Human pluripotent stem cells (hPSCs) are being used for both disease modeling and cell therapy; however, efficient homologous recombination (HR) is often crucial to develop isogenic control or reporter lines. This study showed that limited low-dose irradiation significantly enhanced HR frequency. This simple method allows higher throughput of new, targeted hPSC lines that are crucial to expand the use of disease modeling and to develop novel avenues of cell therapy.
Keywords: Irradiation, Induced pluripotent stem cell, Gene targeting, Clustered regularly interspaced short palindromic repeats, Zinc finger nuclease, Transcription activator-like effector nuclease
Abstract
Human pluripotent stem cells (hPSCs) are now being used for both disease modeling and cell therapy; however, efficient homologous recombination (HR) is often crucial to develop isogenic control or reporter lines. We showed that limited low-dose irradiation (LDI) using either γ-ray or x-ray exposure (0.4 Gy) significantly enhanced HR frequency, possibly through induction of DNA repair/recombination machinery including ataxia-telangiectasia mutated, histone H2A.X and RAD51 proteins. LDI could also increase HR efficiency by more than 30-fold when combined with the targeting tools zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Whole-exome sequencing confirmed that the LDI administered to hPSCs did not induce gross genomic alterations or affect cellular viability. Irradiated and targeted lines were karyotypically normal and made all differentiated lineages that continued to express green fluorescent protein targeted at the AAVS1 locus. This simple method allows higher throughput of new, targeted hPSC lines that are crucial to expand the use of disease modeling and to develop novel avenues of cell therapy.
Significance
The simple and relevant technique described in this report uses a low level of radiation to increase desired gene modifications in human pluripotent stem cells by an order of magnitude. This higher efficiency permits greater throughput with reduced time and cost. The low level of radiation also greatly increased the recombination frequency when combined with developed engineered nucleases. Critically, the radiation did not lead to increases in DNA mutations or to reductions in overall cellular viability. This novel technique enables not only the rapid production of disease models using human stem cells but also the possibility of treating genetically based diseases by correcting patient-derived cells.
Introduction
Human pluripotent stem cells (hPSCs) can be isolated from the inner cell mass of preimplantation embryos as human embryonic stem cells (hESCs) [1] or generated from adult somatic cells by overexpressing pluripotency genes as human induced PSCs (hiPSCs) [2, 3]. They provide an invaluable starting source for producing any human cell type and have many applications including cell therapy, disease modeling, and drug screening [4]. The ability to efficiently edit the cell genome through homologous recombination (HR) is crucial to insert reporter or modifying genes into specific loci or to allow the production of control isogenic lines in disease-modeling studies [5]. However, available gene-targeting methods are limited because of the low frequency at which HR occurs in most cells [6, 7], especially human-derived lines and pluripotent cells [8]. A more efficient method of gene targeting is clearly needed.
Increased HR frequency after induction of DNA double-strand breaks (DSBs) with the I-SceI endonuclease was described two decades ago [9, 10]. More recently, zinc finger nucleases (ZFNs) [11–13], transcription activator-like effector nucleases (TALENs) [14, 15], clustered regularly interspaced short palindromic repeats (CRISPR), and the CRISPR-associated system (Cas) [16–20] have been shown to substantially facilitate gene targeting by generating site-specific DSBs in a specific locus. TALENs and CRISPR are especially efficient for nonhomologous end joining (NHEJ) and producing insertion/deletion (indel) mutations [18, 21]. However, these genome-engineering approaches remain relatively inefficient with regard to HR frequency used to replace DNA with a donor vector, particularly when applied to hPSCs [20].
The safe use of ionizing radiation in therapeutic and diagnostic procedures in medicine is well established [22]. For treatment of cancers and hematopoietic cell transplantation, exposure to high doses of radiation (≥1 Gy) is routinely used to destroy target cells. In contrast, low doses (<1 Gy) do not cause cell death frequently but still induce DNA single-strand breaks (SSBs) and DSBs, which are rapidly repaired by the cellular repair machinery [23, 24]. This suggested to us that a low-radiation dose may produce conditions that enhance the frequency of error-free DNA repair and thus lead to higher rates of successful HR in hPSCs.
Materials and Methods
Human ESC and iPSC Culture
The human female ESC line WA09 (H9) line was obtained from WiCell Research Institute (Madison, WI, http://www.wicell.org). The human female iPSC line CS83iCTR-33n1 (83i) and human male iPSC line CS25iCTR-18n2 (25i) were obtained from the iPSC core facility at Cedars-Sinai Medical Center (Los Angeles, CA). Human ESCs and iPSCs were maintained on mouse embryonic fibroblast (MEF) feeder layers in 5% CO2 at 37°C and passaged using 1 mg/ml collagenase type IV (Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com) every 7 days at normal density (1×) of 1.4 × 105 cells per well (splitting ratio 1:6) onto new MEF feeder layers in a 6-well plate (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). The cells were grown in hESC medium containing Knockout Dulbecco’s modified Eagle’s medium, 17% Knockout Serum Replacer, 5 ng/ml human basic fibroblast growth factor (bFGF), 1× nonessential amino acids, 0.4× insulin-transferrin-selenium (ITS-G), 1× GlutaMax (all from Life Technologies), and 0.1 mM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, http://www.sigmaldrich.com). For nonfeeder cultures, plates (Nunc; Thermo Scientific, Rockford, IL, http://www.piercenet.com) coated with Matrigel (growth factor reduced; BD Biosciences) and mTeSR1 medium (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) were used for maintaining hESCs and hiPSCs. All work was performed with appropriate institutional review board approvals from Cedars-Sinai Medical Center.
Gene Targeting With γ-Ray or X-Ray Radiation Using ZFNs, TALENs, or the CRISPR/Cas9 System
Human ESCs and iPSCs from confluent wells were plated into two wells at a high density (2×) of ∼2.8 × 105 cells per well on DR4 MEF feeder layers in 6-well plates. At 24 hours later, the culture medium was replaced with fresh hESC medium. Cells were transfected by Lipofectamine 2000 (Life Technologies) with donor gene-targeting vector (4 µg per well) and the ZFN-AAVS1 transcripts (∼1.5 μg of mRNA per well, CompoZr AAVS1 targeted integration kit; Sigma-Aldrich), TALEN expression vectors (hAAVS1 1L and hAAVS1 1R, each 3 µg per well; Addgene, Cambridge, MA, http://www.addgene.org), or CRISPR expression vectors (hCas9 and gRNA_AAVS1-T2, each 3 µg per well; Addgene) [20]. Radiation was performed 15 minutes later (or a set time) after (or before) transfection. The plates were sealed with parafilm and were irradiated by 0.4 Gy (or 0.1–4.0 Gy) in a cesium-137 γ-irradiator (0.8 Gy/minute, Gammacell 40; Nordion International Inc., Ottawa, Canada, http://www.nordion.com) or x-irradiator (31 seconds for 0.4Gy, 2.5 mA, 150 kV; CellRad; Faxitron Bioptics, Tucson, AZ, http://www.faxitron.com). A nonirradiated plate of cells was used as a control. At 8 hours later, the culture medium including transfection reagents was changed with fresh medium. Puromycin selection (0.4 µg/ml) was started 48 hours after radiation. During the chemical selection, 50% MEF-conditioned medium in hESC medium was used with 30 ng/ml fresh bFGF. After 6–8 days of puromycin selection, green fluorescent protein-positive/puromycin-resistant (GFP+/PuroR) colonies were analyzed by microscopy for GFP signal and screened by polymerase chain reaction (PCR; PCR screening method is described in the following section). A single dissected piece from each colony was analyzed by PCR, whereas the other pieces were plated into an independent well of a 96-well plate with MEF feeder layers for clonal expansion. PCR-positive clones were expanded until confluence in 1 well of a 12-well plate without feeder cells, at which time genomic DNA was purified for Southern blot as a second screening (the Southern blot procedure is described in the Southern Blot Analysis section). Using γ-irradiation and ZFN nuclease treatment, stable hESC and hiPSC clones correctly targeting GFP expressed in the nucleus were generated. Subsequent experiments used stable hESC clones GN03 and GN17 and stable iPSC clones GN46 and GN47.
PCR Screening in the AAVS1 Locus of Gene-Targeted hESC and hiPSC Clones
Gene-targeted clones at first screening were determined by nested PCR of genomic DNA from a small portion of GFP+/PuroR colonies. Terra PCR Direct Polymerase Mix (Clontech, Mountain View, CA, http://www.clontech.com) was used for cell lysis, and the polymerase was replaced with Takara PrimeSTAR GXL Polymerase (Clontech), with the following PCR reaction in one PCR tube. The nested PCR conditions were 95°C for 5 minutes (lysis reaction), then 15 cycles of 10-second denaturation at 98°C and 15-second annealing at 68°C (−0.5°C per cycle) and 1-minute extension at 68°C, followed by 20 cycles of 10-second denaturation at 98°C and 15-second annealing at 60°C and 1-minute extension at 68°C, plus a final extension at 68°C for 5 minutes by Veriti Thermal Cycler (Applied Biosystems by Life Technologies) with the following primers: 5′-AACTCTGCCCTCTAACGCTG-3′ and 5′-GCGTGAGGAAGAGTTCTTGCAG-3′. The subsequent nested PCR was performed using 5% PCR products from the first PCR and performed with PrimeSTAR GXL buffer for 30 cycles of 98°C for 30 seconds, 60°C for 15 seconds, and 68°C for 1 minute with the nested inner primers 5′-GGACCACTTTGAGCTCTACTG-3′ and 5′-GCTGCCAGATCTCTCGAGG-3′, according to the manual of Takara PrimeSTAR GXL DNA Polymerase (Clontech). The expected PCR product was 925 base pairs (bp). The PCR primers for loading control (PPP1R12C gene; 270 bp) were 5′-CCAGGCTGAGAGCTTTAGAGG-3′ and 5′-AATCCTACCTAACGCACTCCTGGG-3′. Amplicons were sequenced by Genewiz, Inc. (South Plainfield, NJ, http://www.genewiz.com) sequencing services.
Southern Blot Analysis
The PPP1R12C gene in AAVS1 locus has two EcoRV restriction cleavage sites: one site is at the 5′ end of exon 1, and the second site is located at the 3′ end of exon 3 (Fig. 1D). Because the AVVS1 donor vector does not contain an EcoRV restriction site, gene-targeted AAVS1 DNA shifted up the fragment size from 5.4 to 8.9 kilobase pairs (kb) detected by the “external probe” (Fig. 1D). Presence of an 8.9-kb fragment indicates the occurrence of a correct HR event. The 661 bp from the external probe were synthesized by PCR using primers 5′-ACCGTCCGCTTCGAGCG-3′ and 5′-CAGATAGACCAGACTGAGCTATGG-3′ from genomic DNA purified from H9 hESCs. The 1.17 kb of the “internal probe” was purified from the GFP gene of the AAVS1 donor vector by SphI and AgeI restriction-enzyme digestions. Fragments of any other size detected by the internal probe represent random/additional insertions. Genomic DNA was separated on a 0.7% agarose gel after EcoRV restriction digestion, transferred to a nylon membrane (Amersham; GE Healthcare Life Sciences, Pittsburgh, PA, http://www.gelifesciences.com), and hybridized with Amersham AlkPhos Direct Labeling and Detection System (GE Healthcare Life Sciences). Increased stringency of wash conditions removed the nonspecific bands from the membrane containing no radiation samples (Fig. 2D).
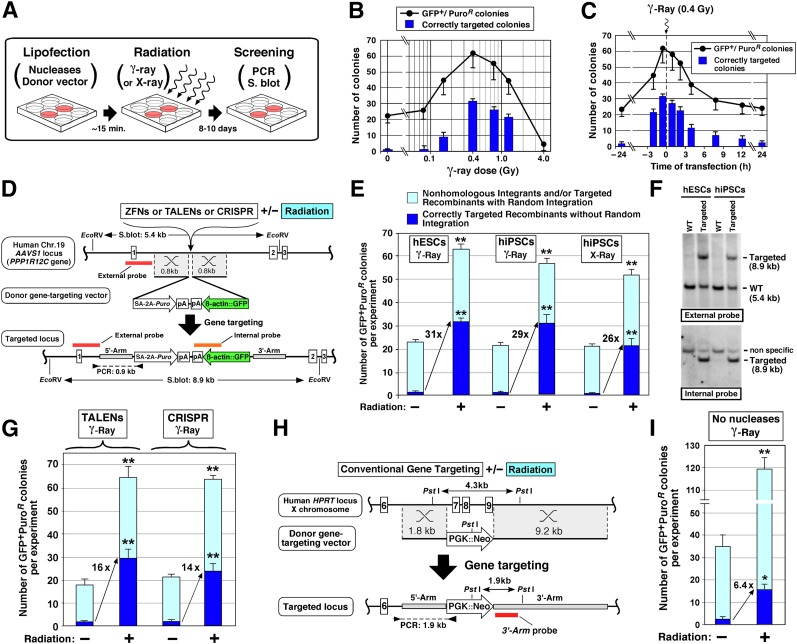
Figure 1.
Enhancing gene-targeting frequency in human pluripotent stem cells by γ- or x-ray radiation. (A): Overview of human pluripotent stem cell gene targeting combined with radiation. (B): Effect of different radiation dose on the formation of correctly targeted clones by ZFN. The number of GFP+/PuroR colonies and correctly targeted colonies were counted after exposure to various doses (0, 0.8, 0.16, 0.4, 0.8, 1.2, and 4.0 Gy) of γ-ray radiation (four independent experiments). Transfection was performed 15 minutes before the radiation. (C): Effect of different times of transfection before or after 0.4 Gy γ-ray radiation by ZFN-mediated targeting. The number of GFP+/PuroR colonies and correctly targeted colonies were counted following different times of transfection (−24, −2, −0.25, +0.5, +2, +4, +8, +12, and +24 hours) relative to radiation (four independent experiments). (D): Gene-targeting strategy for introducing a GFP gene into the AAVS1 locus. Short homologous arms (0.8 kb of 5′- and 3′- homology arms) were chosen for all three engineered nucleases (ZFN, TALEN, and CRISPR systems). A puromycin-resistant (Puro) gene is driven by the endogenous PPP1R12C promoter through a splicing acceptor after homologous recombination (HR). A GFP gene is driven by constitutively active human β-actin promoter (β-actin::GFP). A dashed line between 2 arrowheads shows the location of a PCR-amplified region to confirm HR. Red and orange lines indicate the external and internal DNA probes for Southern blot analysis. (E): Increased correctly targeted recombinants by the ZFN system combined with the optimized low dose of γ- or x-ray radiation (0.4 Gy) in hESCs and hiPSCs. Statistical significance: ∗∗, p < .01. (F): Southern blot analysis of the AAVS1 locus in gene-targeted hESCs and hiPSCs generated with ZFN and low-dose radiation. Additional Southern blot results are shown in Figure 2B and 2D. (G): Increased correctly targeted recombinants by the TALENs or CRISPR systems combined with the optimized low-dose γ-radiation (0.4 Gy) in hiPSCs. Statistical significance: ∗∗, p < .01. (H): Conventional gene targeting strategy for disrupting the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene. Long and short homologous arms (9.2 and 1.8 kb, respectively) were chosen for replacing exons 7–9 with a neomycin-resistance gene driven by a PGK promoter (PGK::Neo). A dashed line between two arrowheads shows the location of a PCR-amplified region to confirm HR. The red line indicates the DNA probes for Southern blot analysis. (I): Increased correctly targeted recombinants by the low dose of γ-ray radiation (0.4 Gy) in hiPSCs. Statistical significance: ∗, p < .05. Abbreviations: Chr., chromosome; CRISPR, clustered regularly interspaced short palindromic repeats; GFP+/PuroR, green fluorescent protein-positive/puromycin-resistant; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; kb, kilobase pairs; min, minutes; PCR, polymerase chain reaction; S. blot, Southern blot; SA, splicing acceptor; TALEN, transcription activator-like effector nuclease; WT, wild type; ZFN, zinc finger nuclease.
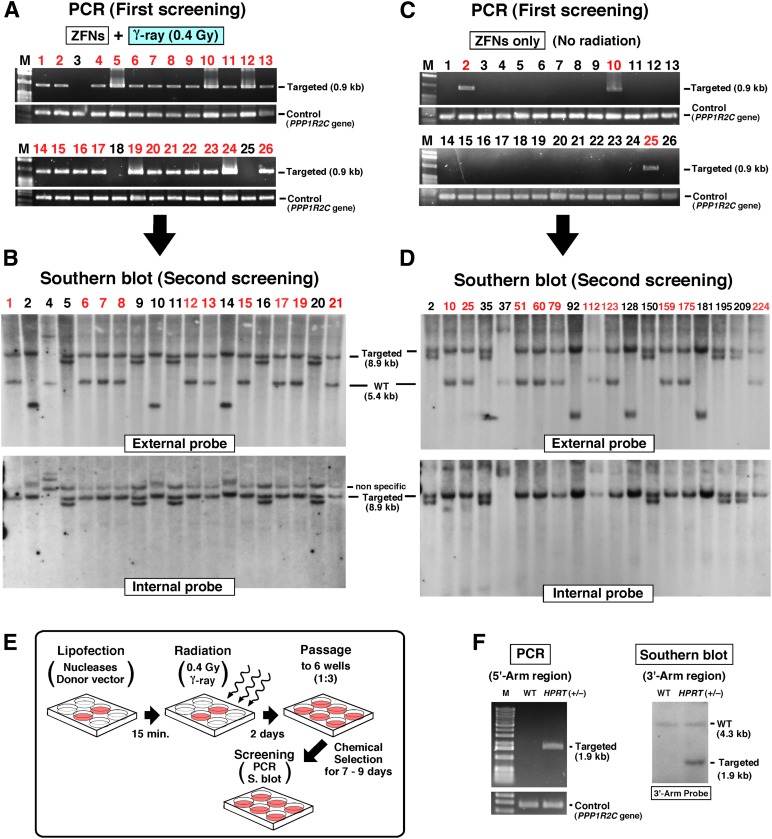
Figure 2.
Confirmation of gene targeting by PCR and Southern blot analyses in irradiated human pluripotent stem cells. (A, C): Representative PCR analysis data for screening gene-targeted clones using primers around the 5′-arm region illustrated in Figure 1D. The data showed a result using ZFNs plus radiation (0.4 Gy of γ-ray) (A) and no-radiation condition (C) in each of 26 GFP+/PuroR colonies. The colonies that had a detected 0.9-kb band were further analyzed by Southern blot. (B, D): Southern blot analysis of PCR-positive clones (the number above the gel image is the clone number). Correctly targeted clones (indicated by the red number at the top of the blot) were carefully confirmed by two kinds probes (internal and external probes, illustrated in Fig. 1D) with the expected 8.9-kb band, which indicates correct homologous recombination (HR) by left and right arms, and no extra band was observed in the same lane. Internal probe from the GFP gene in the donor vector detects any integration to genome so that the internal probe clearly shows a correctly targeted 8.9-kb band or random/additional integration. (E): Overview of conventional gene targeting combined with radiation. (F): Representative PCR and Southern blot results. The position of PCR primers and 3′-arm probe are shown in Figure 1H. Abbreviations: GFP+/PuroR, green fluorescent protein-positive/puromycin-resistant; HPRT, hypoxanthine-guanine phosphoribosyltransferase; kb, kilobase pairs; M, marker; min, minutes; PCR, polymerase chain reaction; S. blot, Southern blot; WT, wild type; ZFN, zinc finger nuclease.
Exome Sequencing
The human iPSC CS02iCTR control line was used for whole-exome sequencing. Early passage (passage 4) was used to reduce possible mosaic genotypes acquired during the passaging process [25–27]. The reprogramming process can also introduce mutations [28]. Mutations that may have occurred during the reprogramming process and that were present in passage 4 cells and in irradiated and nonirradiated clones were subtracted as background. The hiPSCs were passaged into three separate culture plates for (a) an irradiated (0.4 Gy of γ-ray) condition, (b) a nonirradiated condition, and (c) a nonirradiated condition parental sample. At 24 hours later, irradiated and nonirradiated cells were sorted by fluorescence-activated cell sorting (MoFlo; Beckman Coulter, Brea, CA, https://www.beckmancoulter.com) into single cells. A single cell per well was plated into 96-well plates with MEF feeder layers for clonal expansion. During this sorting step, rho-associated protein kinase (ROCK) inhibitor was added in hESC medium (including 30 ng/ml bFGF) for 24 hours. When clonal hiPSCs were expanded to 1 well of a 12-well plate (passage 3 after sorting), genomic DNA was purified from hiPSCs on a nonfeeder plate. The genomic DNA (50 ng) was used for exome sequencing from each of the radiation and control cell lines. 300k exon-targeted amplicons were generated using an ultra-high multiplex PCR in six PCR reactions for each hiPSC clone using the Ion AmpliSeq Exome Kit (Life Technologies). In order to reduce clonality, the post-PCR library protocol was skipped, and exome libraries were quantitated using quantitative PCR before being amplified on Ion Sphere particles using the Ion OneTouch 2 system (Life Technologies). Semiconductor sequencing was performed with an Ion Proton v2 chip using Ion PI XT reagents and the Ion PI Sequencing 200 Kit v2 (Life Technologies). Each hiPSC clone was sequenced to an average depth of coverage of more than 19×. To normalize any differences between radiation, clones were propagated, purified, amplified, and sequenced as experimental pairs. As mentioned, both hiPSC clone exomes were sequenced to an average of 22 ×. In brief, reads were aligned with an Ion Torrent specific alignment program, and single-nucleotide polymorphisms (SNPs) and indels were called using the Ion Torrent server, which has a unique AmpliSeq Exome and Ion Proton-specific error model to identify variants and copy number variations when compared with hg19. Only variants with significant depth of coverage containing at least as many reads as the average coverage of whole exome (e.g., 20×) and with a variant frequencies <30% were assessed. Variants that were rare nonsynomous single-nucleotide variants (SNVs) not identified in the SNP database were then compared among treatment and nontreatment groups. Only variants that were validated by the Integrated Genomics Viewer were selected for further validation including traditional Sanger sequencing validation. Raw sequencing reads are available from the DDBJ Sequence Read Archive under accession number (SRP057851).
Statistical Analysis
Results of experimental points from different experiments are reported as the mean ± SEM. Significance levels were determined by nonpaired Student’s t test analysis, as indicated. Differences were considered significant for p < .05 and p < .01. All other methods are described in the supplemental online data.
Results
Limited Radiation Enhances Homologous Recombination in Pluripotent Stem Cells
Studies have shown that the common integration site of the human nonpathogenic adeno-associated virus (AAV) found in intron 1 of the protein phosphatase 1 regulatory subunit 12C (PPP1R12C) gene, also known as AAV site 1 (AAVS1), is a suitable safe-landing site for stable transgene expression in human cells [29]. Gene targeting at the AAVS1 safe-landing site was accomplished by using lipofection of AAVS1 mRNA ZFNs into hPSCs along with a donor vector designed to incorporate the GFP and puromycin-resistant genes, and then cells were exposed to 0–4.0 Gy radiation (Fig. 1A, 1D). This efficient lipofection method, with ∼20% transfection efficiency, was used because, unlike electroporation, this method maintains high viability of hPSCs and does not require single-cell separation. This approach also avoided cell passaging to reduce additional cell death during transfection, chemical selection, and screening to obtain targeting frequency (Fig. 1A).
Initial quantification by counting GFP+/PuroR colonies showed that the 0.4-Gy radiation dose yielded the highest number of GFP+/PuroR colonies (Fig. 1B, black line). Subsequent Southern blot analysis confirmed that this low dose also provided the maximum number of correctly targeted colonies (Fig. 1B, blue columns). We next determined the optimal timing of hESC transfection and showed that the highest level of integration and correctly targeted colonies occurred at 15 minutes prior to radiation (Fig. 1C). To quantify the enhancement of integration frequency by radiation, hESCs transfected with the donor DNA and the ZFNs at the ideal transfection time (15 minutes before radiation) followed by the optimal radiation condition (0.4 Gy) were compared with no radiation (0 Gy). PCR analysis of the GFP+/PuroR colonies showed that more than 90% of GFP+/PuroR colonies were positive when treated with radiation, compared with 8.2% with no radiation (Table 1, row A; Fig. 2A, 2C). In order to establish how many of these clones had site-specific HR, the PCR-positive clones were further analyzed by Southern blot using external and internal probes (Fig. 2B, 2D). Remarkably, the targeting frequency of correctly targeted clones to the AAVS1 locus was 51% with radiation and ZFNs, which is in marked contrast to 4.3% in ZFN-alone nonirradiated samples (Table 1, row A). Indeed, not only was the targeting frequency more than 10 times greater but also the total number of GFP+/PuroR colonies was approximately 3 times greater, so the final number of correctly targeted clones per experiment had a 31-fold increase (Fig. 1E, left; Table 1, row A). This indicates that the ratio of HR was dramatically enhanced over that of random integration. The presence of improper recombinants in GFP+/PuroR clones seen in the Southern blot (Fig. 2B, 2D) has also been observed by other groups using the ZFN technology targeting the AAVS1 locus [11, 30]. In addition, hiPSCs were tested using the optimized conditions of ZFN transfection with low-dose irradiation (LDI). Results showed similar and significant enhancement in HR targeting frequency compared with no radiation (Fig. 1E, middle; Fig. 1F; Table 1, row B).
Table 1.
Summary of HR-enhancing effects by γ-ray or x-ray in human ESCs and iPSCs
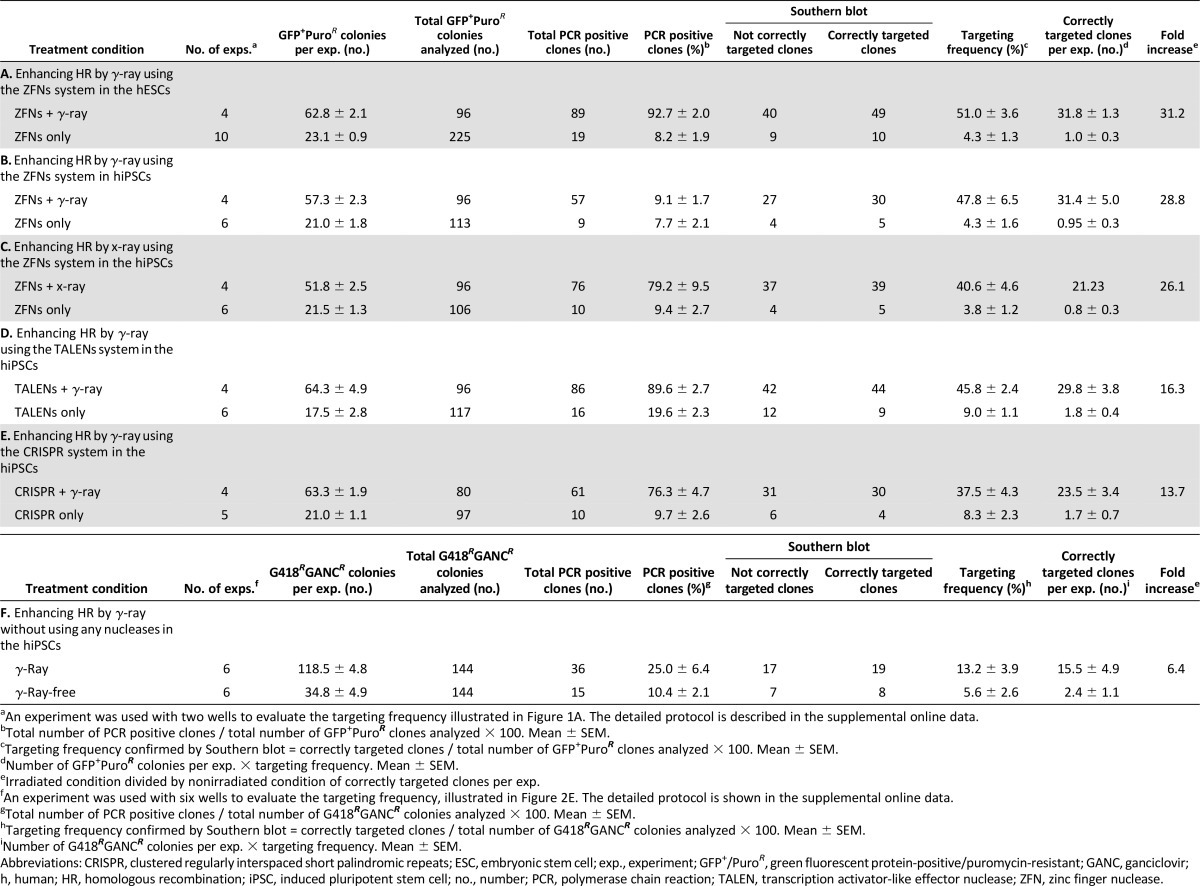
Although γ-radiation worked well for increasing the targeting frequency in both hESCs and hiPSCs, cesium-based irradiators are often restricted by high security and radiation issues. In contrast, a bench top irradiator is a more practical and accessible way of irradiating cells. As such, we used x-ray with a dose and duration similar to that used with γ-ray to irradiate hiPSCs using the ZFN system, which again resulted in successfully increasing the HR targeting frequency compared with no radiation (Fig. 1E, right; Table 1, row C).
In addition to ZFNs, we assessed whether TALENs and CRISPR/Cas9 systems showed enhanced HR frequency with low-dose irradiation. The TALENs or CRISPR systems were used to target the GFP gene into the same AAVS1 locus of hiPSCs, using the optimized radiation and transfection parameters (transfection less than 15 minutes before 0.4-Gy radiation). Analysis of GFP+/PuroR colonies by PCR and Southern blot for correctly targeted clones (Fig. 1G; Table 1, rows D and E) showed that there was a similar enhancing effect of radiation on HR frequency when using TALENs and CRISPR, demonstrating that low-dose γ-radiation enhances HR frequency for multiple engineered nucleases.
Experiments to this point assessed targeting frequency when combining nucleases and radiation. Assessing the targeting frequency in hPSCs using radiation alone compared with the nonirradiated condition could not be assessed because the targeting frequency was too low due to the short 0.8-kb homologous arms in the donor vector, which was sufficient for the ZFN system. In order to overcome this low frequency, we next used a functional targeting vector with long arms (Fig. 1H) for the first demonstration of gene targeting in human ESCs (obtained from Dr. James Thomson) [31]. Quantifying the number of correctly targeted clones at the hypoxanthine-guanine phosphoribosyltransferase (HPRT) locus showed that 0.4 Gy γ-radiation led to significant enhancement (∼6-fold) compared with no radiation (Figs. 1I, 2F; Table 1, row F) in hiPSCs. Consequently, LDI can also increase site-specific HR even with conventional gene targeting, which can eliminate any potential off-target issues introduced by engineered nucleases [32].
LDI Does Not Lead to Mutagenic Events in Human PSCs
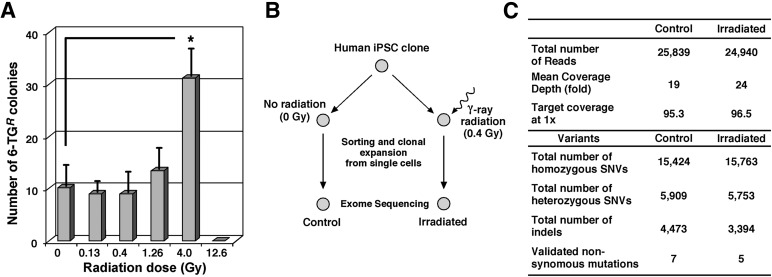
To investigate the mutation frequency caused by γ-radiation, we used an established HPRT assay [33, 34] in which mutations at the HPRT gene were quantified by 6-thioguanine selection. The data clearly showed that no increased mutagenesis was induced until the higher 4.0-Gy dose of radiation was administered, suggesting that the lower doses of radiation, including the ideal 0.4-Gy dose, do not cause genetic abnormalities in this gene within the hiPSC population (Fig. 3A). To analyze all coding-gene mutations, whole-exome sequencing was performed on hiPSCs following 0.4-Gy γ-radiation compared with no radiation. Early passage 4 hiPSCs were used to avoid possible mosaic genotypes acquired during passaging [25–28]. Sorting and clonal expansion from single cells in parallel was used to isolate individual mutations (Fig. 3B). Clones were expanded for three passages, at which time DNA was collected for whole-exome sequencing. Mutations that may have occurred during reprogramming [28] and that were present in passage 4 cells and in irradiated and nonirradiated clones were subtracted as background. Results revealed that radiation produced no gross differences in the number of SNVs and indels between irradiated clones and nonirradiated controls (Fig. 3C). In addition, exome sequencing confirmed that irradiated clones did not have a significant increase in mutation frequency over time, at least in the 1 month of 3 passages. Longer time points still need to be assessed. Selected rare but high-confidence SNV changes predicted to result in nonsynomous amino acid changes in either irradiated or nonirradiated clones were independently validated using Sanger sequencing in parent and other independent lines, suggesting that these mutations were random. Together, these results showed that low-dose 0.4-Gy radiation can significantly increase site-specific HR without causing high mutation levels.
Figure 3.
Analysis of the genomic mutations in irradiated human pluripotent stem cells. (A): Mutation frequency in the HPRT locus. Determination of γ-ray-induced mutations on the HPRT gene was performed using 6-TG selection after exposing the iPSCs to different doses of indicated γ-ray radiation. Cells with an intact or nonmutated HPRT gene will not survive during 6-TG selection, whereas cells that have lost the function of the HPRT gene can survive selection in 6-TG. Note that the low doses (0.13 and 0.4 Gy) of radiation did not increase the number of mutations at the HPRT locus. In the 12.6 Gy column, all iPSCs were eliminated by the high-dose radiation before 6-TG selection. Data presented are from four independent experiments, with statistical significance: ∗, p < .05. (B): Strategy of whole-exome sequencing. The same human iPSC clone (passage 4) underwent radiation or no radiation followed by sorting to single cells and subsequent expansion for whole-exome sequencing (experimental procedure outlined in the supplemental online data). (C): Whole-exome sequencing showed no gross differences in the number of single nucleotide variants or insertions/deletions of single-nucleotide polymorphisms between irradiated clones and control nonirradiated clones. Abbreviations: 6-TGR, 6-thioguanine-resistant; indels, insertion/deletions; iPSC, induced pluripotent stem cell; SNV, single nucleotide variant.
Increased Homologous Recombination Is Associated With Specific Gene Expression Profiles and Altered Protein Phosphorylation
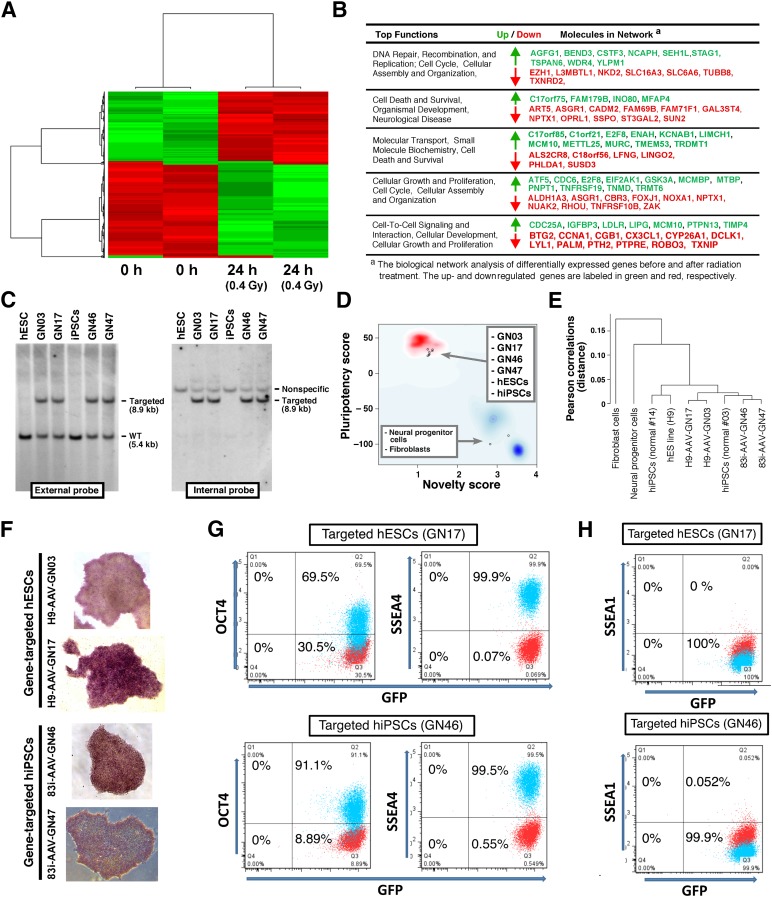
Because previous groups have shown that high-dose irradiation (≥1 Gy) can cause changes at the transcription level [22, 35], we next assessed whether LDI led to any short-term changes in RNA expression using microarrays. Gene expression, measured by supervised two-way hierarchical clustering analysis at 4, 8, and 24 hours after 0.4-Gy radiation, showed no differences at 4 and 8 hours. In contrast, 331 genes were significantly altered in hESCs at 24 hours after 0.4-Gy radiation compared with nonirradiated hESCs at 0 hour (Fig. 4A), as also demonstrated in another study [35]. There were significant changes in genes associated with DNA repair/recombination, cell death and cell cycle, cellular growth, and proliferation pathways (Fig. 4B; supplemental online Table 1).
Figure 4.
Analyses of gene expression and pluripotency after 0.4-Gy γ-ray radiation. (A): Two-way hierarchical clustering analysis of the most variably expressed genes before and after treating hESCs with 0.4-Gy γ-ray radiation . The clustering of samples and genes is based on the Pearson correlation coefficient of the normalized expression of the 703 probes containing more than 0.85 standard deviation either side of the mean across all samples. Data were mean centered. Green and red illustrate relative over- and underexpression, respectively, of the genes. Relative distances between samples and genes are shown in dendrograms above and to the side of the heat map, respectively. The gene list is shown in supplemental online Table 1. (B): The biological network analysis by Ingenuity Pathway Analysis. (C): Southern blot analysis of gene-targeted iPSCs generated by 0.4-Gy radiation in the AAVS1 locus. Original hESC (H9 line) and hiPSC (83i) genomic DNA was included as a negative control. External and internal probes are shown in Figure 1D. (D): Gene expression profiles of four independent targeted clones were analyzed by PluriTest to evaluate pluripotency. Pluripotency and novelty scores of targeted lines are presented. These scores were compared with hESCs, hiPSCs, and differentiated cell samples (neural progenitor and fibroblast cells). (E): A correlation tree of each analyzed sample by PluriTest. (F): Alkaline phosphatase staining of each targeted line. (G): OCT4 and SSEA4 pluripotency gene expression analyzed by flow cytometry in targeted stable hESC (GN17) and hiPSC (GN46) clones. (H): SSEA1 expression analyzed by flow cytometry in targeted stable hESC (GN17) and hiPSC (GN46) clones. Abbreviations: GFP, green fluorescent protein; h, hours; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; kb, kilobase pairs; Q, quartile; WT, wild type.
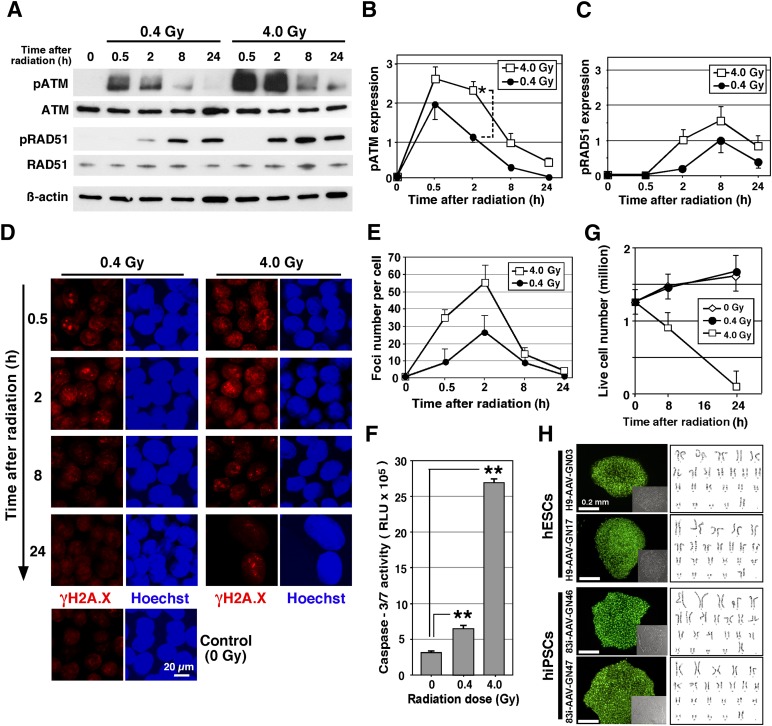
Along with changes in the general RNA expression profile following DNA stress, activation of proteins through phosphorylation by radiation was also detected from 30 minutes to 24 hours. Ataxia telangiectasia mutated (ATM) kinase maintains DNA stability and is activated by DSBs. This activation subsequently regulates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair, or apoptosis. ATM, for instance, directly phosphorylates H2A.X, a histone variant that is an early response protein following DSBs. RAD51 is also activated by phosphorylation after DSBs and plays a critical role in repairing DNA through homologous recombination. After exposure of hESCs to 0.4- and 4.0-Gy γ-radiation, Western blot analysis showed phosphorylation of ATM as early as 30 minutes [36] and phosphorylation of RAD51 beginning at 2 hours (Fig. 5A). Semiquantification of the Western blots highlighted greater phosphorylation following 4.0 Gy relative to 0.4 Gy (Fig. 5B, 5C). After exposure of hESCs to 0.4- and 4.0-Gy radiation, immunocytochemistry showed that a specific antibody of H2A.X, which binds only to the phosphorylated form on serine 139 (γH2A.X), was also detected and localized to foci in the nuclei, which increased over 2 hours and then reversed close to basal levels by 24 hours in both 0.4- and 4.0-Gy radiation conditions (Fig. 5D, 5E). Together these results indicate that the cells exposed to low-dose irradiation can respond with alterations in gene expression and phosphorylation of proteins related to DSB repair, for instance, the phosphorylation of RAD51. It still needs to be defined whether these alterations specifically underlie the increase in HR.
Figure 5.
Analyses of irradiated human pluripotent stem cells. (A): Western blot of ATM-serine 1981 (pATM), total ATM, RAD51-tyrosine 315, and total RAD51 at the indicated time points after cells were exposed to 0.4- or 4.0-Gy γ-ray radiation compared with control, nonirradiated cells. β-actin served as the loading control. To obtain a precise comparison of phosphorylation between 0.4- and 4.0-Gy radiation, an equivalent volume of the proteins was loaded, and antibody signal was used for quantification. (B, C): The relative amount of ATM phosphorylation and RAD51 phosphorylation. The expression level was normalized by β-actin. Statistical significance: ∗, p < .05 (n = 3). (D): Foci formation of phosphorylated H2A.X (γH2A.X) after 0.4- or 4.0-Gy radiation was shown by immunohistochemical analysis after the indicated time by γH2A.X antibody (red). Nuclei were stained by Hoechst (blue). Scale bar = 20 µm. (E): The number of foci stained by γ-H2A.X antibody was counted for each time point. (F): Apoptosis assay that measures caspase-3/7 activity by release of luminescence on activation of caspase-3/7 and cleavage of a target peptide in hESCs irradiated with 0.4- or 4.0-Gy dose. Statistical significance: ∗∗, p < .01. (G): The live cell number of hESCs cultured on a nonfeeder plate was determined by counting cells negative for trypan blue stain 24 hours after 0.4- or 4.0-Gy radiation. Data for panels E–G come from three independent experiments. (H): Analysis of individual gene-targeted clones derived from hESCs and hiPSCs treated by 0.4-Gy radiation. Representative GFP-expressing hESC and hiPSC colonies with inset of corresponding bright field image (left panel) and karyotyping analysis to confirm normal DNA profile (right panel). Abbreviations: ATM, ataxia telangiectasia mutated; h, hours; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; p, phosphorylated; RLU, relative light units.
LDI Increases HR in Human PSCs Without Affecting Cell Viability, Proliferation, or Differentiation Into Major Tissue Lineages
During the course of 24 hours after high-dose irradiation with 4.0 Gy, we noticed the cell nuclei were swollen, indicating the initiation of cell death (Fig. 5D, bottom-right panels). Radiation at both 0.4- and 4.0-Gy levels caused a significant increase in activated caspase-3/7 compared with no radiation; however, the level of apoptosis was lower at 0.4 Gy compared with 4.0-Gy radiation (Fig. 5F). This change in apoptosis corresponds with the similar result related to regulation of apoptosis highlighted in the microarray list (Fig. 4B). Given the low level of detected apoptosis after 0.4-Gy radiation, we wanted to further assess the viability of the cell cultures over time following radiation. Quantifying trypan blue staining showed that approximately 82% of hESCs exposed to 4.0-Gy radiation were dead after 24 hours, whereas hESCs exposed to 0.4 Gy survived and expanded at a rate similar to that seen in nonirradiated control cells (Fig. 5G), indicating that LDI does not significantly reduce overall cell viability.
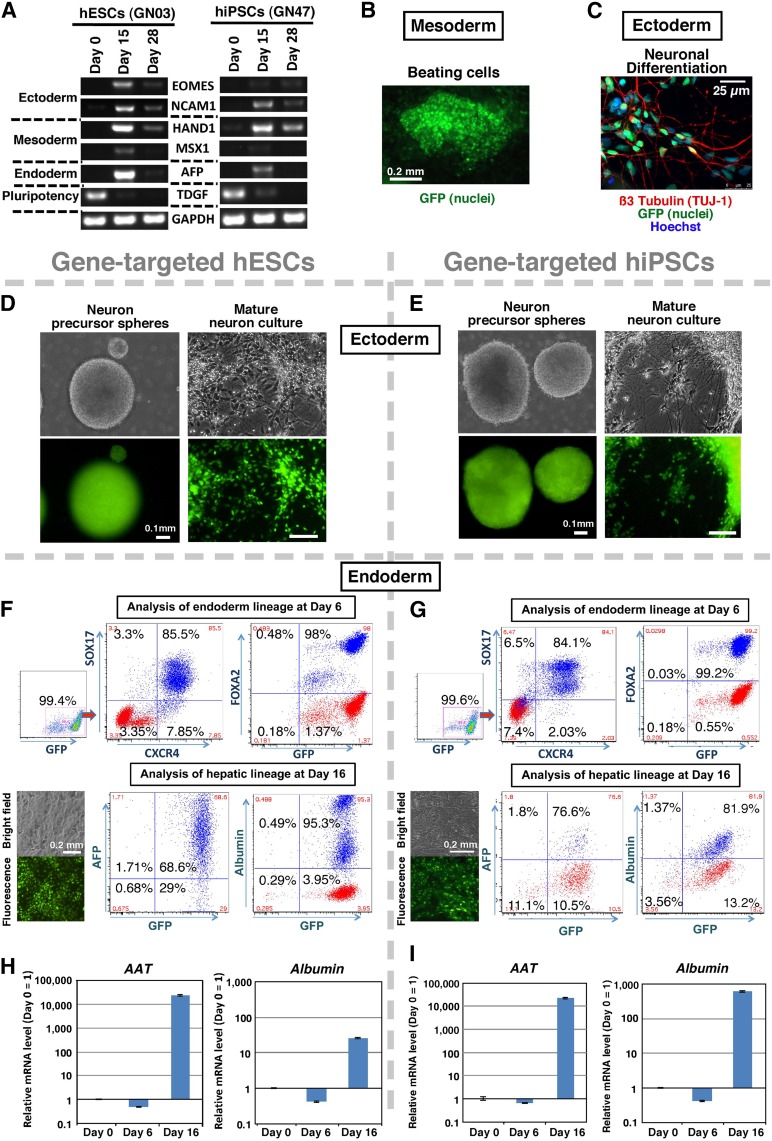
Finally, we investigated potential adverse effects of LDI on the general characteristics and differentiation potential of targeted hPSCs by assessing genomic integrity, pluripotency, stemness, and GFP expression in 4 clones selected from the 49 hESCs and 30 hiPSCs correctly targeted following ZFN transfection and radiation (Table 1, rows A and B). The four clonal lines showed strong GFP expression throughout the selection and expansion processes and retained a normal karyotype (Fig. 5H). Southern blot analysis showed correctly targeted bands in the selected hESCs and hiPSCs (Fig. 4C). A global gene expression PluriTest assay used to define hPSCs [37] revealed a very high pluripotency score and a low novelty score for all four lines that were identical to the original hESC and hiPSC signatures and vastly different from differentiated neural and fibroblast cells, suggesting that pluripotency and novelty were not affected by radiation (Fig. 4D, 4E). The four lines also had alkaline phosphatase activity (Fig. 4F), and flow cytometry confirmed expression of pluripotency markers OCT4 and SSEA4 (Fig. 4G) and no expression of SSEA1 (Fig. 4H). To assess the capacity of the irradiated cells to differentiate into all three germ layers, the targeted lines were spontaneously differentiated as embryoid bodies and PCR-quantified ectoderm, mesoderm, and endoderm markers at days 15 and 28 of differentiation (Fig. 6A). The targeted GFP-expressing hESC and hiPSC clones were further differentiated to lineage-specific mature cells. Beating cardiac cells represented mesoderm, and neural precursor spheres and mature neuron cultures with clear migration and division represented ectoderm (Fig. 6B–6E; supplemental online Movies 1, 2). Hepatic cells expressing early and mature markers, confirmed by flow cytometry and quantitative reverse-transcription PCR, represented endoderm (Fig. 6F–6I). In addition, there was continual robust expression of GFP targeted to the AAVS1 safe-landing site in all lineages in irradiated lines (Fig. 6B–6G). Long-term analysis of LDI-based engineered clones that were passaged for more than 20 generations showed that there was no loss of transgene expression, viability, and differentiation ability. Together, the data clearly showed that radiation combined with ZFN transfection did not hinder the efficient production of hPSC lines with stable transgene expression and normal differentiation potential.
Figure 6.
Analysis of differentiated cells derived from gene-targeted hESC and hiPSC clones generated by 0.4-Gy radiation. (A): Reverse-transcription polymerase chain reaction (RT-PCR) analysis of differentiated gene-targeted hESCs or hiPSCs through embryoid body differentiation. Ectoderm, mesoderm, and endoderm marker expression was confirmed. The GAPDH gene was used as a housekeeping control. (B): Gene-targeted GFP-expressing hESCs (clone GN03) differentiated into beating cardiac cells retained high expression of the GFP signal (supplemental online Movie 1). (C): Gene-targeted hESCs (GN03) differentiated to neurons (ectoderm) were shown to express GFP (green) and β3-tubulin (red) and were counterstained with Hoechst (blue) (supplemental online Movie 2). (D, E): hESC and hiPSC clones after differentiation into neuronal precursor cells and mature neurons. A gene-targeted hESC clone (GN03) (D) and an hiPSC clone (GN47) (E) generated by 0.4-Gy radiation both retained high GFP expression following differentiation into neuronal precursor cells (left) and mature neurons (right). Scale bar = 0.1 mm. (F, G): Analysis of endodermal and hepatocytic lineage in targeted differentiated hESCs and hiPSCs. Flow cytometry analysis shows that targeted hESCs (GN03) and hiPSCs (GN46) with GFP had SOX17, CXCR4, and FoxA2 expression at day 6, and AFP and albumin expression at day 16. Red dots in the flow cytometry image represent isotype control antibody-stained cells. Bright-field and fluorescent images demonstrated that targeted hESC and hiPSC clones retained high GFP expression throughout hepatic differentiation. (H, I): Analysis of liver marker lineage in targeted differentiated hESCs and hiPSCs. Quantitative RT-PCR data show time course of differential expression of α1-antitrypsin (AAT) and albumin genes in targeted hESCs (GN03) (H) and hiPSCs (GN46) (I). Data are represented as mean ± SEM from three independent experiments. Abbreviations: AFP, alpha fetoprotein; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell.
Discussion
The traditional belief that radiation is detrimental to organisms is being reconsidered. High linear energy transfer (LET) radiation with α and neutron particles primarily lead to DNA DSBs, which frequently result in DNA damage and cancer [38]. In contrast, low-LET γ- and x-ray ionizing radiation and a low dose (0.1–0.5 Gy) do not frequently cause DNA DSBs and, rather, can reduce DNA damage, remove cells with DNA damage, activate tumor-suppressor genes, and stimulate detoxification [39–41]. Mouse spermatogonial stem cells receiving a low dose of γ- or x-ray radiation, for example, showed a reduced mutation frequency compared with the nonirradiated condition [42, 43], and a low dose of x-ray total body radiation in humans provided a therapeutic effect by reducing numbers of lung metastases [44]. It must be considered, however, that this positive benefit of LDI could be due to effective DSB repair related to in vivo immunological compensatory mechanisms that can remove mutated and abnormal cells. Furthermore, above 0.5 Gy, negative effects replace these positive effects of radiation.
Current techniques using ZFNs, TALENs, and CRISPR for HR with a donor vector have a low frequency of success, making them time-consuming, expensive, and inefficient to generate larger numbers of targeted lines. In this study, we showed that engineered nuclease-mediated HR was greatly enhanced by low-dose ionizing radiation. Indeed, using ZFNs with irradiation yielded a 51% targeting frequency compared with only 4% with ZFNs alone, demonstrating that LDI enhanced the frequency of correctly targeted clones. This similar enhancing effect was also observed in the TALEN and CRISPR systems. In addition, this enhancement of gene targeting was achieved using an equivalent dose of x-ray, which is a more practical approach for many laboratories. Moreover, LDI enhanced the gene-targeting frequency in a traditional gene-targeting system without the aid of engineered nucleases, demonstrating the power and broader application of this new approach. The DNA breaks caused by LDI can be repaired by both HR and NHEJ mechanisms. LDI-mediated induction of DNA repair machinery and availability of surplus copies of the transfected donor-targeting vector is likely to have favored HR over NHEJ. This notion is supported by the lack of significant genomic sequence change observed in irradiated pluripotent stem cells by exome sequencing. As demonstrated, the dosage of irradiation was a critical parameter for achieving efficient HR-mediated gene targeting. At higher irradiation doses, we observed apoptosis-mediated death of cells within 24 hours, possibly due to irreparable DSBs and activation of the apoptotic pathway. Moreover, irradiation-induced random NHEJ in essential protein coding regions or cis-acting DNA elements could be lethal, and those cells will be subjected to negative selection. The key is to have right amount of irradiation to stimulate the DNA repair machinery and provide a donor DNA vector in the right window to favor HR-mediated DNA repair.
We expect additional reasons for enhancing HR events by LDI. First, cellular DNA repair machinery was activated in response to both 0.4- and 4.0-Gy γ-irradiation. In particular, RAD51 activation has been shown to increase both gene-targeting frequency and DNA damage resistance [45]. Second, radiation-mediated increase in cell membrane permeability could allow efficient transfer of donor-targeting DNA to the nucleus for an HR event [46]. Third, cell cycle arrest caused by irradiation may allow more chance for DNA repair machinery to use the HR process because G(2) but not G(1) cell cycle arrest was reported in γ-irradiated human ESCs [36]. Our data also highlight that the combination of radiation and/or engineered nucleases can significantly enhance the HR frequency without increasing offsite mutations, causing global DNA damage, or significantly reducing cellular viability. Nonetheless, concerns remain about using ionizing radiation; therefore, careful assessment is ongoing for any subtle or long-term differences between the lines targeted with γ- and x-ray radiation compared with nonirradiation.
Whole-genome sequencing recently showed that engineering using TALENs or CRISPR systems did not increase offsite insertions or mutations [25–27]. Recently, however, a more sensitive method showed more than 10-fold off-target events and translocations between bona fide nuclease targets on homologous chromosomes [32]. Increases in the targeting efficiency of both on- and off-targets were dependent on the concentration of CRISPR or TALEN plasmids and the exact gene being targeted. Clearly, this area is complex; however, we suggest that combining CRISPR technology with LDI may allow lower plasmid concentrations while maintaining efficiency, thus decreasing offsite issues. Although we are currently testing this hypothesis, LDI is an important new method for exploring these types of options for everyone in the field doing gene targeting on human PSCs.
The ability to reliably and efficiently target specific genes within human PSCs removes a major roadblock for both disease-modeling studies and therapeutic strategies using hPSCs. The field of in vitro disease modeling will benefit from paired isogenic control cell lines developed by repairing a gene deficit associated with a specific disease and from reverse-engineered cell lines achieved by inserting specific mutations into control pluripotent stem cell lines [18]. In addition, efficient HR of reporter genes downstream of endogenous cell lineage promoters will facilitate the sorting of specific cell types and the identification of cells after transplantation. Finally, the correction of genetic defects by HR will revolutionize cell replacement therapies by permitting autologous transplantation of a patient's own corrected cells. The new, simple technique of combining low-dose radiation with current gene-editing technologies significantly enhances targeting frequency in human PSCs, providing powerful advancement of basic research and disease therapeutics.
Conclusion
We used low-dose irradiation to enhance homologous recombination frequency, possibly through induction of DNA repair machinery in human pluripotent stem cells. We found that both γ- and x-rays can be harnessed for gene-targeting purposes with or without engineered endonucleases. LDI did not introduce mutations in the human iPSCs, as evaluated by exome sequencing. AAVS1 locus-targeted human ESC and iPSC lines were karyotypically normal and made cell lineages of all three germ layers. This new, simple gene-targeting approach provides a powerful platform for basic research and cell therapeutics.
Supplementary Material
Acknowledgments
We thank Dr. James A. Thomson (Department of Cell and Regenerative Biology, University of Wisconsin, Madison, WI) for kindly providing a gene-targeting vector of the human HPRT gene. We thank J.C. Biancotti, D. Talavera, J. Ignatius Irudayam, L. Ornelas, and A. Sahabian for technical assistance; M. Dyer, B. Coullahan, X. Lin, M. Gallad, Z. Xu, and J. Ni of Life Technologies for help with the bioinformatics; and Q. Nguyen and L. Spurka for help with deep exome analysis. We thank W.R. Wilcox for the use of his x-ray unit. We acknowledge Drs. Soshana Svendsen and Oliver Smithies for critical review and editing of the manuscript. The work was supported by the National Institutes of Health (1U24NS078370 and UL1TR000124) and California Institute for Regenerative Medicine (RT2-02040 and RP1-05741) grants to C.N.S.
Author Contributions
S.H.: generation of reagents and performance of most of the experiments, design of experiments, data analysis, manuscript writing; A.S.: characterization of irradiated and gene-targeted hESCs and hiPSCs, manuscript writing; B.M., S.R., and D.S.: differentiation and analysis of gene-targeted hESCs and hiPSCs generated by radiation; H.W.K.: generation of targeting vector; J.T.: performance and analysis of mRNA expression in the irradiated hESCs; V.F.: performance and analysis of mRNA expression and whole exome sequencing in the irradiated hESCs; R.H.B.: performance and analysis of whole exome sequencing in the irradiated hESCs; V.A. and C.N.S.: design of experiments, data analysis, manuscript writing.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induced pluripotent stem cells in medicine and biology. Development. 2013;140:2457–2461. doi: 10.1242/dev.092551. [DOI] [PubMed] [Google Scholar]
- 5.Koller BH, Smithies O. Altering genes in animals by gene targeting. Annu Rev Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 6.Hatada S, Arnold LW, Hatada T, et al. Isolating gene-corrected stem cells without drug selection. Proc Natl Acad Sci USA. 2005;102:16357–16361. doi: 10.1073/pnas.0508263102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatada S, Nikkuni K, Bentley SA, et al. Gene correction in hematopoietic progenitor cells by homologous recombination. Proc Natl Acad Sci USA. 2000;97:13807–13811. doi: 10.1073/pnas.240462897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soldner F, Laganière J, Cheng AW, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choulika A, Perrin A, Dujon B, et al. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smih F, Rouet P, Romanienko PJ, et al. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 13.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 14.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Cho SW, Kim S, Kim JM, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 17.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Q, Lee YK, Schaefer EA, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Q, Regan SN, Xia Y, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokolov MV, Neumann RD. Human embryonic stem cell responses to ionizing radiation exposures: Current state of knowledge and future challenges. Stem Cells Int. 2012;2012:579104. doi: 10.1155/2012/579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choppin GR, Liljenzin J-O, Rydberg J. Radiation Biology and Radiation Protection. Radiochemistry and Nuclear Chemistry. 3rd ed. Oxford, U.K.: Butterworth-Heinemann; 2002. pp. 474–513. [Google Scholar]
- 24.Streffer C, Bolt H, Follesdal D, et al. Low Dose Exposures in the Environment: Dose-Effect Relations and Risk Evaluation (Ethics of Science and Technology Assessment). Vol 23. New York, NY: Springer, 2004. doi:10.1007/978-3-662-08422-9.
- 25.Veres A, Gosis BS, Ding Q, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki K, Yu C, Qu J, et al. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15:31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith C, Gore A, Yan W, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji J, Ng SH, Sharma V, et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 29.Samulski RJ, Zhu X, Xiao X, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou J, Mali P, Huang X, et al. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 32.Frock RL, Hu J, Meyers RM, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canitrot Y, Falinski R, Louat T, et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood. 2003;102:2632–2637. doi: 10.1182/blood-2002-10-3207. [DOI] [PubMed] [Google Scholar]
- 34.Plo I, Nakatake M, Malivert L, et al. JAK2 stimulates homologous recombination and genetic instability: Potential implication in the heterogeneity of myeloproliferative disorders. Blood. 2008;112:1402–1412. doi: 10.1182/blood-2008-01-134114. [DOI] [PubMed] [Google Scholar]
- 35.Wilson KD, Sun N, Huang M, et al. Effects of ionizing radiation on self-renewal and pluripotency of human embryonic stem cells. Cancer Res. 2010;70:5539–5548. doi: 10.1158/0008-5472.CAN-09-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momcilović O, Choi S, Varum S, et al. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells. 2009;27:1822–1835. doi: 10.1002/stem.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller FJ, Schuldt BM, Williams R, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertell R. No Immediate Danger: Prognosis for a Radioactive Earth. London, U.K.: Women’s Press; 1985. [Google Scholar]
- 39.Feinendegen LE, Pollycove M. Biologic responses to low doses of ionizing radiation: Detriment versus hormesis. Part 1. Dose responses of cells and tissues. J Nucl Med. 2001;42:17N–27N. [PubMed] [Google Scholar]
- 40.Tubiana M, Feinendegen LE, Yang C, et al. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology. 2009;251:13–22. doi: 10.1148/radiol.2511080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollycove M, Feinendegen LE. Biologic responses to low doses of ionizing radiation: Detriment versus hormesis. Part 2. Dose responses of organisms. J Nucl Med. 2001;42:26N–32N, 37N. [PubMed] [Google Scholar]
- 42.Russell WL, Kelly EM. Mutation frequencies in male mice and the estimation of genetic hazards of radiation in men. Proc Natl Acad Sci USA. 1982;79:542–544. doi: 10.1073/pnas.79.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilenchik MM, Knudson AG. Radiation dose-rate effects, endogenous DNA damage, and signaling resonance. Proc Natl Acad Sci USA. 2006;103:17874–17879. doi: 10.1073/pnas.0607995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosoi Y, Sakamoto K. Suppressive effect of low dose total body irradiation on lung metastasis: Dose dependency and effective period. Radiother Oncol. 1993;26:177–179. doi: 10.1016/0167-8140(93)90101-d. [DOI] [PubMed] [Google Scholar]
- 45.Yáñez RJ, Porter AC. Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 1999;6:1282–1290. doi: 10.1038/sj.gt.3300945. [DOI] [PubMed] [Google Scholar]
- 46.Stevens CW, Zeng M, Cerniglia GJ. Ionizing radiation greatly improves gene transfer efficiency in mammalian cells. Hum Gene Ther. 1996;7:1727–1734. doi: 10.1089/hum.1996.7.14-1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.