motivation for correction of the methylation levels determined by MB-seq
RRBS has been confirmed in multiple studies as capable of accurate determination of the absolute methylation levels of CpGs (Bock et al., 2010; Harris et al., 2010). Therefore, we tried to compare concordance between the methylation levels of CpGs in MB-seq and RRBS (Supplementary Figure S4A). In MB-seq, methylated DNA fragments were preferentially enriched by 5mC antibody, which resulted in generation of the relative methylation levels; thus, the methylation levels of MB-seq and RRBS could not be directly compared, indicating that the methylation level in MB-seq needs to be corrected. Therefore, we first try to explore relationship between the observed methylation levels on gene body plus its flanking 2 kb regions in MB-seq with those derived from RRBS. Despite being inflated, the methylation levels observed in MB-seq showed an approximately linear relationship with those in RRBS with R2 = 0.8256 (Supplementary Figure S3C and D). This linear correlated regularity suggested that the observed methylation level in MB-seq could be potentially calibrated to the absolute methylation level. Because regularized linear regression models are appropriate for solving an ill-posed problem or preventing over-fitting, we selected three different regression methods (ridge, elastic-net, and lasso regression) in R package glmnet (Friedman et al., 2010) for calibration. To confirm which regression model is suitable for calibration of MB-seq, we performed cross-validation in these three models. Among ∼3 million CpGs covered by at least 10 reads in RRBS, we randomly selected 50% of the CpGs with information of DNA methylation from RRBS and the corresponding one from MB-seq as training data. The remaining half of data from RRBS and the corresponding one from MB-seq were used as testing data. The three regression models were trained using the training data, and resultant models were applied to the testing data of MB-seq to predict the methylation levels. Based on calculated PCC values by correlating the predicted methylation levels in MB-seq with the corresponding absolute levels in the RRBS testing data, we found that ridge regression had the best performance for the calibration (Supplementary Table S3). The detail of our ridge regression model (shown in Supplementary Figure S3E) is described in Materials and methods section.
Robust performance of ridge regression calibration for MB-seq
To evaluate performance of ridge regression for MB-seq, we compared the DNA methylation levels in MB-seq and RRBS in independent cell lines T29 and T29H, both before and after ridge regression. As shown in Supplementary Figures S4A and S5A, before ridge regression, PCC values for correlation of the methylation levels between MB-seq and RRBS were 0.87 and 0.81 for T29 and T29H cell lines, respectively. Following ridge regression, the PCC values were elevated to 0.96 and 0.95 for T29 and T29H, respectively (Supplementary Figures S4B and S5B). Consistent with a previous study (Stevens et al., 2013), the methylation levels calibrated by ridge regression displayed bimodal distribution that is similar to those measured by RRBS. By defining methylation proportion difference <0.25 (Stevens et al., 2013), the methylation level calibrated by ridge regression compared with the one in RRBS is 92.7%, 92.3% concordant for T29 and T29H, respectively (Supplementary Figures S4C and S5C). We further evaluated the performance of calibration of ridge regression in various genomic features for T29 cell line. For each genomic feature (the number of CpGs used is illustrated in Supplementary Figure S4D), we observed a significant improvement in PCC values after ridge regression (with the range improving from 0.74–0.86 to 0.85–0.96; Supplementary Figure S4E). In addition, ridge regression could precisely calibrate the methylation levels in various genomic features of T29H cell line with similarly improved PCC values to T29 (Supplementary Figure S5D and E).
Next, we compared the DNA methylation levels of all CpGs identified by MB-seq before and after ridge regression with the ones from MethylC-seq with sequencing depth ≥10 in T29 cell line. Prior to ridge regression, the DNA methylation levels for MB-seq vs. MethylC-seq were correlated at PCC = 0.774; while after ridge regression, PCC reached 0.905 (Figure 4A and B). The percentage of CpGs with a methylation proportion difference <0.25 between MB-seq and MethylC-seq were 87.9% and 93.6% before and after ridge regression, respectively (Figure 4C). For each genomic feature (the number of CpGs used is illustrated in Figure 4D), we observed significant improvement in concordance of the methylation levels between MB-seq after ridge regression and MethylC-seq. For example, PCC values for MB-seq after ridge regression vs. MethylC-seq within promoters, CGIs, 5′ UTRs, repeats, enhancers, and CDS, improved to 94.58%, 95.69%, 93.79%, 93.02%, 86.60%, and 96.53%, respectively (Figure 4E). All of above findings strongly suggested that MB-seq combined with ridge regression, termed as MBRidge, could achieve the satisfactory performance as MethylC-seq both on genome-wide scale and with various genomic features at single-base resolution. Such high concordance is further demonstrated by genome browser visualization of MethylC-seq, MBRidge, MB-seq, RRBS and MeDIP-seq in representative genomic loci (Supplementary Figure S6).
Figure 4.
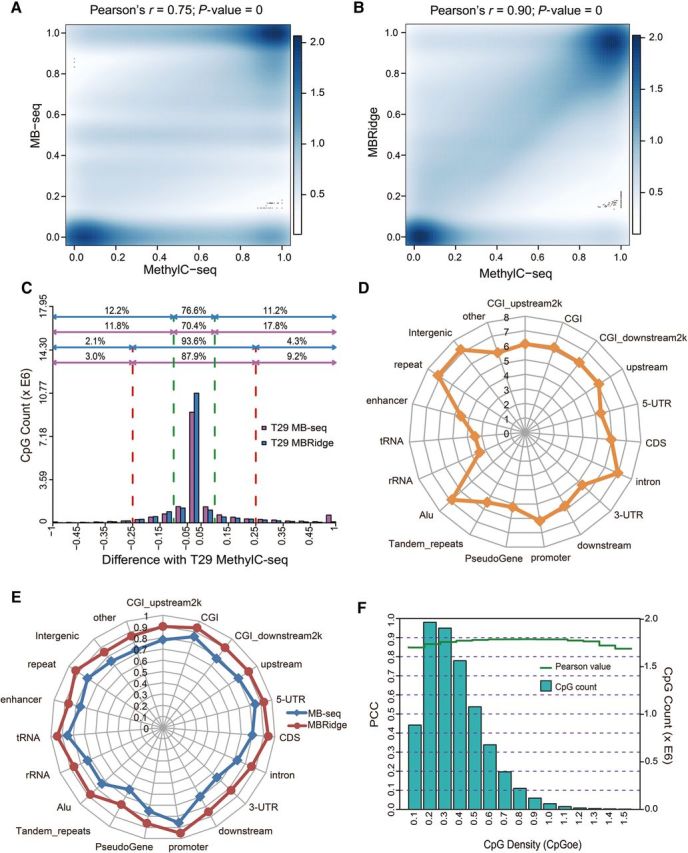
Accuracy evaluation for the ridge regression model. (A and B) Scatter plots of PCC compare MB-seq vs. MethylC-seq (A) and MBRidge vs. MethylC-seq (B). The bar displays a legend/color scale that describes the relative difference in numeric terms between different shades. Black dots exhibit the 100 most ‘sparse’ points plotted over the smoothed density plot. (C) The number of CpGs as a function of the difference in methylation levels between MethylC-seq and MBRidge. By defining the methylation difference <0.25 and 0.10, the number of concordant CpGs between these two methods are 93.6% and 76.6% of the total CpGs, respectively. The corresponding percentages of the number of concordant CpGs between T29 MethylC-seq and T29 MB-seq are 87.9% and 70.4%, respectively. (D) The number of CpGs in each genomic feature identified by MethylC-seq with coverage of at least 10× depth. (E) PCC values from the comparison of MBRidge and MB-seq with MethylC-seq for annotated genomic features. The axes in each radar chart represent annotated genomic features. (F) The concordance between MBRidge and MethylC-seq (measured by PCC) as a function of CpG density (measured by CpGoe value). The left y-axis (cyan bars) indicates the number of CpGs corresponding to the CpG density.
When using MeDIP-generated data, estimations of the DNA methylation level can be confounded by variable density of mCpG sites across a genome (Laird, 2010), which could be problematic when analyzing CpG-poor (low density) regions (Down et al., 2008; Pelizzola et al., 2008). Therefore, we examined performance of MBRidge across different regions with varying CpG density. PCC of the methylation levels in MBRidge vs. MethylC-seq showed that the two methods were highly concordant and they did not vary significantly based on various CpG densities (Figure 4F). Additionally, we found that bias between MBRidge and MethylC-seq did not vary significantly with the differing methylation levels (Supplementary Figure S7). In summary, the strong correlation between results of MBRidge and MethylC-seq regardless of genomic features, the variable CpG densities, and the varying methylation levels, suggest that MBRidge could detect a methylome with comparable capabilities as MethylC-seq.
Evaluating contribution of methylation-related factors in ridge regression
As multiple variables related to methylation were introduced in ridge regression model (see Materials and methods section), we tried to determine contribution of each variable in ridge regression model by a permutation test with comparison of the methylation levels between MBRidge and MethylC-seq. When all variables were integrated, PCC value for the predicted methylation levels in MBRidge vs. MethylC-seq was 0.905. Among all variables, the methylation levels observed in MB-seq (MB level) and the mean methylation levels flanking 100-bp region adjacent to local CpG observed in MB-seq (MB back level) played the most import role in our ridge regression model (Supplementary Figure S8A). This could be supported by the result that PCC could reach to 0.903 when only considering the MB level and the MB back level in our ridge regression model. In a previous study, genomic sequences and features provided a default prediction of methylation status (Stevens et al., 2013). However, in our analysis, PCC was equal to 0.774 when ridge regression was based on genomic features alone with the assumption that the methylation level of CpGs uncovered by MB-seq was zero (Supplementary Figure S8A). Additionally, given that MB-seq reads depth covering individual CpG (MB depth) can be regarded as data from MeDIP-seq, when only including MB depth in our model with genomic features, PCC was 0.784. This implied that the MB level and the MB back level at single-base resolution, derived from bisulfite conversion in MB-seq, were most important in the ridge regression model. It also suggested the infeasibility to accurately predict the absolute methylation level from MeDIP-seq data alone. Taken together, our results strongly suggested that by taking advantage of a priori training model, the ridge regression algorithm could effectively integrate all variables with weighed coefficient factors into training data and produce accurate predictions.
Reaching saturation in MB-seq and achieving higher accuracy in MBRidge
For any given sequencing-based methylation profiling method, how much sequencing data is required remains unclear because it is directly related to the balance between sequencing costs and accuracy of methylation profiling. By gradually analyzing coverage of CpGs in sequencing data from MB-seq#4 (14.57 Gb clean data) (Supplementary Figure S8B), we found that the coverage of CpGs reaches ∼80% of the 28.2 million CpGs in the human genome when sequencing data from MB-seq reached 9 Gb, similar to the one which had been considered as saturation in a previous study using MeDIP-seq (Chavez et al., 2010).
On the other hand, the amount of sequencing data required for achieving satisfactory performance by the ridge regression model is still yet to be explored. Thus, we randomly selected the different amount of reads from RRBS or MB-seq to evaluate the performance of the ridge regression model. By comparing the methylation levels calibrated by our ridge regression on each data set with those in MethylC-seq, we found that the minimum data size required to achieve PCC about 0.9 was 1.4 Gb in RRBS and 7.5 Gb in MB-seq, respectively (Supplementary Figure S8C and D). Based on the saturation-analysis of MB-seq, for a 3 Gb human genome, ∼10.4 Gb clean data from RRBS (1.4 Gb) and MB-seq (9 Gb) enable MBRidge to achieve satisfactory correction.
Aberrant alteration of DNA methylome in a human ovarian cancer model revealed by MBRidge
To understand the role of DNA methylation in ovarian-tumorigenesis, we generated DNA methylomes of both T29 and T29H by MBRidge and identified DMRs according to our developed pipeline (Wang et al., 2013). We observed prominent hypo-methylation on genome-wide scale and almost elements while upstream, CGI, promoter and 5-UTR elements were containing tiny alternating for T29H compared with T29 (Supplementary Figure S9A and B), which is consistent with a previous conclusion regarding carcinogenesis and DNA methylation (Jones, 2012). We found 131790 independent regions (ranging from 9 to 2996 bp with median size of 162 bp; Supplementary Table S4; see Supplementary Methods for more details) were candidate DMRs that had significant differences in the methylation level between two cell lines (t-test P < 0.05 and FDR < 0.05). This was substantially more than 7567 identified previously by RRBS. MBRidge uniquely detected 126936 DMRs, which primarily appeared in repeats, Alu elements, and introns. By performing experimental validation of DMRs using locus-specific bisulfite sequencing, all of the 10 randomly chosen DMRs were in agreement with results of the ridge regression model by displaying the differential levels of DNA methylation between T29 and T29H (Figure 5A and B, Supplementary Figure S10A–H), which further confirmed the accuracy of MBRidge.
Figure 5.
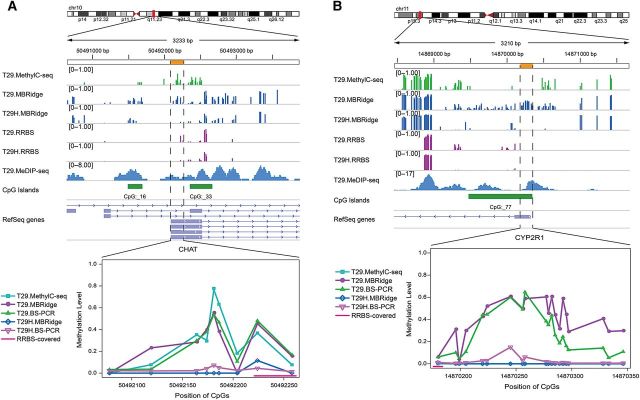
Experimental validation of DMRs between T29 and T29H cell lines identified by MBRidge. Genome browser views and line graphs show the DMRs between T29 and T29H, validated by locus-specific bisulfite sequencing. The line graph shows the methylation levels measured by MethylC-seq, RRBS, bisulfite PCR validation (BS-PCR), and MBRidge. (A) A representative DMR in chr10: 50492078–50492258. MBRidge agrees with BS-PCR and MethylC-seq. (B) A representative DMR in chr11: 14870180–14870346. MBRidge agrees with BS-PCR for both T29 and T29H, while MethylC-seq and RRBS do not detect the region that is enriched by MeDIP-seq in T29 cells.
We found that 4910 DMRs were located in 2 kb flanking sequences (defined as promoters) of transcription start sites (TSSs) in 3992 genes. ClueGO analysis revealed that DMRs were enriched in multiple pathways with hypergeometric P-value <0.05 (Figure 6A), including calcium signaling pathway, type II diabetes mellitus, Wnt signaling pathway, Basal cell carcinoma, Rap1 signaling pathway, AMPK signaling pathway, proteoglycans in cancer, pathways in cancer, glycolysis/gluconeogenesis, Ras signaling pathway, MAPK signaling pathway, and central carbon metabolism in cancer. Most of them were related to tumorigenesis and metabolism (Karnoub and Weinberg, 2008).
Figure 6.
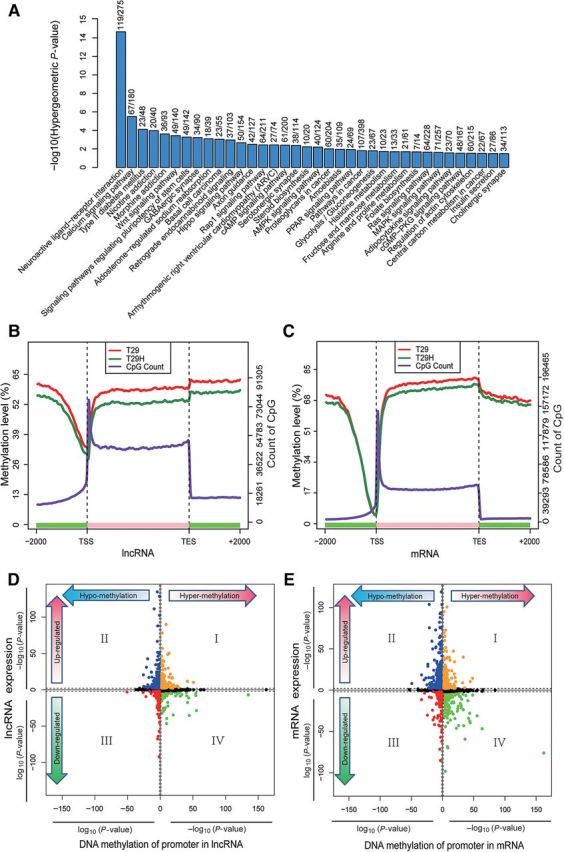
Canonical DNA methylation profiles depicted by MBRidge and the relationship between DNA methylation and gene expression. (A) Barplot shows the hierarchical order of the pathway enrichment of DMR-related genes, based on their enrichment scores (−log [hypergeometric P-value]). The fraction number above each bar indicates the percentage of genes overlapped with DMRs in each term of pathway. (B and C) Canonical DNA methylation profiles across regions in lncRNA (B) and mRNA (C), respectively. The canonical gene structure is defined by three different features, denoted by the x-axis. The length of each feature was normalized and divided into equal numbers of bins. Respective lines denote the average methylation levels for T29 (red) and T29H (forest-green). Two black vertical dashed lines indicate the mean location of transcription start sites (TSS) and transcription end sites (TES), respectively. The blue-violet line indicates the sum of CpG numbers. (D and E) Starburst plots denote the relationship between DMRs and lncRNA (D) and mRNA (E) expressions, respectively. Log10 (P-value) is plotted for DNA methylation (x-axis) and gene expression (y-axis) of each gene. If a mean DNA methylation level or gene expression value is higher (>0) in T29H, −1 is multiplied to log10 (P-value), providing a positive value. The black dashed lines indicate P-value at 0.05 for DMRs and 0.01 for gene expression. Data points in orange and red indicate genes in which significant expression changes are positively correlated with significant methylation changes, while blue and green points represent genes in which significant expression changes are inversely correlated with significant methylation changes. The remaining genes are marked in black.
In contrast, we found that the DMRs located in promoters account for only a small fraction of all DMRs and alteration of methylation beyond promoters are far more dynamic than previously revealed by RRBS (Wang et al., 2014b) (Supplementary Figure S9C). Therefore, we performed functional analysis of the DMRs identified by MBRidge and RRBS by using GREAT tool to analyze functional significance from cis-regulatory regions (McLean et al., 2010), respectively. Intriguingly, we found that DMRs derived from MBRidge were highly enriched in biological functions and pathways mainly associated with carcinogenesis (Supplementary Table S5), while DMRs derived from RRBS were primarily related to functions and pathways in development or differentiation (Supplementary Table S6). This strongly suggests that distal DMRs from TSSs are biologically meaningful for comprehensive functional interpretation of DNA methylation alteration in cancers.
Integrated analysis of the relationship between DNA methylation and gene expression
DNA methylation changes in promoter regions are integral to all aspects of tumorigenesis and have been shown to have important relevance with gene expression (Weisenberger, 2014). Recently, transcriptome profiling has revealed that a significant subset of transcripts longer than 200 nucleotides, located in non-coding regions, known as long non-coding RNAs (lncRNAs), can modulate gene expression, but their DNA methylation patterns remain poorly understood. As a genome-wide approach, MBRidge enabled investigation of methylation patterns in lncRNA loci (downloaded from database NONCODE v4) (Xie et al., 2014). We found that although lncRNA genes exhibited methylation patterns similar to protein-coding genes in the gene-body and flanking regions, they displayed the higher levels of methylation around TSSs (Figure 6B and C). This observation is contrary to the one derived from a previous study based on MeDIP-seq data (Sati et al., 2012). Our observation indicated that the generally higher levels of methylation in TSSs of lncRNAs could account for the generally lower expression of lncRNAs when compared with protein-coding genes (Derrien et al., 2012).
In order to establish cross-association between DNA methylation and gene expression, we performed RNA-seq with T29 and T29H and identified 2191 and 2939 differentially expression transcripts (P < 0.01, FDR < 0.01, fold change ≥2) for protein-coding genes and lncRNA genes, respectively (Robinson et al., 2010). We integrated differentially expressed transcripts and DMRs occurring in their corresponding promoters for both protein-coding and lncRNA genes. Interestingly, in contrast to a previous study (Sati et al., 2012), we observed 465 lncRNA genes in which expression changes were inversely correlated with methylation alterations (Figure 6D, Supplementary Table S7). This also occurs in 614 protein-coding genes (Figure 6E, Supplementary Table S8), suggesting that methylation regulation is similar in lncRNA and mRNA. Functional annotation of these inversely methylation-regulated protein-coding genes revealed that these genes were enriched in tumor associated pathways (Supplementary Figure S9D) including Hippo signaling pathway, transcriptional misregulation in cancer, Calcium signaling pathway, TGF-beta signaling pathway, MAPK signaling pathway, and ECM-receptor interaction. Interestingly, gene ontology analysis of these inversely methylation-regulated lncRNA genes unveiled that these lncRNAs genes were related to significant biological processes (Supplementary Figure S9E) including mitotic prometaphase, negative regulation of protein binding, protein trimerization, renal absorption, G2/M transition of mitotic cell cycle, water-soluble vitamin metabolic process, fructose 1,6-bisphosphate metabolism, regulation of BMP signaling pathway, response to mechanical stimulus, regulation of osteoblast proliferation and protein complex localization. Intriguingly, the important roles of inversely methylation-regulated lncRNAs in cellular proliferation of carcinomas were highlighted (White et al., 2014). Taken together, our result provides a new insight of leveraging relevant knowledge of those lncRNAs in carcinogenesis.
Surprisingly, we also found that the ability of DMRs to inversely regulate gene expression was more significant along with more close between DMRs and TSSs in both protein-coding and lncRNA genes (Supplementary Figure S9F and G). Our discovery was consistent with previous study which reported that methylation closest to TSS rather than flanking was a significant impediment to the polymerase extension in transcriptional initiation (Brenet et al., 2011).
Discussion
An essential step towards unraveling complex role of DNA methylation in phenotype is to generate methylomes with high resolution and accuracy (Pomraning et al., 2009; Laird, 2010). In this study, we developed a novel method to profile DNA methylome, named MBRidge, which integrated innovation in molecular technology (MB-seq) and bioinformatic algorithm (ridge regression model). MBRidge showed high accuracy in generating the absolute methylation levels of each single-CpG, which were highly correlated to the ones from MethylC-seq and RRBS in T29 with PCC values of 0.905 and 0.96, respectively. The accuracy of MBRidge was further confirmed by high accuracy of a methylome in T29H cell line (PCC for MBRidge vs. RRBS: 0.95), which has aberrantly induced by H-RasV12 to mediate carcinogenesis. Furthermore, MBRidge is a cost- and time-efficient method for profiling the DNA methylome. The ridge regression model required 9 and 1.4 Gb clean data from MB-seq and RRBS to reach satisfied accuracy for measuring a human DNA methylome, respectively (Wang et al., 2012) which cost a small fraction (∼10%) of MethylC-seq. Finally, a satisfied reproducibility in comparison 1 µg with 50 ng of input genomic DNA in MB-seq suggesting its potential for wide application.
MB-seq in our MBRidge has substantial advantages over previous methods as elucidated as the following. Although previous studies have utilized unmethylated or methylated adapters in methylome sequencing (Down et al., 2008; Meissner et al., 2008; Ball et al., 2009; Lister et al., 2009), these regular adapters were unsuitable for our MB-seq approach due to the following reasons: (i) if unmethylated DNA fragments were ligated with the regular methylated adapters in advance, they would be pulled down non-specifically in 5mC-antibody-enrichment step in MB-seq; (ii) cytosine in the regular unmethylated adapters would be converted to Us by sequential bisulfite treatment in MB-seq, resulting in failure in Illumina sequencing. In MB-seq, as an optimal strategy, C-hydroxylmethylated Illumina multiplexing adapters were firstly introduced since they cannot be enriched by 5mC antibodies, and 5hmCs in adapters can hardly be converted to Us in the bisulfite treatment (Huang et al., 2010; Jin et al., 2010). In contrast, recently developed method MeDIP-Bseq, has two additional steps, including random-primed PCR and restriction enzyme digestion. Furthermore, our MB-seq protocol can produce strand-specific library, resulting in generation of directional reads of either Bisulfite Watson (BSW) or Bisulfite Crick (BSC). MeDIP-Bseq protocol reported previously, however, performs two rounds of PCR, which resulted in yielding BSW and BSC reads, as well as their reverse complementary reads (BSWRC and BSCRC). The discrepancy between our MB-seq and MeDIP-Bseq has immediate impact on the bioinformatic analysis in terms of time-consumption and mapping accuracy. Consequently, searching complexity of bisulfite read mapping procedure in MeDIP-Bseq is double compared with that in MB-seq because the Watson and Crick strands of bisulfite-treated DNA sequences are not complementary to each other. Finally, benefiting from the bisulfite treatment in MB-seq, we determined methylation status of each CpG, and thereby significantly increased resolution and reduced false positives compared with MeDIP-Bseq.
The performance of ridge regression in our MBRidge can significantly improve with regard to resolution and accuracy compared with that by using other affinity-enrichment based methods and their corresponding bioinformatic tools. MEDME and BayMeth can transform the observed read counts of MBD-seq or MeDIP-seq data into the regional methylation levels with low resolution (∼100 bp) (Riebler et al., 2014). MEDIPS only obtains methylation scores (1–1000), rather than the methylation levels with single-base resolution (Chavez et al., 2010). These scores are not directly comparable with the methylation levels derived from other non-affinity-enrichment based methods. Batman obtains the methylation levels of each single CpG from MeDIP-seq data using long-term calibration (Bock, 2012), but the methylation levels are generally underestimated (Riebler et al., 2014). MethylCRF provides the methylation levels at single-CpG resolution on a genome-wide scale, but it exhibited a PCC value of 0.77 in comparison with MethylC-seq when using the H1 cell line, and the accuracy of this method may be further reduced when it is applied to aberrant DNA methylomes such as those from cancers (Stevens et al., 2013).
We found that accuracy of the DNA methylation levels derived from MBRidge will be unaffected by biases derived from variation in CpG densities and the methylation levels, suggesting that MBRidge is a promising method for detection of methylomes in species with low global level of DNA methylation. This is in stark contrast to MethylC-seq, whose accuracy is generally correlated with sequencing depth and is relatively uneconomic to profile methylome on species with the ultra-low global DNA methylation level. Recently, MeDIP-Bseq provided the first unequivocal evidence of cytosine methylation in Drosophila, which has long been thought to lack cytosine methylation (Takayama et al., 2014). Therefore, we prospect that MBRidge could be used to detect methylomes in species such as the silkworm (Xiang et al., 2010) and Locusta migratoria (Wang et al., 2014a), which have large genome size but low CpG methylation levels.
Despite the advantages presented in this study, MBRidge still has potential to be further improved. Firstly, by using bisulfite treatment alone, MBRidge cannot distinguish 5hmC from 5mC. Secondly, MBRidge is dependent on the actual methylation levels from RRBS for correcting the methylation level derived from MB-seq. Given the fact that NIH Roadmap Epigenomics Project's current release of the Human Epigenome Atlas deposited 108 RRBS data sets for 67 tissues or cell lines with only 5 of these samples being MethylC-seq data sets, by performing MB-seq on these samples, our new method, MBRidge could convert the data into single-CpG resolution, genome-wide methylomes, and thus significantly increase value of the existing datasets.
In case study, MBRidge was used to evaluate the DNA methylation levels in a normal human ovarian epithelial cell line, T29, and its oncogenic H-RasV12-mediated counterpart, T29H. We revealed that only a fraction of DMRs between T29 and T29H occur in CGI and promoter regions, while the remaining DMRs arise at other regions such as repeats and Alu elements. This indicated that MBRidge can detect genome-wide alteration of DNA methylation, thereby providing an interpretation of the regulatory mechanism involved in DMRs located proximally and distally to TSSs. In our study, even in the promoter region, alterations to DNA methylation enforced reverse regulation of gene expression depending on the distance from the proximal DMR to TSSs in local genes. This is similar to a previous observation in which specific histone modifications achieved proper regulatory function in correlation with their distance from TSSs (Chai et al., 2013). Although it is generally believed that DNA methylation in promoters can reversibly regulate gene expression in local genes, our results suggest this assumption need to be carefully interpreted in context with the position of DNA methylation in related to the distance to TSSs; therefore, it is worth to explore spatial regulatory function of DNA methylation in the future. Furthermore, by our analysis of DMR integrated with local gene expression, both DMRs-regulated protein-coding gene and lncRNA were enriched in several cellular pathways (including metabolism, calcium signaling, cancer related pathways, and cell cycles), which could serve to decipher the roles of DNA methylation to the sequential proteomic alteration and phenotype induced by oncogenic HRAS in T29H cell lines as previously reported (Liu et al., 2004; Young et al., 2005). In summary, MBRidge can provide an effective approach to obtaining single-CpG resolution and a complete DNA methylome. As a cost-efficient and accurate approach, we envisage MBRidge could be potentially utilized in large scale genome-wide DNA methylation research, such as studies of epigenomic plasticity within populations, clinical epigenomics, and ecological epigenomics.
Materials and methods
Hydroxylmethylated Illumina adapters
5-Hydroxymethyl-dC-CE phosphoramidite was purchased from Glen Research (Sterling), and hydroxylmethylated oligonucleotides were synthesized by Invitrogen. Both hydroxylmethylated adapter1 (sequence 5′-pho-GATCGGAAGAGCACACGTCT-3′) and adapter2 (sequence 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′), in which all the Cs were hydroxylmethylated, were annealed following the protocol previously described (Zheng et al., 2011).
Sample preparation
Human ovarian epithelial cell lines T29 and T29H were provided by Dr Jinsong Liu (MD Anderson Cancer Center, University of Texas) and cell lines were cultured as previously described (Young et al., 2005). We extracted genomic DNA from the cell lines by using a Qiagen DNeasy Blood & Tissue Kit (Qiagen), and 20 mg/ml RNase (Qiagen) was used to degrade any contaminated RNA in the DNA samples. We verified DNA integrity by agarose gel electrophoresis, and DNA was quantified using Quant-iT PicoGreen dsDNA Kits on a Qubit 2.0 fluorometer (Invitrogen).
Preparation of MB-seq library
From T29 cell line, various input amounts of genomic DNA (1 µg, 500 ng, 200 ng, and 50 ng) were fragmented to ∼100–500 bp using a Covaris E210 sonicator (Covaris Inc.). Fragmented DNA was end-repaired, A-tailed, and ligated to C-hydroxylmethylated Illumina multiplexing adapters following the standard protocol of TruSeq DNA Sample Prep kits (Illumina). Methylated DNA was pulled down using the method of Weber et al. (2005), but with some modifications. Briefly, adapter-ligated DNA was denatured at 95°C and then immunoprecipitated overnight at 4°C with 2 µl 5-MeC-mAb (MeDIP, Calbiochem) in a final volume of 100 µl IP buffer (comprising 20 mM sodium phosphate pH 7, 280 mM NaCl, and 0.1% Triton X-100). We washed 20 µl Dynabeads (M-280 Sheep anti-Mouse IgG, Invitrogen) with 1% PBS-BSA buffer according to the manufacturer's instructions, and added this to the MeDIP-DNA mixture using a slow rotation during incubation at 4°C for 2 h. The Dynabead-MeDIP-DNA mixture was then washed three times with 800 µl 1× IP buffer at 4°C, and for 15 min each time. MeDIP enriched DNA was recovered from Dynabead-MeDIP-DNA mixture by phenol-chloroform extraction followed by ethanol precipitation. To evaluate MeDIP recovery efficiency, we detected the resulting MeDIP-DNA by using qPCR with SYBR according to methods described in a previous study, and results are shown in Supplementary Table S1 (Li et al., 2010a). MeDIP-DNA was then bisulfite-treated using EpiTect Bisulfite Kit (Qiagen) with two rounds of the standard conversion. The bisulfite-treated product was amplified for 12 cycles with Illumina multiplexing PCR primer 1.0 and 2.0 (final concentration of 0.5 µM) in KAPA2G Robust HotStart ReadyMix (Kapa Biosystems, Inc., Woburn, MA, USA) by using large-scale amplification (8 × 25 µl) with 25 µl of reaction volume for every 4 µl of the bisulfite-treated template. Subsequently, the PCR products were purified by QIAquick PCR Purification Kit (Qiagen, Germany) and subjected to a final size-selection step on a 2% low melting agarose gel, and we excised ethidium bromide-stained gel slices containing fragments within the range of 270–370 bp. Excised DNA was once again purified with QIAquick and quantified by Quant-iT PicoGreen dsDNA Kits before it was sequenced using an Illumina Hiseq 2000 with a TruSeq SBS Kit v3-HS (200-cycles). The same protocol was used to construct an MB-seq library from the genomic DNA of T29H cells.
Ridge regression model
The ridge regression model was implemented using ridge regression, a type of regularized linear regression (Hoerl and Kennard, 2000). Ridge regression is ideal for data with several predictors that have non-zero coefficients and are drawn from a normal distribution (Friedman et al., 2010). Ridge regression performs particularly well when each of multiple predictors has small effect, and it prevents coefficients of linear regression models with many correlated variables from being poorly determined and exhibiting high variance. Ridge regression shrinks the coefficients of correlated predictors equally towards zero. For example, given k identical predictors, each would receive identical coefficients equal to 1/kth of the value that any single predictor would receive if fitted alone (Friedman et al., 2010). Thus, ridge regression does not force coefficients to vanish and cannot select a model containing only the most relevant and predictive subset of predictors. The ridge regression (2) estimator solves the regression problem in (1) using ℓ2 penalized least squares:
| (1) |
where is the vector of observed phenotypes, is a column vector of n ones and µ is a common intercept, X is a n × p matrix of markers, β is the vector of the regression coefficients of the markers, and is the vector of the residual errors with var
| (2) |
where is the ℓ2-norm (quadratic) loss function (i.e. residual sum of squares), is the ith row of X, is the ℓ2-norm penalty on β, and λ ≥ 0 is the tuning (penalty, regularization, or complexity) parameter that regulates the strength of the penalty (linear shrinkage) by determining the relative importance of the data-dependent empirical error and the penalty term. The larger the value of λ, the greater is the amount of shrinkage. As the value of λ is dependent on the data, it is determined using cross-validation, a data-driven method.
According to previous studies (Down et al., 2008; Pelizzola et al., 2008; Chavez et al., 2010; Nair et al., 2011; Riebler et al., 2014), most features related to methylation are approximately linear where the methylation levels are absolute, with the exception of genomic annotations, for which methylation displays distinct genomic feature-specific characteristics (Li et al., 2010b; Stevens et al., 2013). We introduced multiple features related to methylation as the predictors of ridge regression as follows: the methylation levels observed in MB-seq (MB level), mCpG numbers (MB mCG), read densities covering individual CpG (MB depth), the mean methylation levels flanking 100-bp region adjacent to local CpG observed in MB-seq (MB back level), mean number of mCpGs within windows of 200 bp centered on local CpG (MB back mCG), genomic CpG density, GC content, and CpG-OE value within windows of 200 bp centered on local CpG. We used an R package, glmnet, to implement the ridge regression algorithm, and cross-validation was used to estimate the model's prediction. Data (predictors and true methylation) were divided into training and testing datasets. Among ∼3 million CpGs (definite CpGs) with at least 10× coverage in RRBS, we randomly selected 50% of CpGs to be used as the training data; the remaining 50% became the testing data. The model was trained using the training data before being applied to the testing data and datasets of CpGs covered by MB-seq but uncovered by RRBS. We then calculated the PCC between the predicted methylation values and RRBS measured levels in the testing data. This procedure was repeated 1000 times and the mean PCC was computed to represent the prediction accuracy of the model. For each genomic feature, we trained and tested ridge regression separately. For CpGs that were annotated with multiple features, we combined the methylation predictions by averaging the corresponding ridge predictions and giving each prediction equal precedence. The methylation levels were predicted on a genome-wide scale and the methylation levels of definite CpGs were inherited from RRBS. Specifically, all CpGs uncovered by MB-seq were assumed to be unmethylated (i.e. with a methylation level of zero) and were excluded from calibration because, with sufficient sequencing depth, MeDIP-based methods are sensitive even to very low methylation (see Supplementary Figure S3A). Predicted methylation levels that were <0 and >1 were forced to 0 and 1, respectively, based on the principles of ridge regression (representing background noise and over-saturation of the sequencing derived antibody enrichment, respectively). In T29 and T29H, outliers such as these accounted for only ∼0.55% and ∼0.71% of all CpGs, respectively. The complete ridge regression model is illustrated in Supplementary Figure S3D.
System evaluation of MB-seq and MBRidge
We performed MethylC-seq and MeDIP-seq for T29 cell lines, and systemly compared the performance of our method to MethylC-seq, RRBS and MeDIP-seq at genome-wide and different genomic features. For additional details, see the Supplementary material.
Integrated analysis of DNA methylome and transcriptome
Integrated analysis and validation of differential DNA methylome and differential transcriptome were performed between T29 and T29H cell lines. For additional details, see the Supplementary material.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by grants from the National High Technology Research and Development Program of China (2012AA02A201, 2012AA02A202), China-Canada Collaboration Project from the Ministry of Science and Technology of China (2011DFA30670), the National Natural Science Foundation of China (31171236/C060503), and the AstraZeneca Innovation Centre China.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank Huiqian Chen (Institute of Genomic Medicine, Wenzhou Medical University) for language editing of the manuscript. We thank Qiang Huang and Ling-Yun Wu (Academy of Mathematics and Systems Science, Chinese Academy of Sciences) for scientific suggestions on bioinformatics analysis.
References
- Ball M.P., Li J.B., Gao Y., et al. (2009). Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S.B., Jones P.A. (2011). A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer 11, 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S. (2010). Taking the measure of the methylome. Nat. Biotechnol. 28, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. (1998). The host defence function of genomic methylation patterns. Novartis Found. Symp. 214, 187–195; discussion 195–199, 228–232. [DOI] [PubMed] [Google Scholar]
- Bock C. (2012). Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 13, 705–719. [DOI] [PubMed] [Google Scholar]
- Bock C., Tomazou E.M., Brinkman A.B., et al. (2010). Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 28, 1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenet F., Moh M., Funk P., et al. (2011). DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6, e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai X., Nagarajan S., Kim K., et al. (2013). Regulation of the boundaries of accessible chromatin. PLoS Genet. 9, e1003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L., Jozefczuk J., Grimm C., et al. (2010). Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 20, 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B.C., Houseman E.A., Marsit C.J., et al. (2009). Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 5, e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., et al. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T.A., Rakyan V.K., Turner D.J., et al. (2008). A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 26, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., Hastie T., Tibshirani R. (2010). Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Harris R.A., Wang T., Coarfa C., et al. (2010). Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 28, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerl A.E., Kennard R.W. (2000). Ridge regression: biased estimation for nonorthogonal problems. Technometrics 42, 80–86. [Google Scholar]
- Huang Y., Pastor W.A., Shen Y., et al. (2010). The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Ladd-Acosta C., Wen B., et al. (2009). The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhang J., Wang J.J., et al. (2013). Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.G., Kadam S., Pfeifer G.P. (2010). Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 38, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. [DOI] [PubMed] [Google Scholar]
- Karnoub A.E., Weinberg R.A. (2008). Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek S., Renz D., Doerfler W. (1993). DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 12, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunde-Ramamoorthy G., Coarfa C., Laritsky E., et al. (2014). Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing. Nucleic Acids Res. 42, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird P.W. (2010). Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203. [DOI] [PubMed] [Google Scholar]
- Lan X., Adams C., Landers M., et al. (2011). High resolution detection and analysis of CpG dinucleotides methylation using MBD-Seq technology. PLoS One 6, e22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Ye M., Li Y., et al. (2010a). Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 52, 203–212. [DOI] [PubMed] [Google Scholar]
- Li Y.R., Zhu J.D., Tian G., et al. (2010b). The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 8, e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Ecker J.R. (2009). Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 19, 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Dowen R.H., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Mukamel E.A., Nery J.R., et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang G., Thompson-Lanza J.A., et al. (2004). A genetically defined model for human ovarian cancer. Cancer Res. 64, 1655–1663. [DOI] [PubMed] [Google Scholar]
- Maunakea A.K., Nagarajan R.P., Bilenky M., et al. (2010). Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., et al. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T.S., Gu H., et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S.S., Coolen M.W., Stirzaker C., et al. (2011). Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 6, 34–44. [DOI] [PubMed] [Google Scholar]
- Nichol K., Pearson C.E. (2002). CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res. 12, 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelizzola M., Koga Y., Urban A.E., et al. (2008). MEDME: An experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 18, 1652–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning K.R., Smith K.M., Freitag M. (2009). Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47, 142–150. [DOI] [PubMed] [Google Scholar]
- Riebler A., Menigatti M., Song J.Z., et al. (2014). BayMeth: improved DNA methylation quantification for affinity capture sequencing data using a flexible Bayesian approach. Genome Biol. 15, R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D. (2005). DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610. [DOI] [PubMed] [Google Scholar]
- Robertson K.D., Wolffe A.P. (2000). DNA methylation in health and disease. Nat. Rev. Genet. 1, 11–19. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sati S., Ghosh S., Jain V., et al. (2012). Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 40, 10018–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee J.S., Schubeler D., Ng H.H. (2010). Tackling the epigenome: challenges and opportunities for collaboration. Nat. Biotechnol. 28, 1039–1044. [DOI] [PubMed] [Google Scholar]
- Schmid C.W. (1998). Does SINE evolution preclude Alu function? Nucleic Acids Res. 26, 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D., Lee B.H., Ting A.H. (2010). MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 38, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M., Cheng J.B., Li D.F., et al. (2013). Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 23, 1541–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Dhahbi J., Roberts A., et al. (2014). Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 24, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sun J., Wu H., et al. (2012). Systematic assessment of reduced representation bisulfite sequencing to human blood samples: a promising method for large-sample-scale epigenomic studies. J. Biotechnol. 157, 1–6. [DOI] [PubMed] [Google Scholar]
- Wang T., Liu Q., Li X.F., et al. (2013). RRBS-analyser: a comprehensive web server for reduced representation bisulfite sequencing data analysis. Hum. Mutat. 34, 1606–1610. [DOI] [PubMed] [Google Scholar]
- Wang X., Fang X., Yang P., et al. (2014a). The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 5, 2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li G.L., Mao F.B., et al. (2014b). Ras-induced epigenetic inactivation of the RRAD (Ras-related associated with diabetes) gene promotes glucose uptake in a human ovarian cancer model. J. Biol. Chem. 289, 14225–14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., et al. (2005). Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862. [DOI] [PubMed] [Google Scholar]
- Weisenberger D.J. (2014). Characterizing DNA methylation alterations from The Cancer Genome Atlas. J. Clin. Invest. 124, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.M., Cabanski C.R., Silva-Fisher J.M., et al. (2014). Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 15, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H., Zhu J.D., Chen Q.A., et al. (2010). Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 28, 516–520. [DOI] [PubMed] [Google Scholar]
- Xie W., Barr C.L., Kim A., et al. (2012). Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C.Y., Yuan J., Li H., et al. (2014). NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 42, D98–D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T., Mei F., Liu J., et al. (2005). Proteomics analysis of H-RAS-mediated oncogenic transformation in a genetically defined human ovarian cancer model. Oncogene 24, 6174–6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Advani A., Melefors O., et al. (2011). Titration-free 454 sequencing using Y adapters. Nat. Protoc. 6, 1367–1376. [DOI] [PubMed] [Google Scholar]
- Ziller M.J., Gu H., Muller F., et al. (2013). Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.