The cardiogenic potential of cardiac c-Kit+ cells through class I histone deacetylase (HDAC) inhibition was augmented and their therapeutic potency in the chronic heart failure (CHF) animal model evaluated. The low cell engraftment suggests that paracrine effects might account for the beneficial effects of c-Kit+ cells in CHF. Selective inhibition of class I HDACs induced expression of cardiac markers in c-Kit+ cells and partially augmented their efficacy for CHF repair.
Keywords: Stem cell transplantation, Epigenetic process, Myocardial infarct, Congestive heart failure
Abstract
Cardiac c-Kit+ cells have a modest cardiogenic potential that could limit their efficacy in heart disease treatment. The present study was designed to augment the cardiogenic potential of cardiac c-Kit+ cells through class I histone deacetylase (HDAC) inhibition and evaluate their therapeutic potency in the chronic heart failure (CHF) animal model. Myocardial infarction (MI) was created by coronary artery occlusion in rats. c-Kit+ cells were treated with mocetinostat (MOCE), a specific class I HDAC inhibitor. At 3 weeks after MI, CHF animals were retrogradely infused with untreated (control) or MOCE-treated c-Kit+ cells (MOCE/c-Kit+ cells) and evaluated at 3 weeks after cell infusion. We found that class I HDAC inhibition in c-Kit+ cells elevated the level of acetylated histone H3 (AcH3) and increased AcH3 levels in the promoter regions of pluripotent and cardiac-specific genes. Epigenetic changes were accompanied by increased expression of cardiac-specific markers. Transplantation of CHF rats with either control or MOCE/c-Kit+ cells resulted in an improvement in cardiac function, retardation of CHF remodeling made evident by increased vascularization and scar size, and cardiomyocyte hypertrophy reduction. Compared with CHF infused with control cells, infusion of MOCE/c-Kit+ cells resulted in a further reduction in left ventricle end-diastolic pressure and total collagen and an increase in interleukin-6 expression. The low engraftment of infused cells suggests that paracrine effects might account for the beneficial effects of c-Kit+ cells in CHF. In conclusion, selective inhibition of class I HDACs induced expression of cardiac markers in c-Kit+ cells and partially augmented the efficacy of these cells for CHF repair.
Significance
The study has shown that selective class 1 histone deacetylase inhibition is sufficient to redirect c-Kit+ cells toward a cardiac fate. Epigenetically modified c-Kit+ cells improved contractile function and retarded remodeling of the congestive heart failure heart. This study provides new insights into the efficacy of cardiac c-Kit+ cells in the ischemic heart failure model.
Introduction
Congestive heart failure (CHF) after myocardial infarction (MI) is a major public health issue worldwide. CHF therapy remains a challenging task: nearly 50% of people diagnosed with CHF die within 5 years. Over the past few years, stem cell transplantation has risen as a new therapeutic strategy for treating ischemic cardiac disease [1, 2]. Among the tested cell types, cardiac c-Kit+ progenitor cells have shown potential as a therapeutic agent [3, 4]. Notwithstanding the preclinical use of cardiac c-Kit+ cell therapy, the low cardiogenic potential of the transplanted cells could present a major obstacle for treatment, with cell transplantation resulting in limited repair and moderate cardiac function improvement. Therefore, a new strategy is needed to improve the cardioprotective potential of these cells.
Recently, epigenetic manipulations have risen as a new molecular tool for cell reprogramming. A number of studies have revealed that chromatin organization influences cell phenotype by regulating the expression of specific genes such as transcription factors. Epigenetic mechanisms were implicated in the regulation of self-renewal, pluripotency, and differentiation of stem cells [5–9]. Thus, the differentiation of embryonic stem cells (ESCs) into cardiomyocytes is accompanied by a programmed temporal alteration in chromatin structure [10]. In addition, dynamic and coordinated epigenetic regulations were observed during differentiation of ESCs to mesodermal cells, cardiac progenitor cells, and, finally, cardiomyocytes [11].
Chromatin remodeling can be induced by several mechanisms, including histone modifications such as methylation and acetylation [12]. Histone deacetylases (HDACs) catalyze the deacetylation of α-acetyl lysine that resides within the NH2-terminal tail of core histones. Removing acetyl groups from histones typically causes condensation of chromatin and a decrease in gene expression [13, 14]. HDACs are a family of 18 molecules grouped into 4 classes. Among them, class I comprises 4 HDAC family members, HDAC1, -2, -3, and -8, which are expressed ubiquitously and display high enzyme activity toward histone substrate [15]. Class I HDACs were found to interact with lineage-specific transcription factors, suggesting that these enzymes might play a role in controlling specific transcriptional programs [16]. Histone deacetylation inhibition results in hyperacetylation and changes in chromatin density that, in turn, regulates gene expression in a cell-specific manner and induces phenotype changes, including cell reprogramming [17]. Thus, pan class I and II HDAC inhibitors such as trichostatin A (TSA) and valproic acid (VPA) were shown to redirect bone marrow-derived mesenchymal cells toward a cardiac fate [18, 19].
HDACs are the enzymes of a ubiquitous nature. They regulate a broad spectrum of cellular mechanisms, including differentiation, cell cycle, DNA repair, gene expression, oxidative stress, and autophagy [20–22]. The administration of compounds that nonspecifically inhibit multiple HDAC isoforms (pan-HDAC inhibitors) induces global changes in multiple cellular processes, including the needless disturbance of normal physiological functions. Although promising in clinical tests, these compounds have exhibited significant toxicities that might limit their potential. It might be possible to reduce some of the toxicity by targeting individual HDAC isoforms that are critically involved in particular processes [23].
We tested the effects of mocetinostat (MOCE), a highly specific inhibitor of class I HDACs, on cardiac explant-derived c-Kit+ cells. Moreover, we evaluated the beneficial effects of epigenetically modified c-Kit+ cells in heart failure treatment.
Materials and Methods
An expanded materials and methods section is available in the supplemental online data.
Animals
Two-month-old Sprague-Dawley male rats were used. The animal studies were performed in accordance with federal regulations and international accreditation standards (Association for Assessment and Accreditation of Laboratory Animal Care International no. 1115) were overseen by the Banner Sun Health Research Institute institutional animal care and use committee (protocol no. 12-01) operating under the “Guide for Care and Use of Laboratory Animals.” Humane animal care was performed in compliance with the “Guide for the Care and Use of Laboratory Animals.” The rats were assigned randomly to 4 groups: (a) CHF animals retrograde coronary vein (RCV) were infused with cell-free, serum-free Dulbecco’s modified Eagle’s medium (vehicle), n = 8; (b) CHF animals were RCV infused with untreated c-Kit+ cells (CHF/c-Kit), n = 8; (c) CHF animals were RCV infused with epigenetically modified c-Kit+ cells (CHF/MOCE-c-Kit), n = 8; and (d) sham-operated rats, n = 8. The group sample sizes were calculated according to 80% statistical power, a significance level of 0.05, and change in left ventricular end-diastolic pressure (LVEDP) >40%.
Myocardial Infarction
MI was created by ligation of the left coronary artery (LAD), as described previously by our laboratory [24]. The rats were anesthetized using a cocktail of ketamine, xylazine, and acepromazine (50 mg/kg, 15 mg/kg, and 2 mg/kg, respectively). The animals were prepared using aseptic methods, intubated, and ventilated before undergoing left thoracotomy to expose the heart. The heart was expressed, and the LAD coronary artery was ligated using a 5-0 TiCron suture (Covidien, Jersey City, NJ, http://www.covidien.com) per standard protocols. The lungs were briefly hyperinflated, the chest was closed using 2-0 silk suture, and the rodents were allowed to recover with a pain management regimen of buprenorphine. The sham-operated animals underwent the same surgical procedure, excluding LAD occlusion, and were allowed to recover with a pain management regiment of buprenorphine. The rats were given a 1.1-mg/kg dose of 72-hour buprenorphine SR Lab (ZooPharm, Boulder, CO, http://www.wildpharm.com) for pain management and a 2.0-ml dose of lactated Ringers solution for supplemental hydration after recovery from anesthesia. After the recovery period, the animals were monitored twice daily.
RCV Infusion of c-Kit+ Cells
RCV c-Kit+ cell infusion was conducted as previously described by our laboratory [25]. Twenty-one days after the initial MI surgery, the rats were randomly assigned to cell- or vehicle-infused groups. Before cell delivery, scar presence was confirmed visually. The right external jugular was cannulated using a polyethylene-25 catheter, which was then advanced into the right atrium. One million green fluorescent protein (GFP)-labeled c-Kit+ cells were suspended in 400 µl of vehicle (cell-free, serum-free medium) and infused for 30–60 seconds to the right atrium, while simultaneously and temporarily occluding the pulmonary artery and inferior and superior venae cavae. This same procedure was used to infuse 400 µl of vehicle to the control CHF group.
Explant Culture and c-Kit+ Cell Isolation
c-Kit+ cells were isolated from cardiac explants generated from 2-month-old Sprague-Dawley male rats. Cardiac explant outgrowth was generated, as previously described [24]. After 21 days in culture, c-Kit+ cells were separated from the cell outgrowth using magnetic beads (Miltenyi Biotec, Carlsbad, CA, http://www.miltenyibiotec.com) and cultured as described (supplemental online Fig. 1A, 1B) [25]. The purity of c-Kit+ cell population was confirmed by flow cytometry; approximately 90% of the cells were positive for a c-Kit marker after sorting as made evident by fluorescence-activated cell sorting screening (supplemental online Fig. 1C). For in vivo experiments, c-Kit+ cells were labeled with GFP lentivirus vector (Clontech Laboratories, Inc., Mountain View, CA, http://www.clontech.com). GFP expression was verified by fluorescent microscopy; the efficiency of GFP expression in c-Kit+ cells before transplantation was >90% (supplemental online Fig. 1D).
Cardiac Function Measurement
Hemodynamic parameters were recorded at 3 weeks after RCV using an in vivo closed chest pressure-volume catheter (Millar Instruments, Houston, TX, http://www.millar.com) [24]. The data were analyzed using PVAN, version 3.6, software (Millar Instruments). To ensure the accuracy of the cardiac function measurements, the conductance catheter was calibrated before each data set acquisition, as previously described [26].
RNA Isolation and Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction
RNA extraction and quantitative reverse transcription-polymerase chain reaction (RT-PCR) were conducted as described previously [24]. Specific primers were synthesized by Life Technologies (Carlsbad, CA, http://www.lifetechnologies.com; supplemental online Table 1). CYPA was used as a reference gene. Data analysis was performed using StepOne software, version 2.1 (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com), and the comparative Ct (ΔΔCt) quantitative method.
Flow Cytometry
Flow cytometry was conducted as described previously [24]. Cell events were detected using a FACS Calibur flow cytometer equipped with an argon laser (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com). Data were analyzed using CellQuest software (BD Biosciences).
HDAC Activity
HDAC activity was measured in c-Kit+ cells treated with 2 µM MOCE for 7 days using the colorimetric HDAC Assay Kit (Upstate, Charlottesville, VA) according to the manufacturer’s instructions. Untreated c-Kit+ cells were used as a control.
Western Blot
Western blot analysis was performed as described previously [24]. The band intensity was determined using FluorChem 8900 software (Alpha Innotech Corp.).
Chromatin Immunoprecipitation Assay
Immunoprecipitation of chromatin was performed using the EZ-ChIP kit (EMD Millipore, Billerica, MA, http://www.emdmillipore.com) according to the manufacturer’s instructions. c-Kit+ cells were treated with 2 µM MOCE for 7 days. Untreated cells were used as a control; 1 × 106 cells were cross-linked using 1% formaldehyde for 10 minutes at room temperature and lysed in SDS lysis buffer containing protease inhibitor cocktail (kit components). DNA was sheared by sonication to lengths between 200 and 1,000 base pairs. Chromatin was precipitated by overnight incubation at 4°C with anti-acetylated histone H3 primary antibody (AcH3K9; EMD Millipore) followed by pelleting with antibody-histone complex using Protein G Agarose (Life Technologies). Normal rabbit IgG was used instead of primary antibody as a negative control. The histone complex was eluted from Protein G Agarose, and cross-linking was reversed by incubating histones at 65°C for 4 hours. DNA was recovered and subjected to quantitative PCR analysis with primers specific to promoter regions of rat Nanog, Sox2, Nkx2.5, GATA4, and Myh6 genes (supplemental online Table 1). The relative DNA levels were normalized to the corresponding input DNA levels.
Collagen Assay
The total collagen amount was measured in cross-sections using Sirius Red Fast Green Collagen Staining Kit (Chondrex, Redmond, WA, http://www.chondrex.com). Absorbance of collagen (540 nm) and noncollagenous (605 nm) protein was assessed using the BioTek Synergy HT Microplate Reader (Winooski, VT, http://www.biotek.com). The collagen percentage was calculated as a ratio of optical density 540 (OD540) collagen to OD605 noncollagenous protein.
Imaging
Fluorescent images were captured using the Leica TCS SPE confocal system configured with the Leica DM 2500 microscope (Leica Microsystems, Heerbrugg, Switzerland, http://www.leica.com). Excitation maximums of 488 nm, 532 nm, and 405 nm were used for image acquisition. The images were processed using LAS AF software (Leica Microsystems). Nonfluorescent images were captured using an Olympus IX-51 microscope equipped with a DP72 device camera and processed using DP2-BSW software (Olympus Corp., Tokyo, Japan, http://www.olympus-global.com).
Statistical Analysis
The results are reported as the mean ± SE. The Mann-Whitney rank sum test was used to compare the differences between untreated and MOCE-treated c-Kit+ cells or the differences between animal groups. Statistical analysis was conducted using SigmaStat, version 3.5, software (Systat Software, Inc., San Jose, CA, http://www.sigmaplot.com). A value of p < .05 was considered statistically significant.
Results
Cardiac Explant-Derived c-Kit+ Cells Express Class I HDACs
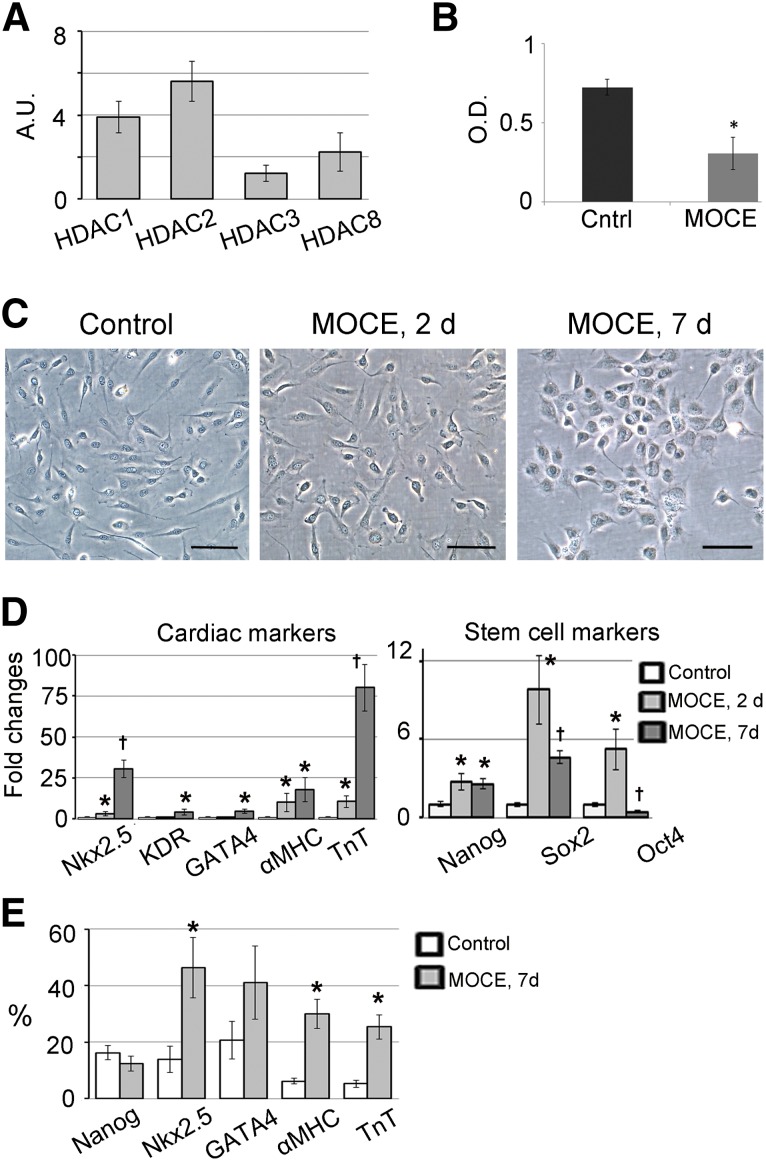
Adult cardiac c-Kit+ cells were isolated from total cardiac explant-derived cells, expanded, and characterized, as previously described by our laboratory [24]. Previous studies have demonstrated that epigenetically active small molecules, such as pan-HDAC inhibitors, might facilitate the redirection of adult cellular functions [18, 19]. We used a selective class I HDAC inhibitor to increase the c-Kit+ cells’ plasticity and redirect them toward a cardiac fate. First, we showed that unmodified (control) c-Kit+ cells expressed class I HDAC genes, including HDAC1, -2, -3, and -8 genes (Fig. 1A). To suppress class I HDAC activity, c-Kit+ cells were treated with the selective class I HDAC inhibitor MOCE. Administration of 2 µM MOCE for 7 days resulted in an approximately 60% reduction of HDAC activity in c-Kit+ cells (Fig. 1B).
Figure 1.
Class I HDAC inhibition stimulates cardiac marker expression in c-Kit+ cells. c-Kit+ cells were treated with 2 µM MOCE for 2 or 7 days as indicated. (A): Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis demonstrated expression of HDAC1, -2, -3, and -8 genes in c-Kit+ cells. Arbitrary units were calculated as [2(−∆Ct)] × 100. ∆Ct was calculated as Cttarget gene – Cthousekeeping gene. CYPA was used as a housekeeping gene; n = 5 per group. (B): MOCE decreased HDAC activity in c-Kit+ cells. The activity was measured after 7 days of MOCE administration; n = 5; ∗, p < .05 compared with control. (C): Phase-contrast images of untreated and MOCE/c-Kit+ cells. Scale bars = 50 µm. (D): qRT-PCR analysis of stem- and cardiac-related gene expression in untreated (control) and MOCE/c-Kit+ cells (MOCE/c-Kit+ cells); n = 5; ∗, p < .05 compared with control; †, p < .05 compared with MOCE, 2 days. (E): Fluorescence-activated cell sorting analysis of MOCE/c-Kit+ cells; n = 5; ∗, p < .05 compared with control. Abbreviations: αMHC, α-myosin heavy chain; A.U., arbitrary units; Cntrl, control; d, day; HDAC, histone deacetylase; MOCE, mocetinostat; O.D., optical density units; TnT, troponin T.
Class I HDAC Inhibition Stimulates Cardiac Marker Expression in c-Kit+ Cells
To characterize the effects of class I HDAC inhibition on c-Kit+ cell phenotype, cells were treated with MOCE for 2 or 7 days. Untreated c-Kit+ cells exhibit the elongated spindle shape morphology typical for myogenic cells. No significant changes in cell morphology were observed after 2 days of MOCE treatment; however, after 7 days, the cells appeared to increase in size and round up (Fig. 1C). Quantitative RT-PCR analysis showed that MOCE resulted in upregulation of the genes associated with pluripotency, including Nanog (2.7-fold), Sox2 (9.8-fold), and Oct4 (5.2-fold) after 2 days of treatment. No additional upregulation was detected for the Nanog gene at 7 days (2.7-fold compared with day 0), and Sox2 and Oct4 gene expression had significantly decreased at 7 days compared with that at 2 days (4.6-fold and 0.4-fold, respectively, compared with day 0; Fig. 1D). In contrast, we observed a gradual increase in the expression levels of cardiac transcription factors Nkx2.5, KDR, and GATA4 at 7 days (30.5-fold, 4-fold, and 4.5-fold, respectively, compared with day 0) and cardiac structural genes, α-myosin heavy chain (αMHC) and cardiac troponin T (TnT; 17.8-fold and 80-fold, respectively, at 7 days compared with day 0), after MOCE treatment (Fig. 1D).
The gene expression data for cardiac-related genes were confirmed by flow cytometry analysis. After 7 days of MOCE treatment, flow cytometry analysis demonstrated an increase in the number of Nkx2.5+ cells (from 14% ± 5% to 46% ± 11%), αMHC+ cells (from 5% ± 1% to 25% ± 4%), and TnT+ cells (from 6% ± 1% to 30% ± 5%; Fig. 1E; supplemental online Fig. 3). In addition, the number of GATA4+ cells tended to increase (from 21% ± 6% to 41% ± 13%). Despite upregulation of gene expression, no differences were found in the number of Nanog+ cells (Fig. 1E).
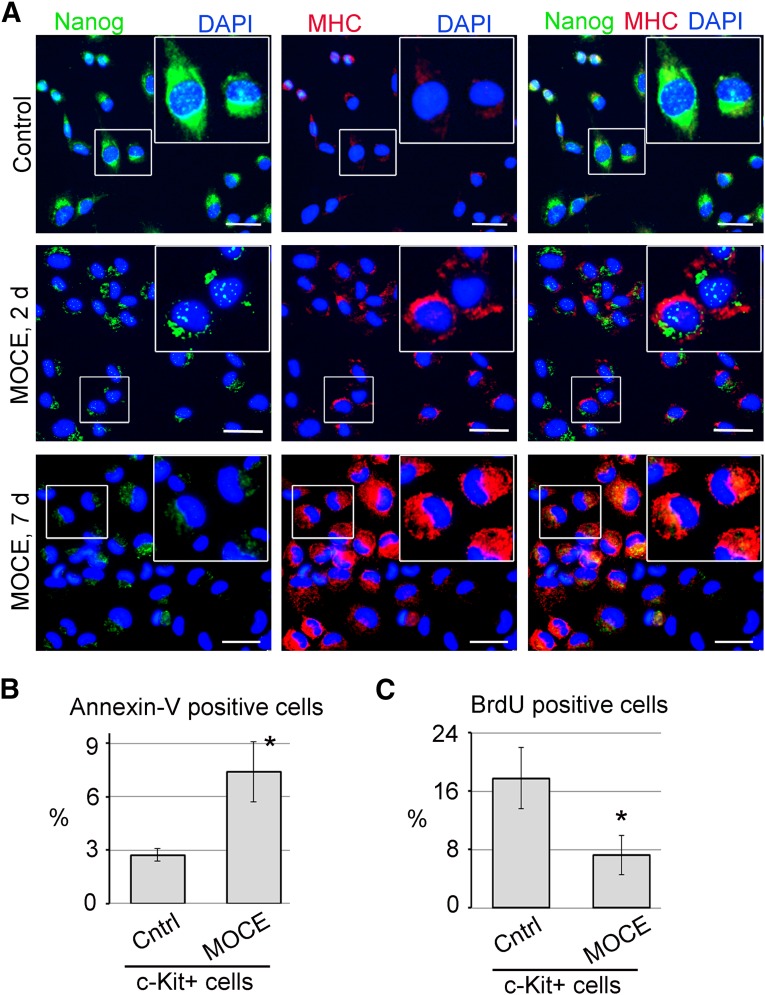
Immunofluorescence staining revealed a marked increase in αMHC expression in MOCE/c-Kit+ cells at day 2, which was more significant at day 7 compared with the untreated controls (Fig. 2A). No well-developed sarcomeric structures were observed in MOCE/c-Kit+ cells, indicating the immature state of these cells. Immunostaining with Nanog demonstrated dual nuclear and cytoplasmic localization of this marker in untreated c-Kit+ cells and after 2 days of MOCE treatment, indicating an early differentiation state [27]. In contrast, after 7 days of MOCE treatment, we observed mostly cytoplasmic Nanog localization potentially suggestive of its inactive form [27] (Fig. 2A).
Figure 2.
Effects of class I histone deacetylase inhibition on c-Kit+ cells. (A): c-Kit+ cells were treated with 2 µM MOCE for 2 or 7 days, as indicated. The cells were double-labeled with anti-Nanog (green) and anti-αMHC (red) antibodies. Nuclei were stained with DAPI (blue). The upper right section of each panel shows the enlargement of the box-selected area. Scale bars = 20 µm. (B, C): Apoptosis and cell proliferation were measured in c-Kit+ cells after 7 days of MOCE treatment. Untreated c-Kit+ cells were used as a control; n = 5; ∗, p < .05 compared with control. (B): Apoptotic cells were labeled with fluorescein isothiocyanate (FITC)-conjugated annexin V and analyzed by fluorescence-activated cell sorting. Necrotic cells were excluded by propidium iodide (PI) staining. The level of apoptosis was calculated as a percentage of annexin V-positive/PI-negative cells. (C): Proliferating cells were identified by BrdU incorporation, followed by FITC-conjugated anti-BrdU labeling. Cell proliferation was calculated as a percentage of BrdU-positive cells. Abbreviations: BrdU, bromodeoxyuridine; Cntrl, control; d, day; DAPI, 4′,6-diamidino-2-phenylindole; MHC, myosin heavy chain; MOCE, mocetinostat.
Subsequently, we found that cardiac marker induction on MOCE treatment is accompanied by an elevation in cell apoptosis level from 2.7% ± 0.4% in untreated cells to 7.4% ± 1.7% in MOCE/c-Kit+ cells (Fig. 2B; supplemental Fig. 2A). In addition, we observed a decrease in a number of bromodeoxyuridine-positive (BrdU+) proliferating cells from 17.8% ± 4% to 7.2% ± 2.7% for untreated versus MOCE-treated c-Kit+ cells (Fig. 2C; supplemental Fig. 2B). These results indicate that inhibition of class I HDAC with MOCE stimulate expression of cardiac-specific markers in c-Kit+ cells.
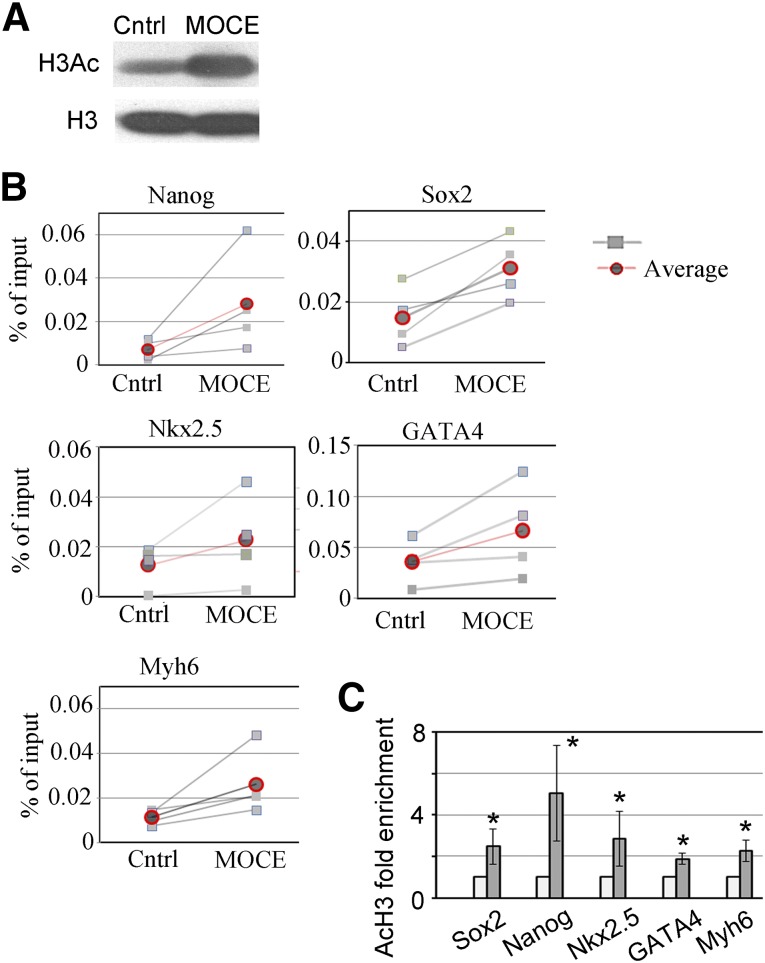
Class I HDAC Inhibition Enhanced Histone H3 Acetylation at Promoter Regions of Pluripotency and Cardiac-Specific Genes
To examine the epigenetic changes associated with those HDACs’ inhibition, we measured the acetylation status of histone H3, a modification associated with an open chromatin structure [28], in control and MOCE-treated cells (MOCE/c-Kit+ cells). A global increase in the acetylated histone H3 level was observed in c-Kit+ cells treated with MOCE compared with untreated control (Fig. 3A). Next, we tested whether MOCE treatment alters the acetylation status of histone H3 in promoter regions of cardiac- and stem cell-specific genes. A chromatin immunoprecipitation assay was conducted using antibody specific to acetylated histone H3 (H3AcK9) and analyzed with primers to promoter regions of cardiomyocyte-specific genes, Nkx2.5, GATA4, and myosin heavy chain (Myh6). We also analyzed the histone H3 acetylation status of Nanog and Sox2 gene promoters, because both are stem cell markers. We found that the H3Ac association with promoter regions of the studied cardiac- and stem cell-specific genes significantly increased in c-Kit+ cells with MOCE administration (Fig. 3B, 3C), suggesting that histone H3 acetylation might account for the observed upregulation in expression of the indicated cardiac and stem cell genes (Fig. 1D).
Figure 3.
Effects of class I histone deacetylase (HDAC) inhibition on epigenetic landscape of c-Kit+ cells. (A): Western blot analysis showed upregulation in H3Ac protein level in MOCE/c-Kit+ cells compared with control. The total level of histone H3 remained the same in MOCE/c-Kit+ cells. (B, C): Effects of HDAC inhibition on acetylation status of histone H3 in promoter regions of stem cell- and cardiac-specific genes. (C): Summary of chromatin immunoprecipitation assay; n = 5; ∗, p < .05 compared with control. Abbreviations: Cntrl, control; H3, histone H3; H3Ac, acetylated histone H3; MOCE, mocetinostat.
Retrograde Infusion of Epigenetically Modified c-Kit+ Cells Improves Cardiac Function of CHF Rats
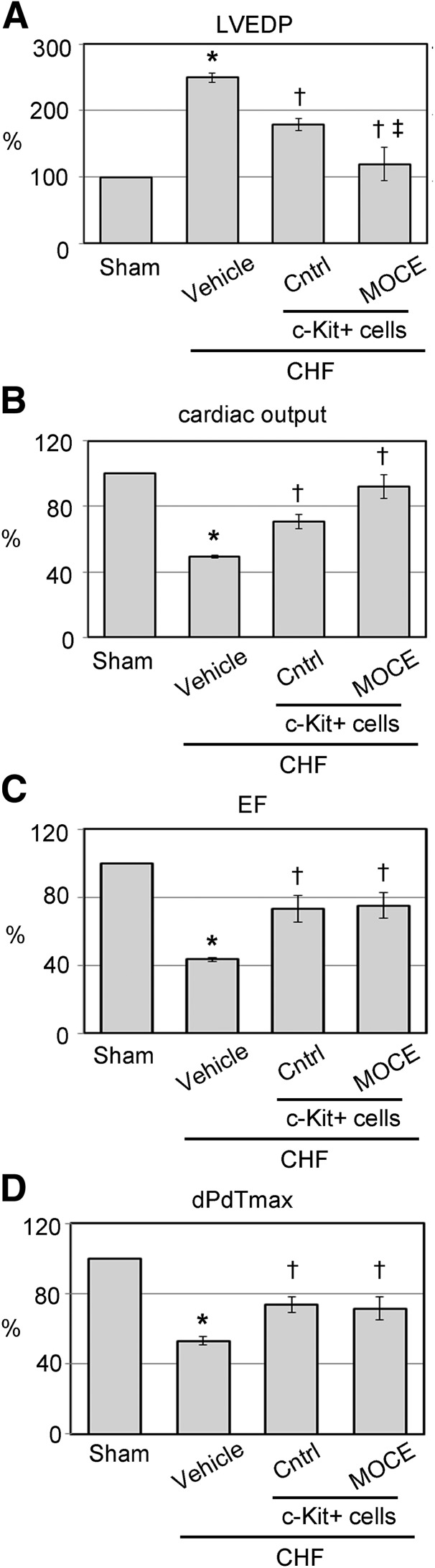
To compare the regenerative potential of control and epigenetically modified c-Kit+ cells in vivo, MIs were created by LAD occlusion [25]. Three weeks after MI surgery, the rats that had developed CHF were selected for cell transplantation. The CHF rats were selected from the pool of infarcted animals according to a scar size >30% [24, 29]. Approximately 35% of the infarcted rats were classified as CHF and used in the subsequent experiments. Non-CHF rats were excluded from the present study. One million of control or MOCE/c-Kit+ cells were delivered to the hearts by RCV infusion [25]. For tracking purposes, the cells were transfected with GFP-expressing lentiviral vector before transplantation [25]. Cardiac function and histological tissue evaluations were conducted at 3 weeks after RCV infusion. At 3 weeks after MI, we observed a significant loss of cardiac function in the CHF group compared with the sham rats. Compared with the sham rats, the CHF rats’ LVEDP had increased from 6.8 ± 2.5 mmHg to 29.4 ± 5.9 mmHg, the ejection fraction (EF) had decreased from 61.6% ± 11.3% to 31.8% ± 7.7%, and the peak rate of pressure increase (dPdt/max) had decreased to 3,923% ± 156 mmHg/sec from 7,390 ± 177 mmHg/sec (Fig. 4; supplemental online Table 2), confirming heart failure [30]. Transplantation of the CHF rats with either control or MOCE/c-Kit+ cells resulted in significant improvement in cardiac function, including LVEDP, EF, cardiac output, and dPdt/max (Fig. 4; supplemental online Table 2). Next, we compared the effects of control and MOCE/c-Kit+ cells on the cardiac function of the CHF rats. The rats infused with MOCE/c-Kit+ cells exhibited a 33% larger reduction in LVEDP and 30% higher increase in cardiac output compared with the rats infused with control cells (LVEDP, 12.2 ± 1.8 mmHg vs. 8.6 ± 0.7 mmHg; cardiac output, 30 ± 2 ml/min vs. 39 ± 3 ml/min for control vs. MOCE/c-Kit+ cells, respectively). For the other cardiac parameters, smaller differences were observed; rats transplanted with MOCE/c-Kit+ cells showed a 3% higher increase in EF and dPdt/max compared with rats transplanted with control cells (EF, 47% ± 2% vs. 48% ± 2%; dPdt/max, 5,440 ± 318 vs. 5,267 ± 480 mmHg/sec for control vs. MOCE/c-Kit+ cells, respectively; Fig. 4; supplemental online Table 2).
Figure 4.
Retrograde perfusion of epigenetically modified c-Kit+ cells improves cardiac function of CHF rats. Key hemodynamic parameters of CHF hearts transplanted with untreated (control) or MOCE/c-Kit+ cells. Hemodynamic parameters are presented as the percentage of change of the cardiac parameter in the test group compared with that in the sham group (assuming sham cardiac function is 100%). Percentage of change was calculated as (PTest group – Psham)/Psham × 100%: LVEDP (A); cardiac output (B); EF (C); dPdt/max (D). Data are presented as mean ± SE; n = 8; ∗, p < .05 compared with sham; †, p < .05 compared with vehicle-infused CHF; ‡, p < .05 compared with CHF infused with control c-Kit+ cells. Abbreviations: CHF, congestive heart failure; Cntrl, control; dPdt/max, peak rate of pressure increase; EF, ejection fraction; LVEDP, left ventricle end diastolic pressure; MOCE, mocetinostat.
In Vivo Differentiation of RCV-Delivered Untreated and MOCE-Treated c-Kit+ Cells
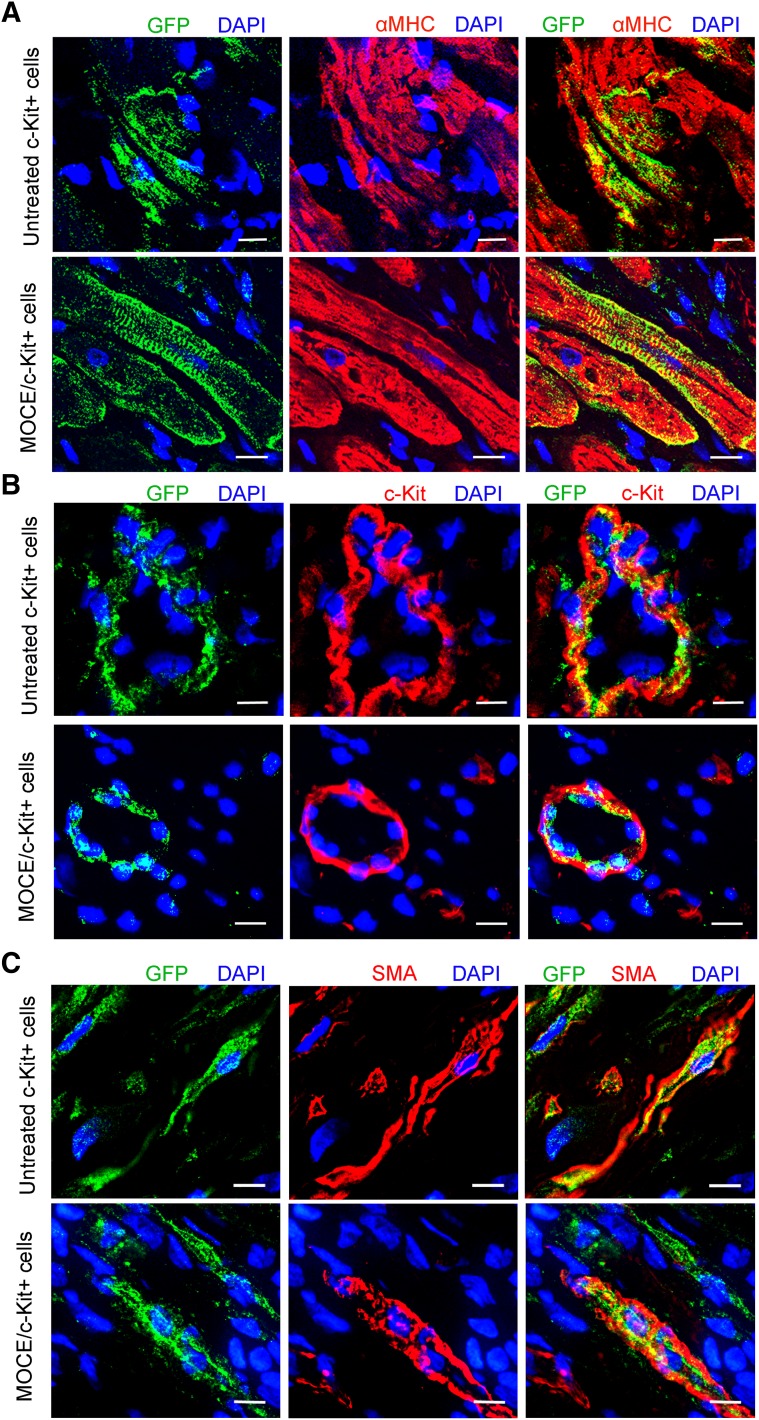
To track transplanted cells in vivo, c-Kit+ cells were transduced with a GFP-carrying lentiviral vector [25]. On transplantation, the exogenous transplanted cells were identified in the myocardium by GFP labeling (Fig. 5; supplemental online Fig. 4). CHF hearts infused with culture medium (vehicle) were used as a negative control. At 21 days after RCV infusion, most MOCE/c-Kit+ cells, similar to the control cells, resided mainly in the scar border zone (supplemental online Fig. 4). Immunohistochemical analysis revealed cells coexpressing GFP and αMHC in the scar border zone of CHF rats transplanted with either control or MOCE/c-Kit+ cells, suggesting cardiomyocyte differentiation of the transplanted cells (Fig. 5A). Most observed GFP+/αMHC+ cells appeared smaller than adult cardiomyocytes, suggesting a possible immature state of those cells. In addition, some GFP+ cells coexpressed c-Kit marker in vivo, indicating that some of the transplanted cells remained undifferentiated (Fig. 5B). These GFP+/c-Kit+ cells were mainly found close to the epicardial layer. We rarely found cells that were double positive for GFP and the myofibroblast marker smooth muscle actin outside the of scar area in both cell transplanted groups (Fig. 5C). Finally, no colocalization was found between GFP and the proliferation marker Ki67 in animals transplanted with untreated or MOCE-treated c-Kit+ cells, suggesting that cells do not actively proliferate at 21 days after RCV infusion (data are not shown). Therefore, no significant differences were noted in the differentiation pattern of transplanted control and MOCE/c-Kit+ cells in vivo.
Figure 5.
In vivo differentiation of unmodified and MOCE-treated c-Kit+ cells delivered by the retrograde coronary vein (RCV) to the congestive heart failure heart. Three weeks after RCV infusion, engrafted cells were identified in heart sections by GFP marker. Representative microscopic images of left ventricular infarct zone show colocalization of GFP with cardiac markers as follow: αMHC (A), c-Kit (B), and SMA (C). Yellow indicates colocalization of GFP and corresponding specific marker. Scale bars = 10 µm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; αMHC, α-myosin heavy chain; MOCE, mocetinostat; SMA, smooth muscle actin.
Effects of Untreated and MOCE-Treated c-Kit+ Cells on Post-MI Cardiac Remodeling
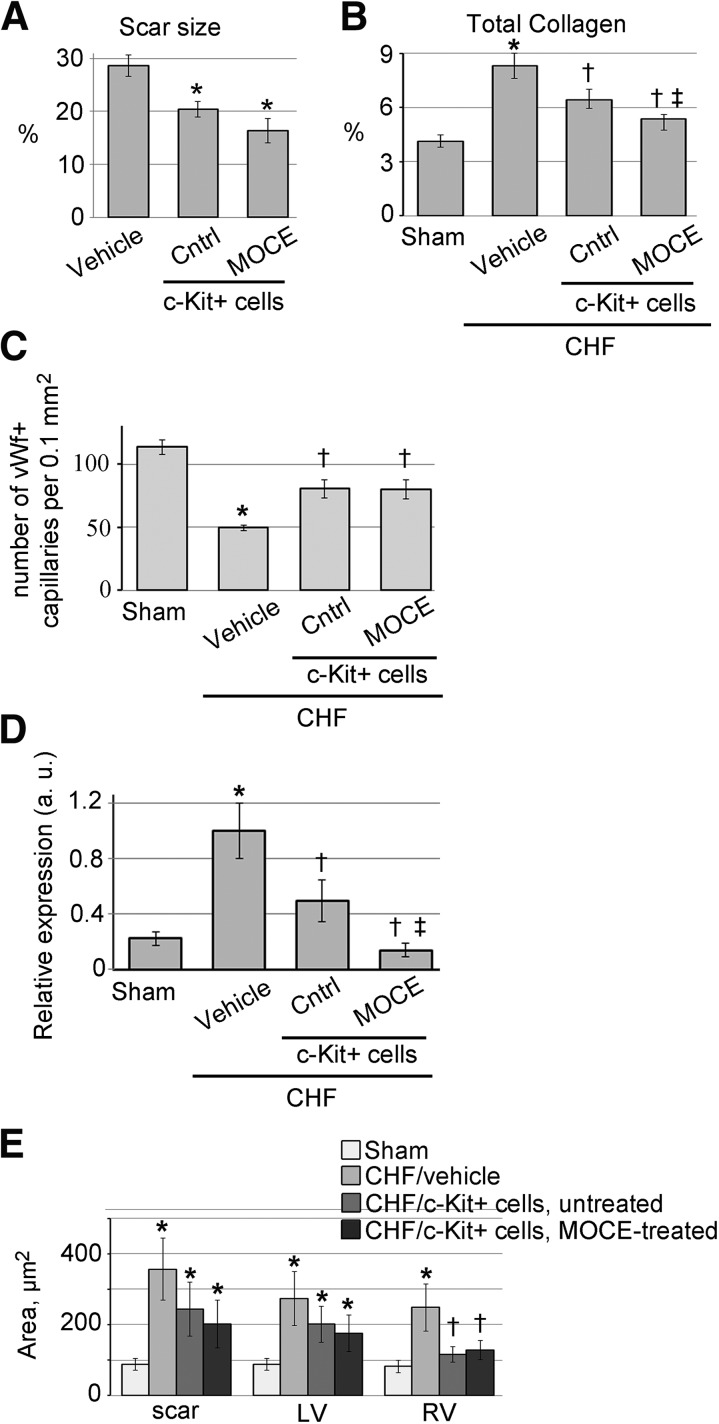
We tested the effects of c-Kit+ cell transplantation on CHF heart remodeling and compared the efficiency of control and MOCE/c-Kit+ cells. At 21 days after RCV infusion, the scar size was significantly smaller in the CHF rats treated with either control or MOCE/c-Kit+ cells compared with vehicle-infused CHF rats (Fig. 6A). A trend toward a decrease in scar size was observed in the CHF rats treated with MOCE/c-Kit+ cells compared with those treated with control cells (18.6% ± 0.9% vs. 20.4% ± 1.4%, respectively; Fig. 6A).
Figure 6.
Effects of untreated and MOCE-treated c-Kit+ cells on CHF remodeling. CHF hearts were retrograde coronary vein (RCV) infused with vehicle, untreated (control), or MOCE/c-Kit+ cells and analyzed at 21 days after RCV. (A): Scar size was measured in trichrome-stained CHF heart sections. CHF hearts were infused with media (vehicle), control, or MOCE/c-Kit+ cells. ∗, p < .05 compared with CHF or vehicle; n = 6 per condition. (B): Total collagen percentage was measured in sham or CHF hearts infused with vehicle, control, or MOCE/c-Kit+ cells. The collagen amount was normalized to the total protein amount; n = 5; ∗, p < .05 compared with sham; †, p < .05 compared with vehicle-infused CHF; ‡, p < .05 compared with CHF infused with control c-Kit+ cells. (C): A number of vWf+ capillaries were quantified in 5 random microscopic fields; n = 4; ∗, p < .05 compared with sham; †, p < .05 compared with CHF. (D): Quantitative reverse transcription-polymerase chain reaction analysis of interleukin-6 gene expression; n = 5; ∗, p < .05 compared with sham; †, p < .05 compared with vehicle-infused CHF; ‡, p < .05 compared with CHF infused with control c-Kit+ cells. (E): Quantitative analysis of cardiomyocyte area of CHF hearts infused with control or MOCE/c-Kit+ cells. The cardiomyocyte cross-sectional area was measured in the scar border zone, LV, and RV; n = 3; ∗, p < .05 compared with sham; †, p < .05 compared with vehicle-infused CHF. Abbreviations: a.u., arbitrary units; CHF, congestive heart failure; Cntrl, control; LV, left ventricle; MOCE, mocetinostat; RV, right ventricle; scar, scar border zone; vWf, von Willebrand factor.
Furthermore, after 21 days, both cell treatments resulted in a significant decrease in total collagen amount compared with the amount in vehicle-infused CHF rats (Fig. 6B). In addition, the total collagen amount was smaller in the CHF rats infused with MOCE/c-Kit+ cells compared with the rats infused with control cells.
Next, we compared the effects of control and MOCE/c-Kit+ cells on angiogenesis in CHF hearts. Compared with vehicle-infused CHFs, a significantly higher number of von Willebrand factor-positive (vWf+) capillaries were found in the infarct border zone of rats transplanted with control and MOCE/c-Kit+ cells (Fig. 6C). No differences were found in vWf+ capillary density between rats transplanted with control and MOCE/c-Kit+ cells.
Cardiomyocyte hypertrophy was observed in CHF rats in the scar border zone and the rest of the left and right ventricles (Fig. 6E; supplemental online Table 3). In both cell-infused CHF groups, the average area of right ventricular cardiomyocytes was significantly smaller. Compared with vehicle-treated CHF rats, however, no differences were noted in the cardiomyocyte area of CHF rats infused with MOCE/c-Kit+ cells compared with controls (Fig. 6E; supplemental online Table 3).
Furthermore, the expression level of the interleukin-6 (IL-6) gene, a marker of decompensated heart failure, was determined in heart tissue samples. Inflammatory cytokines, such as IL-6, contribute to the progression of CHF by a variety of mechanisms [31]. IL-6 reduction is associated with better outcomes in CHF patients and serves as a biomarker of CHF progression [32–34]. As expected, the IL-6 expression level was significantly higher in the CHF heart tissues than in the sham tissues. Infusion of either unmodified or MOCE-treated c-Kit+ cells significantly decreased the IL-6 expression level. Moreover, IL-6 expression was significantly lower in the hearts infused with MOCE/c-Kit+ cells compared with the hearts infused with unmodified c-Kit+ cells (Fig. 6D).
In summary, our data have shown that rats infused with MOCE/c-Kit+ cells had less interstitial fibrosis and decreased LVEDP compared with rats infused with control c-Kit+ cells.
Discussion
The present study was designed to augment the cardiogenic potential of c-Kit+ cells using epigenetic modifications. We applied the selective class I HDAC inhibitor, MOCE, to induce expression of cardiac markers. To elucidate the mechanisms of gene expression, we examined the MOCE-induced changes in the histone H3 status of cardiac and stem cell-related genes. Next, we measured the effects of epigenetically modified c-Kit+ cells on CHF hearts. Our major findings included (a) inhibition of class I HDAC with MOCE-stimulated expression of cardiac-specific markers in c-Kit+ cells; (b) histone H3 acetylation might account for the observed upregulation in the expression of cardiac and stem cell genes after MOCE treatment; (c) the rats infused with MOCE/c-Kit+ cells demonstrated a lower LVEDP compared with rats infused with control cells; and (d) rats infused with MOCE/c-Kit+ cells demonstrated slower CHF progression than rats infused with control c-Kit+ cells. This last conclusion was based on two observations: (a) the total collagen amount was smaller in the CHF rats infused with MOCE/ c-Kit+ cells; and (b) IL-6 expression was lower in hearts infused with MOCE/c-Kit+ cells than in hearts infused with unmodified c-Kit+ cells.
Ischemic myocardium injury, such as MI, results in a significant loss of cardiomyocytes, scar formation, and loss of contractile function, which ultimately leads to heart failure. Stem cell-based therapies have the potential, not only to attenuate heart failure symptoms, but also to replenish the loss of cardiomyocytes. Cardiac differentiation of the donor stem cells is essential for the functional benefits that these cells could provide [35]. Cardiac explant-derived c-Kit+ cells were previously characterized by our laboratory, and other investigators, as a potential cell source for treating ischemic myocardium [24]. c-Kit+ cells exhibit mesenchymal features; they are multipotent and able to give rise to myocardial, smooth muscle, and endothelial cells in vitro and in vivo [36]. Many recent studies have demonstrated that administration of c-Kit+ cells attenuates cardiac pathologies in animal models of both acute MI and CHF [37, 38]. However, the cardiomyogenic capacity of c-Kit+ cells is limited; a low level and incomplete cardiac differentiation of these cells in vitro and a lack of robust differentiation of transplanted cells into cardiomyocytes in vivo were observed [39]. It was suggested that the beneficial effects of transplanted adult stem cells, including c-Kit+ cells, are mainly attributed to paracrine factors secreted by these cells, rather than by direct differentiation of transplanted stem cells into cardiac cell types and engraftment into the muscle [39, 40].
Therefore, in the present study, we aimed to augment the cardiomyogenic potential of c-Kit+ cells using an epigenetically active molecule, a selective class I HDAC inhibitor, MOCE. A variety of epigenetically active small molecules were previously used to reprogram bone marrow-derived progenitor cells into cardiovascular precursors. Small molecules include modifiers of DNA methylation inhibitors such as 5-azacytidin [41, 42] or pan-histone deacetylation inhibitors such as phenyl butyrate, TSA, and VPA [18, 43], or a combination of these drugs [19]. However, owing to the ubiquitous nature of pan-inhibitors, their administration disrupts multiple isoforms, including those uninvolved in cell differentiation. In contrast, isoform-selective inhibitors can improve efficacy and reduce the toxicities associated with the inhibition of multiple isoforms [23]. In the present study, we showed that selective class I HDAC inhibition in c-Kit+ cells led to an increase in acetylation of histone H3 at the promoter regions of cardiac and pluripotency genes. Histone H3 acetylation was accompanied by a transient increase in expression of pluripotency genes, a progressive increase in expression of cardiac genes, and an increase in the number of cells positive for cardiac markers. The differences in the time course suggest class I HDAC inhibition induces activation of various molecular pathways that are likely interdependent. MOCE-modified cells continued to express stem cell and pluripotency markers, suggesting an immature state of these cells [44]. It is also possible that MOCE/c-Kit+ cells represent mixed population of cells of various differentiation stages. Overall, the effects of selective class I HDAC inhibition on cardiac differentiation were comparable with previously described effects of broad spectrum HDAC inhibitors, such as the inhibitors TSA, VPA [18, 19], and phenyl butyrate [43] and the methylation inhibitor 5-azacytidin [45].
We further tested the effects of epigenetically modified c-Kit+ cells on CHF rats and compared the functional efficiency of unmodified and MOCE/c-Kit+ cells. To deliver cells to the CHF heart, we used a retrograde coronary venous infusion (RCV) technique recently developed in our laboratory [25]. We observed a significant enhancement in contractile cardiac function in CHF rats infused with control or MOCE/c-Kit+ cells compared with vehicle-infused CHFs. In addition, both cell-treated CHF groups showed significant retardation in heart remodeling compared with the vehicle-infused CHF animals.
Comparing cell-transplanted groups, we found that the LVEDP was significantly lower in CHF animals infused with MOCE/c-Kit cells than in animals infused with control c-Kit+ cells. We also observed a significant decrease in both the total collagen amount and the IL-6 level in CHF rats infused with MOCE/c-Kit+ cells compared with rats infused with control cells. These data suggest that MOCE/c-Kit+ cells are more efficient in reducing interstitial fibrosis and retarding heart failure progression compared with unmodified cells. However, despite the induced cardiomyogenesis of MOCE/c-Kit+ cells in vitro, histological analysis revealed no robust differences in the engraftment and differentiation patterns of the control and MOCE/c-Kit+ cells in vivo. These results contrast with those from a previous study by Zhang et al., demonstrating that conditioning of c-Kit+ cells with the pan-HDAC inhibitor, TSA, significantly increased Kit+ cell-derived cardiomyocytes and enhanced post-MI functional heart recovery compared with untreated c-Kit+ cells [46]. It is possible that this inconsistency of our data with their report resulted from differences in the animal model. Zhang et al. used an acute MI model and delivered cells immediately after MI. In contrast, in the present study, the cells were delivered 3 weeks after MI.
Our findings suggest that the enhanced therapeutic response of CHF with MOCE/c-Kit+ cells might possibly result from potential modulations in paracrine effect rather than cardiomyogenesis in vivo. This hypothesis is consistent with previous work demonstrating improved paracrine signaling of bone marrow stem cells treated with HDAC inhibitors [18]. The lack of robust neo-cardiomyogenesis in CHF rats infused with MOCE/c-Kit+ cells can be explained, at least in part, by suggesting that MOCE treatment does not improve cell retention in CHF host myocardium. As was recently shown, the number of transplanted cells detected in host myocardium declines rapidly with time: only 2%–3% of transplanted cells were retained in the myocardium after 35 days [39, 47, 48]. Therefore, the low retention rate of transplanted cells can potentially diminish the benefits of induced cell cardiac differentiation in vitro. Further studies are required to discover novel techniques to enhance cell retention and to elucidate the mechanisms of CHF remodeling retardation by epigenetically modified c-Kit+ cells.
A limitation of the present study might have been the lack of dose escalation for each type of cells administered to treat CHF. Previous reports have suggested a possible dependence of the efficacy of post-MI stem cell therapy on the dose of transplanted cells [46]. Therefore, we could not rule out the possibility that an increase in the number of infused cells might increase the difference in therapeutic efficacy of control versus MOCE-treated c-Kit+ cells.
Conclusion
This study has shown that selective class 1 HDAC inhibition is sufficient to redirect c-Kit+ cells toward a cardiac fate. Epigenetically modified c-Kit+ cells improved contractile function and retarded remodeling of the CHF heart; however, an improvement in the heart regeneration capacity of these cells has yet to be demonstrated. The present study has provided new insights into the efficacy of cardiac c-Kit+ cells in ischemic heart failure model.
Supplementary Material
Acknowledgments
This study was supported by NIH Grant R01-AG027263 and the Sun Health Foundation.
Author Contributions
L.Z.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; H.N.-G.: collection and assembly of data, data analysis and interpretation, manuscript writing; L.F., S.P.-S.: collection and assembly of data, data analysis and interpretation; M.A.G.: financial support, administrative support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Donndorf P, Strauer BE, Haverich A, et al. Stem cell therapy for the treatment of acute myocardial infarction and chronic ischemic heart disease. Curr Pharm Biotechnol. 2013;14:12–19. [PubMed] [Google Scholar]
- 2.Schulman IH, Hare JM. Key developments in stem cell therapy in cardiology. Regen Med. 2012;7(suppl):17–24. doi: 10.2217/rme.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 4.Rota M, LeCapitaine N, Hosoda T, et al. Diabetes promotes cardiac stem cell aging and heart failure, which are prevented by deletion of the p66shc gene. Circ Res. 2006;99:42–52. doi: 10.1161/01.RES.0000231289.63468.08. [DOI] [PubMed] [Google Scholar]
- 5.Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- 6.Atkinson S, Armstrong L. Epigenetics in embryonic stem cells: Regulation of pluripotency and differentiation. Cell Tissue Res. 2008;331:23–29. doi: 10.1007/s00441-007-0536-x. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 8.Karantzali E, Schulz H, Hummel O, et al. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: Transcriptomic and epigenetic analysis. Genome Biol. 2008;9:R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paige SL, Thomas S, Stoick-Cooper CL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wamstad JA, Alexander JM, Truty RM, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 13.de Ruijter AJ, van Gennip AH, Caron HN, et al. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glozak MA, Sengupta N, Zhang X, et al. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichert N, Choukrallah MA, Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol Life Sci. 2012;69:2173–2187. doi: 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villagra A, Sotomayor EM, Seto E. Histone deacetylases and the immunological network: Implications in cancer and inflammation. Oncogene. 2010;29:157–173. doi: 10.1038/onc.2009.334. [DOI] [PubMed] [Google Scholar]
- 18.Burba I, Colombo GI, Staszewsky LI, et al. Histone deacetylase inhibition enhances self renewal and cardioprotection by human cord blood-derived CD34 cells. PLoS One. 2011;6:e22158. doi: 10.1371/journal.pone.0022158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajasingh J, Thangavel J, Siddiqui MR, et al. Improvement of cardiac function in mouse myocardial infarction after transplantation of epigenetically-modified bone marrow progenitor cells. PLoS One. 2011;6:e22550. doi: 10.1371/journal.pone.0022550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaser KB. HDAC inhibitors: Clinical update and mechanism-based potential. Biochem Pharmacol. 2007;74:659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Rosato RR, Grant S. Histone deacetylase inhibitors: Insights into mechanisms of lethality. Expert Opin Ther Targets. 2005;9:809–824. doi: 10.1517/14728222.9.4.809. [DOI] [PubMed] [Google Scholar]
- 22.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 23.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: The next step? Cancer Lett. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Zakharova L, Nural-Guvener H, Nimlos J, et al. Chronic heart failure is associated with transforming growth factor beta-dependent yield and functional decline in atrial explant-derived c-Kit+ cells. J Am Heart Assoc. 2013;2:e000317. doi: 10.1161/JAHA.113.000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zakharova L, Nural-Guvener H, Feehery L, et al. Retrograde coronary vein infusion of cardiac explant-derived c-Kit+ cells improves function in ischemic heart failure. J Heart Lung Transplant. 2014;33:644–653. doi: 10.1016/j.healun.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Pacher P, Nagayama T, Mukhopadhyay P, et al. Measurement of cardiac function using pressure-volume conductance catheter technique in mice and rats. Nat Protoc. 2008;3:1422–1434. doi: 10.1038/nprot.2008.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elatmani H, Dormoy-Raclet V, Dubus P, et al. The RNA-binding protein Unr prevents mouse embryonic stem cells differentiation toward the primitive endoderm lineage. Stem Cells. 2011;29:1504–1516. doi: 10.1002/stem.712. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Gaballa MA, Goldman S. Overexpression of endothelium nitric oxide synthase reverses the diminished vasorelaxation in the hindlimb vasculature in ischemic heart failure in vivo. J Mol Cell Cardiol. 1999;31:1243–1252. doi: 10.1006/jmcc.1999.0956. [DOI] [PubMed] [Google Scholar]
- 30.Francis J, Weiss RM, Wei SG, et al. Progression of heart failure after myocardial infarction in the rat. Am J Physiol Regul Integr Comp Physiol. 2001;281:R1734–R1745. doi: 10.1152/ajpregu.2001.281.5.R1734. [DOI] [PubMed] [Google Scholar]
- 31.Mann DL. Inflammatory mediators and the failing heart: Past, present, and the foreseeable future. Circ Res. 2002;91:988–998. doi: 10.1161/01.res.0000043825.01705.1b. [DOI] [PubMed] [Google Scholar]
- 32.Deswal A, Petersen NJ, Feldman AM, et al. Cytokines and cytokine receptors in advanced heart failure: An analysis of the cytokine database from the Vesnarinone trial (VEST) Circulation. 2001;103:2055–2059. doi: 10.1161/01.cir.103.16.2055. [DOI] [PubMed] [Google Scholar]
- 33.Orús J, Roig E, Perez-Villa F, et al. Prognostic value of serum cytokines in patients with congestive heart failure. J Heart Lung Transplant. 2000;19:419–425. doi: 10.1016/s1053-2498(00)00083-8. [DOI] [PubMed] [Google Scholar]
- 34.Gullestad L, Ueland T, Vinge LE, et al. Inflammatory cytokines in heart failure: Mediators and markers. Cardiology. 2012;122:23–35. doi: 10.1159/000338166. [DOI] [PubMed] [Google Scholar]
- 35.Behfar A, Yamada S, Crespo-Diaz R, et al. Guided cardiopoiesis enhances therapeutic benefit of bone marrow human mesenchymal stem cells in chronic myocardial infarction. J Am Coll Cardiol. 2010;56:721–734. doi: 10.1016/j.jacc.2010.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gambini E, Pompilio G, Biondi A, et al. C-kit+ cardiac progenitors exhibit mesenchymal markers and preferential cardiovascular commitment. Cardiovasc Res. 2011;89:362–373. doi: 10.1093/cvr/cvq292. [DOI] [PubMed] [Google Scholar]
- 37.Sanganalmath SK, Bolli R. Cell therapy for heart failure: A comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ Res. 2013;113:810–834. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang XL, Rokosh G, Sanganalmath SK, et al. Intracoronary administration of cardiac progenitor cells alleviates left ventricular dysfunction in rats with a 30-day-old infarction. Circulation. 2010;121:293–305. doi: 10.1161/CIRCULATIONAHA.109.871905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chimenti I, Smith RR, Li TS, et al. Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circ Res. 2010;106:971–980. doi: 10.1161/CIRCRESAHA.109.210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirotsou M, Jayawardena TM, Schmeckpeper J, et al. Paracrine mechanisms of stem cell reparative and regenerative actions in the heart. J Mol Cell Cardiol. 2011;50:280–289. doi: 10.1016/j.yjmcc.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smits AM, van Vliet P, Metz CH, et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: An in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc. 2009;4:232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- 42.Pasha Z, Haider HK, Ashraf M. Efficient non-viral reprogramming of myoblasts to stemness with a single small molecule to generate cardiac progenitor cells. PLoS One. 2011;6:e23667. doi: 10.1371/journal.pone.0023667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vecellio M, Meraviglia V, Nanni S, et al. In vitro epigenetic reprogramming of human cardiac mesenchymal stromal cells into functionally competent cardiovascular precursors. PLoS One. 2012;7:e51694. doi: 10.1371/journal.pone.0051694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bearzi C, Leri A, Lo Monaco F, et al. Identification of a coronary vascular progenitor cell in the human heart. Proc Natl Acad Sci USA. 2009;106:15885–15890. doi: 10.1073/pnas.0907622106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Thal MA, Krishnamurthy P, Mackie AR, et al. Enhanced angiogenic and cardiomyocyte differentiation capacity of epigenetically reprogrammed mouse and human endothelial progenitor cells augments their efficacy for ischemic myocardial repair. Circ Res. 2012;111:180–190. doi: 10.1161/CIRCRESAHA.112.270462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Chen B, Zhao Y, et al. Inhibition of histone deacetylase-induced myocardial repair is mediated by c-kit in infarcted hearts. J Biol Chem. 2012;287:39338–39348. doi: 10.1074/jbc.M112.379115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong KU, Guo Y, Li QH, et al. c-kit+ Cardiac stem cells alleviate post-myocardial infarction left ventricular dysfunction despite poor engraftment and negligible retention in the recipient heart. PLoS One. 2014;9:e96725. doi: 10.1371/journal.pone.0096725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuleri KH, Feigenbaum GS, Centola M. et al. Autologous mesenchymal stem cells produce reverse remodelling in chronic ischaemic cardiomyopathy. Eur Heart J. 2009;30:2722–2732. doi: 10.1093/eurheartj/ehp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.