Abstract
Intracellular pattern-recognition receptors NOD1 and NOD2 are capable of sensing common structural units of bacterial walls. Recognition triggers specific immune signalling pathways and leads to pro-inflammatory cytokine upregulation and adequate immune response. We investigated whether two functional polymorphisms in NOD1 and NOD2 exert an effect on susceptibility to (STD patients) and severity of (female patients visiting the fertility clinic) Chlamydia trachomatis infection in 807 Dutch Caucasian women. A significant association of the NOD1 +32656 GG insertion variant with protection against infection with C. trachomatis has been detected [p: 0.0057; OR: 0.52]. When comparing C. trachomatis-positive women without symptoms to C. trachomatis-positive women with symptoms, and to C. trachomatis-positive women with TFI, we observed an increasing trend in carriage of the GG allele [Ptrend: 0.0003]. NOD2 1007fs failed to reveal an association. We hypothesize that the underlying mechanism might be a functional effect of the GG insertion on IFN-beta-dependent regulation of immune response in the genital tract. The research is part of an ongoing effort of identifying key polymorphisms that determine the risk of TFI and effectively translating them into the clinical setting for the purpose of optimizing diagnostic management of women at risk for developing TFI.
Keywords: Chlamydia trachomatis, NOD1, NOD2, polymorphism, host genetics, translational research
Investigation of whether two functional polymorphisms in NOD1 and NOD2 exert an effect on susceptibility to (STD patients) and severity of (female patients visiting the fertility clinic) C. trachomatis infection in 807 Dutch Caucasian women.
Investigation of whether two functional polymorphisms in NOD1 and NOD2 exert an effect on susceptibility to (STD patients) and severity of (female patients visiting the fertility clinic) C. trachomatis infection in 807 Dutch Caucasian women.
INTRODUCTION
Chlamydia trachomatis is the most common sexually transmitted bacterial infection, and it is strongly associated with tubal infertility (Brunham et al., 1985; Sellors et al., 1988; Mabey 2014). The course of C. trachomatis infection varies between individuals—only certain people are successfully infected and only part of those infected develops more severe disease as a result of an uncleared infection and prolonged accompanying inflammation (Kinnunen et al., 2002; Darville and Hiltke 2010). Substantial evidence for the contribution of variation of host immunogenetic factors in the clinical course of infection with this pathogenic bacterium has been accumulated, mainly in the last decade (Wang et al., 2005; den Hartog et al., 2006; Bailey et al., 2009; Morré, Ouburg and Pena 2009; Jiang et al., 2012; Al-Kuhlani et al., 2014). These factors appear at this point to be the most promising biological indicators of complicated infection with Chlamydiae (Ouburg et al., 2009; Lal et al., 2013; Malogajski et al., 2013; Branković, Malogajski and Morré 2014).
Nucleotide-binding oligomerization domain (NOD) receptors are intracellular proteins that participate in diverse innate immune processes (Proell et al., 2008). Receptors NOD1 and NOD2 were found to play a crucial role in priming immune responses to intracellular pathogens (Inohara et al., 2001; Chamaillard et al., 2003; Inohara and Nunez 2003), but also in initiating autophagy of infected cells, thereby demonstrating their multifaceted role in host defence (Cooney et al., 2010; Travassos et al., 2010; Homer et al., 2012). Research consistently shows evidence of the involvement of NOD receptors and their polymorphisms in a multitude of diseases—various infections, inflammatory bowel disease (IBD), asthma, as well as cancers (McGovern et al., 2005; Lu et al., 2010; Oosting et al., 2010; Wang et al., 2012). NOD1 (also known as CARD4) and NOD2 (CARD15) are the most extensively researched members of the NLR family (Correa, Milutinovic and Reed 2012). NOD1 recognizes gamma-d-glutamyl-meso-diaminopimelic acid, specific for peptidoglycans found predominantly in walls of Gram-negative bacteria (Chamaillard et al., 2003; Uehara et al., 2006). NOD2 is capable of sensing muramyl dipeptide, a minimal peptidoglycan motif ubiquitous in walls of both Gram-negative and Gram-positive bacteria, making it a more general pathogen detector (Chamaillard et al., 2003; Girardin et al., 2003). Both receptors signal the presence of lipopolysaccharides, found in outer membranes of Gram-negative bacteria (Inohara et al., 2001; Marriott et al., 2005). Expression of NOD2 is mainly observed in monocytes (Bonen and Cho 2003), while NOD1 appears to be active in a wider range of cell types that represent potential points of first entry for pathogens (Park et al., 2007; Kufer et al., 2008).
Activation of the pathogen recognition receptor (PRR) signalling pathways results in NF-κB activation and subsequent immune response (Takeuchi and Akira 2010). A number of studies have implicated the involvement of NOD1- and NOD2-induced immune responses in genital tract infection with C. trachomatis (Derbigny, Kerr and Johnson 2005; Opitz et al., 2005; Welter-Stahl et al., 2006). However, whether functional polymorphisms of NOD1 and NOD2 impact C. trachomatis infection and clinical course has hardly been explored. One study investigated the role of NOD2 1007fs polymorphism in tubal pathology upon C. trachomatis infection and reported a tendency for the heterozygotes; however, the numbers were too small and mutant homozygotes were not present among samples (den Hartog et al., 2006). NOD1 +32656 T > GG deletion–insertion polymorphism affects the receptor's recognition of bacterial motifs. Different studies associated each of its alleles with a diverse set of diseases. GG insertion variant appeared to confer higher susceptibility to erosive oesophagitis in Helicobacter pylori-infected individuals (Oikawa et al., 2012) and was associated with asthma in Australian individuals (Hysi et al., 2005). Deletion allele (T) was found to heighten the risk of IBD's early onset in British Caucasian population (McGovern et al., 2005).
To our knowledge, the role of the NOD1 +32656 T > GG and NOD2 1007fs polymorphisms in susceptibility to C. trachomatis infection and the severity, i.e. the risk of developing tubal factor infertility (TFI; i.e. Fallopian tube obstruction) has not been studied before. We hypothesize that NOD1 and NOD2 proteins might play a role in the course of C. trachomatis infections. To analyse this, we used two clinically well-defined Dutch Caucasian cohorts.
METHODS
Study population
Susceptibility cohort
Out of 1150 female patients visiting the STD outpatient clinic in Amsterdam, The Netherlands (between 2000 and 2002), we selected all Dutch Caucasian women (n = 727, age 18–30). Questionnaires were collected in regard to urogenital complaints, varying from increased discharge, having bloody discharge during and/or after coitus, recent lower abdominal pain (not gastrointestinal or menstruation-related) and/or dysuria. A flow diagram of the cohort used for the current study is presented in panel (A) of Fig. 1. The Medical Research Involving Human Subjects Act (WMO, Dutch Law) stated that official approval of the study by the Medical Ethical Committee does not apply to our anonymous human material collected (MEC Letter reference: # 10.17.0046). The local medical ethical committee also approved this study, based on the fact that in the Netherlands ethical approval is not required for a retrospective, de-identified clinical samples. Nevertheless, since we performed host genetic marker studies in relation to Chlamydial infection, we still made sure all participants signed informed consent forms.
Figure 1.
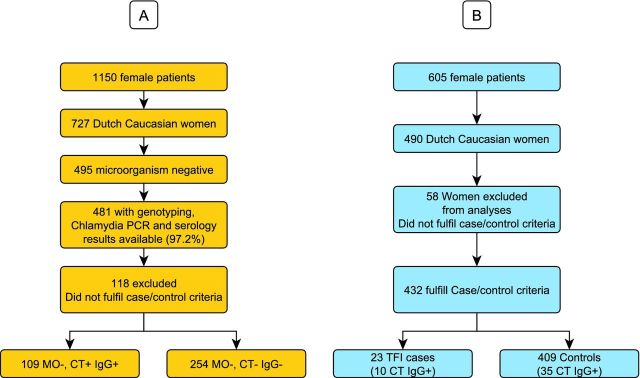
Flow-chart overview of the cohort selection, sample numbers and microbiology, C. trachomatis DNA analysis and IgG serology analysis. (A) Susceptibility cohort; (B) Severity cohort. [CT+ = positive for C. trachomatis DNA; CT− = negative for C. trachomatis DNA; IgG+ = positive for C. trachomatis-specific IgG antibodies; IgG− = negative for C. trachomatis-specific IgG antibodies; MO− = microorganism (N. gonorrhoeae, T. vaginalis, HSV1/2, C. albicans) negative; TFI+ = positive for TFI; TFI− = negative for TFI.]
Severity cohort
Our severity group consisted of 605 female patients visiting the fertility clinic in the University Medical Center Groningen (UMCG) between 2007 and 2012. In all women, Chlamydia IgG antibody testing (CAT) and hysterosalpingography and/or laparoscopy had been part of the fertility work-up. Data were retrospectively collected by chart review and anonymized for the researchers. Among the 605 subfertile patients (20–41years) 490 were Dutch Caucasian. Two independent investigators, who were unfamiliar with the CAT results, scored the laparoscopy reports to assess the grade of tubal pathology (0 = no abnormalities; 1 = any peritubal and/or periovarian adhesions and/or proximal or distal occlusion of at least one tube; 2 = extensive periadnexal adhesions and/or proximal or distal occlusion of at least one tube; 3 = extensive periadnexal adhesions and/or distal occlusion of at least one tube; 4 = extensive periadnexal adhesions and/or distal occlusion of both tube).
The hysterosalpingography results were scored as 0 = normal; 1 = unilateral occlusion proximal; 2 = unilateral occlusion distal; 3 = bilateral occlusion proximal; and 4 = bilateral occlusion distal. For the present study, TFI was defined as extensive peri-adnexal adhesions and/or distal occlusion of at least one tube at laparoscopy (Laparoscopy Grades 3 and 4). By applying this definition, 23 cases with TFI were identified. Controls were defined as women negative for TFI [as determined by laparoscopy (grade 0 and/or hysterosalpingography grade 0: n = 409)]. Women who did not fulfil the criteria or had missing data for cases and controls were excluded from the study (n = 58). A flow diagram of the cohort used for the current study is presented in panel (B) of Fig. 1.
In the Netherlands, no ethical board approval is required for retrospective chart review and collection of anonymized data. Couples attending the fertility clinic in the UMCG are informed about possible use of their anonymized data for research purposes, and a ‘no objection procedure’ is followed. Only patients who had not objected were included in the present study. Available clinical material (sera) from included patients was used. No additional or new clinical material was collected for the purpose of this study.
Laboratory analyses
Susceptibility cohort
A cervical swab was taken for the detection of C. trachomatis DNA using PCR. Peripheral venous blood was collected for the analysis of IgG antibodies against C. trachomatis. Infections with the microorganisms Candida albicans, Neisseria gonorrhoeae, Trichomonas vaginalis or herpes simplex virus (HSV) 1 or 2 may result in symptoms similar to C. trachomatis infection; therefore, the patients’ infection status for these microorganisms has been determined (van Doornum et al., 2001). Only the patient samples negative for all of these microorganisms were used in our analyses. Mycoplasma genitalium infection may result in similar symptoms; however, M. genitalium infections are relatively rare in the Netherlands and therefore not included in these analyses. In addition, C. trachomatis serovars were assessed in all C. trachomatis-positive samples, as described previously, to confirm true C. trachomatis positivity (Morré et al., 1998).
Severity cohort
Blood had been drawn from all patients for CAT. Chlamydia trachomatis IgG MOMP serology was determined by Medac C. trachomatis IgG p Elisa (Medac, Germany). Spare serum was cryopreserved.
Single nucleotide polymorphism detection
Two polymorphisms genotyped in this study were NOD1 T/GG (+32656 T > GG, partially identified as rs6958571) and NOD2 1007fs (rs2066847, SNP13, Leu1007fsinsC, 2936insC). We used an assay-by-design from Applied Biosystems to detect the NOD1 +32656 T > GG polymorphism. Detection of the NOD2 1007fs mutation was performed using real-time PCR under standard conditions.
Statistical methods
We defined the following groups.
Susceptibility
(1) Chlamydia trachomatis positive: women who are both C. trachomatis DNA and IgG positive; (2) Chlamydia trachomatis negative: women who are both C. trachomatis DNA and IgG negative; (3) As a subgroup of group 1: C. trachomatis positive with or without symptoms. In all groups women infected with other microorganisms were excluded from the analyses to generate clear groups for the association between symptoms or the lack of symptoms and the NOD SNs analysed.
Severity
(1) Chlamydia trachomatis positive and negative, respectively: CT IgG serology-positive or -negative women, respectively; (2) TFI positive and negative: TFI-positive or -negative women; (3) Chlamydia trachomatis positive with TFI: CT IgG serology-positive women with TFI; (4) Chlamydia trachomatis positive without TFI: CT IgG serology-positive women without TFI. Before conducting the severity analyses, the CT IgG positivity in cases and controls was assessed to show the expected relation between CT serology positivity and the severity of tubal pathology. Finally, trend analyses were performed to test the hypothesis the polymorphisms would show a trend when comparing CT+ women without symptoms to those with symptoms to those with TFI.
All groups were tested for Hardy–Weinberg equilibrium to check for Mendelian inheritance. Fisher's exact and χ2 tests were used where appropriate and P-values < 0.05 were considered statistically significant.
RESULTS
Genotype distributions were in Hardy–Weinberg equilibrium.
Full overview of genotype distributions for all the subgroups based on the C. trachomatis status and their presence or absence of symptoms are presented in Table 1.
Table 1.
Distribution of the NOD1 +32656 T > GG and NOD2 1007fs genotypes in Dutch Caucasian women (negative for tested non-C. trachomatis microogranisms) with or without clearly defined C. trachomatis infection (CT+ = positive for C. trachomatis DNA and C. trachomatis-specific antibodies; CT− = negative for C. trachomatis DNA and C. trachomatis-specific IgG antibodies; N = number of samples with both serological and genotyping results; S = symptomatic; AS = asymptomatic; TFI = TFI; sample numbers per SNP may differ, because genotypes could not be obtained for all samples.)
| NOD1 +32656 T > GG | NOD2 1007fs insC | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | n | T/T | % | T/GG | % | GG/GG | % | n | −/− | % | −/C | % | C/C | % |
| CT+ | 109 | 73 | 67 | 32 | 29 | 4 | 4 | 109 | 105 | 96 | 4 | 4 | 0 | 0 |
| CT+ S | 50 | 27 | 54 | 20 | 40 | 3 | 6 | 50 | 47 | 94 | 3 | 6 | 0 | 0 |
| CT+ AS | 59 | 46 | 78 | 12 | 20 | 1 | 2 | 59 | 58 | 98 | 1 | 2 | 0 | 0 |
| CT− | 254 | 130 | 51 | 115 | 45 | 9 | 4 | 254 | 245 | 96 | 9 | 4 | 0 | 0 |
| TFI+ | 21 | 10 | 48 | 10 | 48 | 1 | 4 | 23 | 23 | 100 | 0 | 0 | 0 | 0 |
| TFI− | 391 | 237 | 61 | 134 | 34 | 20 | 5 | 405 | 393 | 97 | 10 | 2 | 2 | 0 |
| CT+TFI+ | 9 | 5 | 56 | 4 | 44 | 0 | 0 | 10 | 10 | 100 | 0 | 0 | 0 | 0 |
| CT+TFI− | 33 | 19 | 58 | 13 | 39 | 1 | 3 | 35 | 35 | 100 | 0 | 0 | 0 | 00 |
Susceptibility
NOD1 +32656 T > GG
Carriage of the NOD1+32656 GG insertion was significantly lower in C. trachomatis-positive women (33%) compared to C. trachomatis-negative women (49%) [p: 0.0057, OR: 0.52, 95% CI: 0.32–0.83] (Table 1). Chlamydia trachomatis-positive women carrying the GG insertion were more likely to have symptoms [p: 0.013, OR: 3.01, 95% CI: 1.32–6.91]. Homozygous GG carriage increased risk of symptoms [OR: 3.70] in CT-positive women, although this did not reach statistical significance.
NOD2 1007fs
Carriage of the NOD2 1007fs polymorphism did not show a significant association with susceptibility to C. trachomatis infections (Table 1). Carrying the C insertion allele and symptoms in C. trachomatis-positive women showed a risk effect similar to that observed in NOD1 *GG carriage [OR: 3.70]; although this did not reach statistical significance.
Severity
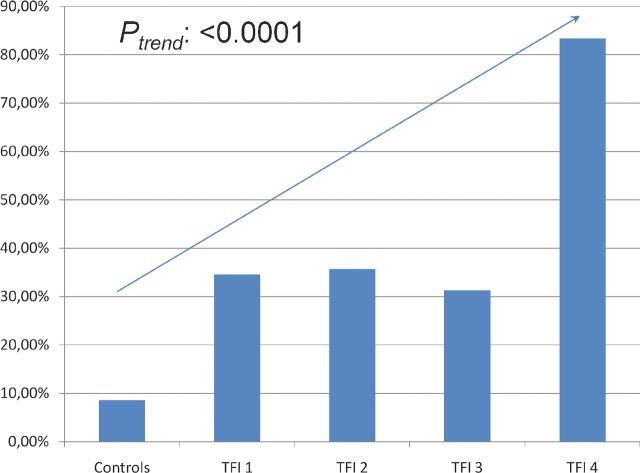
Presence of C. trachomatis IgG antibodies significantly increased in more severe TFI [Fig. 2; Ptrend: <0.0001], confirming the expected relation between past C. trachomatis infections and development of tubal pathology.
Figure 2.
Distribution of C. trachomatis positivity in controls and TFI 1–4 groups (with increasing severity of TFI). In all other analyses in this study, TFI was defined as extensive peri-adnexal adhesions and/or distal occlusion of at least one tube at laparoscopy (Laparoscopy Grades 3 and 4).
NOD1 +32656 T > GG
Carriage of the NOD1+32656 GG insertion was significantly more frequent in women diagnosed with TFI (59,4%) compared to women negative for TFI (39,4%) [p: 0.0383, OR: 2.25, CI: 1.08–4.69].
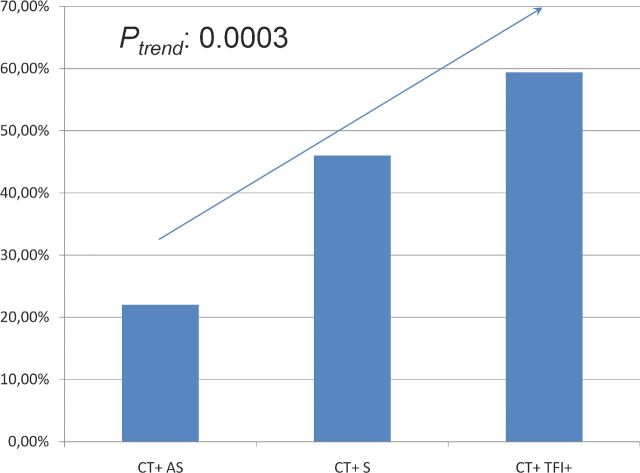
When comparing C. trachomatis-positive women without symptoms to C. trachomatis-positive women with symptoms, and to C. trachomatis-positive women with TFI, we observed an increasing trend in carriage of the NOD1 GG allele (Fig. 3; Ptrend: 0.0003).
Figure 3.
Carriage of the NOD1 *GG allele in C. trachomatis-positive women without symptoms (AS), with symptoms (S) and with TFI.
DISCUSSION
Our results show a significantly reduced carriage of the NOD1 +32656 GG insertion allele in C. trachomatis-positive women compared to the C. trachomatis-negative women, indicating a protective effect against C. trachomatis infections. There was a significant association of GG carriage with the occurrence of symptoms during an infection. Presence of symptoms was also significantly more frequently seen in NOD1 GG homozygotes among C. trachomatis-positive women. The NOD2 C insertion allele and symptoms in C. trachomatis-positive women showed the same direction of association.
Analysis of our severity cohort revealed, however, a deleterious effect of the GG insertion polymorphism in patients with TFI. Unlike in the susceptibility cohort, it was significantly more frequent in C. trachomatis-specific IgG-positive women diagnosed with TFI compared to women negative for C. trachomatis-specific IgG antibodies. NOD2 1007fs did not show associations with either susceptibility to C. trachomatis infection or the subsequent TFI in our study.
Our results contribute to the incomplete body of knowledge on the role of NOD1 receptor in infections with C. trachomatis. This is, to our knowledge, the first study of the effects of NOD1 +32656 T > GG in C. trachomatis. Despite the fact that several studies confirmed that NOD1 indeed senses and induces cellular changes in response to this pathogen, precise mechanisms and scope of its involvement in this particular infection are not fully understood (Welter-Stahl et al., 2006). It has been established that an upregulation of pro-inflammatory cytokine genes upon Chlamydiae infection occurs in mice primary fibroblasts, but not in those from NOD1-deficient mice. The study did not confirm that the presence of a functional NOD1 receptor also increased cytokine secretion in C. trachomatis infection (Welter-Stahl et al., 2006). Opitz and colleagues demonstrated that C. pneumoniae induced a NOD1 (and NOD2)-mediated NF-κB activation in HEK293 cells. In endothelial cells, NOD1 played a dominant role in triggering a C. pneumoniae-mediated inflammatory process (Opitz et al., 2005). Opitz et al., demonstrated that infection with both active and heat-inactivated C. pneumoniae resulted in increased NOD1-associated RIP2 expression, which induces NF-κB expression, and that NOD1 induced cytokine expression after C. pneumoniae infection is abolished with NOD1-specific siRNAs. These results indicate that NOD1 is involved in Chlamydia infections (Opitz et al., 2005). Another study has shown that NOD1 and NOD2 are expressed in a cloned murine fallopian tube epithelial cell line (Derbigny, Kerr and Johnson 2005). What lacked so far are studies investigating the exact associations of different functional NOD polymorphisms in relation to infection with Chlamydiae.
In vitro studies of NOD1 recognition of pathogenic bacteria and subsequent cytokine induction point to its role in maintaining intestinal defence against invasive Gram-negative species, but their precise role is not always as clear in in vivo models (Kim, Lee and Kagnoff 2004; Travassos et al., 2005).
NOD1 has been identified as the maximal inducer of the secretion of interferon-beta (IFN-beta), typical of murine genital C. muridarum infection (Prantner, Darville and Nagarajan 2010). Research on the effect of IFN-beta shows its pleiotropic nature and variability across different bacterial infections and in different tissues. The effect of IFN-beta in Chlamydia-infected mice is, however, adverse—secretion of IFN-beta results in the inhibition of Chlamydia-specific CD4 T-cell responses and the delay of C. muridarum clearance (Nagarajan et al., 2008). Type I interferons (IFN-alpha and IFN-beta) were also demonstrated to enhance susceptibility to Listeria monocytogenes (Auerbuch et al., 2004; Carrero, Calderon and Unanue 2004; O'Connell et al., 2004). However, in a study of a lung model infected with C. pneumoniae, IFN-beta appeared to be acting synergistically with IFN-gamma and helped resolve the infection (Rothfuchs et al., 2006). Similar was found in H. pylori infection in a murine model, where IFN-beta enhanced Th1 differentiation as a result of NOD1 stimulation, which in turn led to a heightened IFN-gamma secretion (Watanabe et al., 2010).
Hence, the protective effect of the NOD1 +32656 GG insertion variant on susceptibility to C. trachomatis encountered in our study might be consistent with the findings of these studies. Insertion would then likely disrupt the native functionality of NOD1, leading to diminished IFN-beta secretion. This would prevent its inhibition of Chlamydia-specific T-cell responses, thereby resulting in an effective immune reaction to the infection and a more rapid clearance. Specific IFN-gamma-producing T-cells can successfully clear C. trachomatis infection (Miyairi, Ramsey and Patton 2010); hence, it is relevant how a NOD1-dependent immune response might influence this type of immune response. Conversely, in women with TFI, a full-fledged immune reaction in the upper reproductive tract may damage the tubes; therefore, the ascended spreading of C. trachomatis in the reproductive tract would put the female GG allele carriers in a greater risk of TFI. This would explain how a same variant may lead to different consequences of the infection in lower and upper genital tract. Higher occurrence of symptoms in carriers of the GG insertion would also be consistent with this hypothesis—a more elaborate immune reaction to C. trachomatis would manifest in the form of more pronounced symptoms. Also, the fact that different CD4 T-cell subsets take dominance in different parts of the genital tract in response to C. trachomatis infections may also be contributing to the diversity in outcomes of the infection (Marks, Tam and Lycke 2010). A number of studies used to construct our hypothesis has however been done on murine models. Further research on the roles of the NOD1 +32656 T > GG polymorphism, IFN-beta secretion, specific T-cell function in relation to human urogenital C. trachomatis information is therefore needed.
Moreover, NOD1 +32656 GG insertion appears to not only alter the sensing capacity of NOD1, it auto-modulates the translation of the protein, with GG/GG carriers expressing much lower NOD1 levels compared to T/T carriers (Oikawa et al., 2012). Considering the proposed effect of NOD1-induced IFN-beta secretion on C. trachomatis infection in urogenital tract, the insertion would therefore exert protection against the infection via two mechanisms. This warrants further studies on the NOD1-induced immunity in response to urogenital C. trachomatis.
The fact that Chlamydia and NOD1 are in different compartments, in the inclusion and in the cytosol, respectively, is in contrast to our results and hypothesis. Currently no direct binding between Chlamydia and NOD has been proven. Therefore, it might be hypothesized that Chlamydia indirectly activates NOD. Degradation of host proteins by the chlamydial protease- or proteasome-like activity factor (CPAF) may result in the formation of danger-associated molecular patterns which could then activate the NOD-like receptor inflammasome (Bauernfeind and Hornung 2013; Wen, Miao and Ting 2013). This hypothesis might explain the observed association between NOD and Chlamydia infections. Recent studies however have demonstrated that CPAF does not cleave as many host proteins as previously expected, and that CPAF may be limited to the inclusion and not secreted into the host cytosol (Chen et al., 2012; Bavoil and Byrne 2014; Snavely et al. 2014). These results indicate that the actual biological mechanisms through which Chlamydia is associated with NOD remains to be elucidated.
Analysis of NOD2 1007fs in susceptibility to and severity of C. trachomatis infection did not demonstrate an association in our study. Therefore, we did not manage to confirm the assumption that this functional polymorphism profoundly affects the C. trachomatis sensing capabilities. Den Hartog and colleagues observed a trend of NOD2 1007fs carriage in tubal pathology patients, but the sample numbers were too small for statistical associations (den Hartog et al., 2006).
NOD2 mutations resulting in truncated Leucine Rich Repeats (LRR) regions lead to Crohn's disease, arguably due to the impaired sensing of pathogens and disruption in the intestinal protection barrier, followed by invasion of the intestinal lining and prolonged inflammation, leading into IBDs (Li et al., 2004; Vignal et al., 2007). The inability of monocytes to register pathogens might ultimately result in an exaggerated adaptive response (Ogura et al., 2001). However, how the mutations in the LRR-coding section affect their carriers in the presence of C. trachomatis infection has not been extensively studied to date.
It should be noted that the NOD2 1007fs mutant allele is rare in the general population. In our susceptibility cohort, there were no patients homozygous for this allele, and only 4% heterozygotes were observed. The severity cohort had only 0.49% mutant homozygotes. We did not observe any effects of mutation carriage when comparing C. trachomatis cases versus controls, nor in comparisons based on the presence or absence of symptoms within these two groups. Providing the NOD2 1007fs mutation was recessive in its nature, the absence of any observed effect in a C. trachomatis-positive cohort that lacks the homozygotes would not be surprising, since it could be compensated in heterozygote's phenotype by the wild-type allele. However, several previous studies have found that the compound heterozygote carriage for this and one other polymorphism (SNP12) resulted in a heightened risk of developing Crohn's disease (Hugot et al., 2001; Heresbach et al., 2004). Even though we failed to confirm the association of NOD2 1007fs in our study, we encourage further research involving a larger study group, preferably with more other polymorphisms analysed simultaneously.
Given the partially converging roles of NOD1 and NOD2 in sensing certain pathogens, it can be useful to consider investigating whether combinations of their polymorphisms exhibit synergistic effects. Therefore, a greater number of polymorphisms would need to be researched. There is a potential for implementing the knowledge on these factors and using them as genetic traits in order to improve individual risk predictions by existing clinical models, as part of the paradigm shift within health care towards personalized medicine (Harvey et al., 2012; Lal et al., 2013; Malogajski et al., 2013). More work in this field, however, still has to be done. A study by Sanders and colleagues (2013) increased the predictive capacities of a post-meningitis hearing loss prediction model by adding a number of single nucleotide polymorphisms (SNPs) in different PRR genes (Sanders et al., 2013). Women carrying two or more SNPs in PRR genes involved in recognizing that C. trachomatis show more than 2-fold higher risk of developing tubal pathology after C. trachomatis infection compared to women with less than two PRR SNPs (den Hartog et al., 2006). The studies show the potential of host genetic markers as indicators of risk of complication from C. trachomatis infection in women. Host genetic markers could be applied for improving clinical management of women at risk and making more salient decisions on which patients to refer to laparoscopies for diagnosing tubal pathology, which are invasive and costly diagnostic procedures.
In order to identify immunogenetic factors with strongest predictive value, next-generation sequencing and genome-wide association studies (GWAS) are a favourable alternative and can be used next to the candidate gene approach. New mouse models (Su et al., 2014) and new analysis methods such as Pathway of Distinction Analysis (PODA) (Roberts et al., 2014) will help identify novel genes linked to the susceptibility to and severity of C. trachomatis disease.
Further research on the effects of common SNPs in PRR receptors in the risk of tubal damage and TFI as a result of uncleared C. trachomatis infection may be a step towards successful advances in personalized medicine. Improving diagnostic protocols for subfertility by enabling risk group stratification of women at the highest risk of complications due to persistent genital C. trachomatis is a promising direction for a future development, aimed at addressing important health needs.
In conclusion, the NOD1 +32656 GG insertion variant appears to protect against the infection with C. trachomatis, whereas it acts as a risk factor in developing TFI in women with a past C. trachomatis infection. Also, the GG insertion leads to a higher occurrence of symptoms in C. trachomatis-positive women.
Our finding supports the involvement of NOD1 in C. trachomatis recognition and subsequent responses. Taking into account previously published research, it appears that NOD1 has a vital role in bacterial sensing in humans and other species, but specific actions elicited by each of these receptors should preferably be examined for different bacterial species or groups separately. Variations in these pathogen-specific roles that might be observed in different PRR receptors should not be surprising, considering the known diversity of bacterial mechanisms of pathophysiology. NOD receptors are evidently important for recognizing those pathogenic bacteria that manage to surpass the extracellular or endosomal TLR sensing, as well as obligatory intracellular bacteria, such as C. trachomatis. Diversity of PRR receptors enables not only a greater spectrum of sensing, but also better fine-tuning of immune responses, establishing the basis for specific, effective defence.
Acknowledgments
The authors want to thank Ing. Jolein Pleijster (Laboratory of Immunogenetics, Department of Medical Microbiology and Infection Control, VU University Medical Center, Amsterdam, The Netherlands) for excellent technical laboratory analyses. They also want to thank the Public Health Laboratory, Cluster Infectious Diseases, Public Health Service Amsterdam, Amsterdam, the Netherlands) for the STD cohort included in this study and Prof.dr. J.A. Land for the tuba factor infertility cohort enrolled in this study.
FUNDING
Partial funding was obtained from or the work was in line with the following grants: Dutch NGI Life Sciences Pre-Seed Grant Reg.Nr. 93611006, USA NIH R21 Grant Prime award number 1R21AI.098660-01 with subaward number 0025996 (120407-1) and the European EuroTransBio Grant, Reference number 110012 ETB.
Conflict of interest statement. None declared.
REFERENCES
- Al-Kuhlani M, Rothchild J, Pal S, et al. TRAIL-R1 is a negative regulator of pro-inflammatory responses and modulates long-term sequelae resulting from Chlamydia trachomatis infections in humans. PLoS ONE. 2014;9:e93939. doi: 10.1371/journal.pone.0093939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, et al. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–33. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RL, Natividad-Sancho A, Fowler A, et al. Host genetic contribution to the cellular immune response to Chlamydia trachomatis: heritability estimate from a Gambian twin study. Drugs Today (Barcelona) 2009;45(Supplement B):45–50. [PubMed] [Google Scholar]
- Bauernfeind F, Hornung V. Of inflammasomes and pathogens—sensing of microbes by the inflammasome. EMBO Mol Med. 2013;5:814–26. doi: 10.1002/emmm.201201771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoil PM, Byrne GI. Analysis of CPAF mutants: new functions, new questions (the ins and outs of a chlamydial protease) Pathog Dis. 2014;71:287–91. doi: 10.1111/2049-632X.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen DK, Cho JH. The genetics of inflammatory bowel disease. Gastroenterology. 2003;124:521–36. doi: 10.1053/gast.2003.50045. [DOI] [PubMed] [Google Scholar]
- Branković I, Malogajski J, Morré SA. Biobanking and translation of human genetics and genomics for infectious diseases. Appl Translational Genom. 2014;3:30–5. doi: 10.1016/j.atg.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham RC, Maclean IW, Binns B, et al. Chlamydia trachomatis: its role in tubal infertility. J Infect Dis. 1985;152:1275–82. doi: 10.1093/infdis/152.6.1275. [DOI] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200:535–40. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–7. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Chen AL, Johnson KA, Lee JK, et al. CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Pathog. 2012;8:e1002842. doi: 10.1371/journal.ppat.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Correa RG, Milutinovic S, Reed JC. Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in innate immunity and inflammatory diseases. Bioscience Rep. 2012;32:597–608. doi: 10.1042/BSR20120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville T, Hiltke TJ. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J Infect Dis. 2010;201:S114–25. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog J, Ouburg S, Land J, et al. Do host genetic traits in the bacterial sensing system play a role in the development of Chlamydia trachomatis-associated tubal pathology in subfertile women. BMC Infect Dis. 2006;6:122. doi: 10.1186/1471-2334-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbigny WA, Kerr MS, Johnson RM. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J Immunol. 2005;175:6065–75. doi: 10.4049/jimmunol.175.9.6065. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–72. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Harvey A, Brand A, Holgate ST, et al. The future of technologies for personalised medicine. New Biotechnol. 2012;29:625–33. doi: 10.1016/j.nbt.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Heresbach D, Gicquel-Douabin V, Birebent B, et al. NOD2/CARD15 gene polymorphisms in Crohn's disease: a genotype–phenotype analysis. Eur J Gastroen Hepat. 2004;16:55–62. doi: 10.1097/00042737-200401000-00009. [DOI] [PubMed] [Google Scholar]
- Homer CR, Kabi A, Marina-Garcia N, et al. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J Biol Chem. 2012;287:25565–76. doi: 10.1074/jbc.M111.326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Hysi P, Kabesch M, Moffatt MF, et al. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14:935–41. doi: 10.1093/hmg/ddi087. [DOI] [PubMed] [Google Scholar]
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–82. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Chen FF, et al. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem. 2001;276:2551–4. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- Jiang J, Karimi O, Ouburg S, et al. Interruption of CXCL13-CXCR5 axis increases upper genital tract pathology and activation of NKT cells following chlamydial genital infection. PLoS ONE. 2012;7:e47487. doi: 10.1371/journal.pone.0047487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Lee SJ, Kagnoff MF. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by Toll-like receptors. Infect Immun. 2004;72:1487–95. doi: 10.1128/IAI.72.3.1487-1495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen AH, Surcel HM, Lehtinen M, et al. HLA DQ alleles and interleukin-10 polymorphism associated with Chlamydia trachomatis-related tubal factor infertility: a case-control study. Hum Reprod. 2002;17:2073–8. doi: 10.1093/humrep/17.8.2073. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Kremmer E, Adam AC, et al. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008;10:477–86. doi: 10.1111/j.1462-5822.2007.01062.x. [DOI] [PubMed] [Google Scholar]
- Lal JA, Malogajski J, Verweij SP, et al. Chlamydia trachomatis infections and subfertility: opportunities to translate host pathogen genomic data into public health. Public Health Genomi. 2013;16:50–61. doi: 10.1159/000346207. [DOI] [PubMed] [Google Scholar]
- Li J, Moran T, Swanson E, et al. Regulation of IL-8 and IL-1beta expression in Crohn's disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–25. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- Lu WG, Zou YF, Feng XL, et al. Association of NOD1 (CARD4) insertion/deletion polymorphism with susceptibility to IBD: a meta-analysis. World J Gastroentero. 2010;16:4348–56. doi: 10.3748/wjg.v16.i34.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabey D. Epidemiology of sexually transmitted infections: worldwide. Medicine. 2014;42:287–90. [Google Scholar]
- Malogajski J, Brankovic I, Verweij SP, et al. Translational potential into health care of basic genomic and genetic findings for human immunodeficiency virus, Chlamydia trachomatis, and human papilloma virus. Biomed Res Int. 2013;2013:892106. doi: 10.1155/2013/892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks E, Tam MA, Lycke NY. The female lower genital tract is a privileged compartment with IL-10 producing dendritic cells and poor Th1 immunity following Chlamydia trachomatis infection. PLoS Pathog. 2010;6:e1001179. doi: 10.1371/journal.ppat.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott I, Rati DM, McCall SH, et al. Induction of Nod1 and Nod2 intracellular pattern recognition receptors in murine osteoblasts following bacterial challenge. Infect Immun. 2005;73:2967–73. doi: 10.1128/IAI.73.5.2967-2973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Hysi P, Ahmad T, et al. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum Mol Genet. 2005;14:1245–50. doi: 10.1093/hmg/ddi135. [DOI] [PubMed] [Google Scholar]
- Miyairi I, Ramsey KH, Patton DL. Duration of untreated chlamydial genital infection and factors associated with clearance: review of animal studies. J Infect Dis. 2010;201:S96–S103. doi: 10.1086/652393. [DOI] [PubMed] [Google Scholar]
- Morré SA, Ossewaarde JM, Lan J, et al. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J Clin Microbiol. 1998;36:345–51. doi: 10.1128/jcm.36.2.345-351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré SA, Ouburg S, Pena AS, et al. The EU FP6 EpiGenChlamydia Consortium: contribution of molecular epidemiology and host–pathogen genomics to understanding Chlamydia trachomatis-related disease. Drugs Today (Barcelona) 2009;45:7–13. [PubMed] [Google Scholar]
- Nagarajan UM, Prantner D, Sikes JD, et al. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun. 2008;76:4642–8. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya SA, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–45. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Asano N, Imatani A, et al. Gene polymorphisms of NOD1 and interleukin-8 influence the susceptibility to erosive esophagitis in Helicobacter pylori infected Japanese population. Hum Immunol. 2012;73:1184–9. doi: 10.1016/j.humimm.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Oosting M, Berende A, Sturm P, et al. Recognition of Borrelia burgdorferi by NOD2 s central for the induction of an inflammatory reaction. J Infect Dis. 2010;201:1849–58. doi: 10.1086/652871. [DOI] [PubMed] [Google Scholar]
- Opitz B, Förster S, Hocke AC, et al. Nod1-mediated endothelial cell activation by Chlamydophila pneumoniae. Circ Res. 2005;96:319–26. doi: 10.1161/01.RES.0000155721.83594.2c. [DOI] [PubMed] [Google Scholar]
- Ouburg S, Lyons JM, Land JA, et al. TLR9 KO mice, haplotypes and CPG indices in Chlamydia trachomatis infection. Drugs Today (Barcelona) 2009;45:83–93. [PubMed] [Google Scholar]
- Park JH, Kim YG, Shaw M, et al. Nod1/RICK and TLR signaling regulate chemokine and antimicrobial innate immune responses in mesothelial cells. J Immunol. 2007;179:514–21. doi: 10.4049/jimmunol.179.1.514. [DOI] [PubMed] [Google Scholar]
- Prantner D, Darville T, Nagarajan UM. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol. 2010;184:2551–60. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, et al. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS ONE. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CH, Franklin C, Molina-Gonzales S, et al. In: A genome wide association scan reveals pathway-wide genomic differences between cases of scarring trachoma and controls. Schachter J, Byrne GI, Chernesky MA, et al., editors. Pacific Grove, San Francisco, CA, USA: International Chlamydia Symposium; June 22–27, 2014. pp. 501–4. Paper presented at the Thirteenth International Symposium on Human Chlamydial Infections. [Google Scholar]
- Rothfuchs AG, Trumstedt C, Mattei F, et al. STAT1 regulates IFN-alpha beta- and IFN-gamma-dependent control of infection with Chlamydia pneumoniae by nonhemopoietic cells. J Immunol. 2006;176:6982–90. doi: 10.4049/jimmunol.176.11.6982. [DOI] [PubMed] [Google Scholar]
- Sanders M, de Jonge R, Terwee C, et al. Addition of host genetic variants in a prediction rule for post meningitis hearing loss in childhood: a model updating study. BMC Infect Dis. 2013;13:340. doi: 10.1186/1471-2334-13-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellors JW, Mahony JB, Chernesky MA, et al. Tubal factor infertility: an association with prior chlamydial infection and asymptomatic salpingitis. Fertil Steril. 1988;49:451–7. doi: 10.1016/s0015-0282(16)59772-6. [DOI] [PubMed] [Google Scholar]
- Snavely EA, Kokes M, Dunn JD, et al. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis. 2014;71:336–51. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Wang X, Jones E, et al. In: Host genetics and upper genital tract disease in Chlamydia muridarum infected mice: a forward genetic approach with translational implications. Schachter J, Byrne GI, Chernesky MA, et al., editors. Pacific Grove, San Francisco, CA, USA: International Chlamydia Symposium; June 22–27, 2014. pp. 261–4. Paper presented at the Thirteenth International Symposium on Human Chlamydial Infections. [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LAM, Girardin SE, et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem. 2005;280:36714–8. doi: 10.1074/jbc.M501649200. [DOI] [PubMed] [Google Scholar]
- Uehara A, Fujimoto Y, Kawasaki A, et al. Meso-diaminopimelic acid and meso-lanthionine, amino acids specific to bacterial peptidoglycans, activate human epithelial cells through NOD1. J Immunol. 2006;177:1796–804. doi: 10.4049/jimmunol.177.3.1796. [DOI] [PubMed] [Google Scholar]
- van Doornum GJJ, Schouls LM, Pijl A, et al. Comparison between the LCx Probe System and the COBAS AMPLICOR System for detection of Chlamydia trachomatis and Neisseria gonorrhoeae infections in patients attending a clinic for treatment of sexually transmitted diseases in Amsterdam, The Netherlands. J Clin Microbiol. 2001;39:829–35. doi: 10.1128/JCM.39.3.829-835.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignal C, Singer E, Peyrin-Biroulet L, et al. How NOD2 mutations predispose to Crohn's disease. Microbes Infect. 2007;9:658–63. doi: 10.1016/j.micinf.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Wang C, Tang J, Geisler WM, et al. Human leukocyte antigen and cytokine gene variants as predictors of recurrent Chlamydia trachomatis infection in high-risk adolescents. J Infect Dis. 2005;191:1084–92. doi: 10.1086/428592. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhang L, Jiang JM, et al. Association of NOD1 and NOD2 genes polymorphisms with Helicobacter pylori related gastric cancer in a Chinese population. World J Gastroentero. 2012;18:2112–20. doi: 10.3748/wjg.v18.i17.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Fichtner-Feigl S, et al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. 2010;120:1645–62. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter-Stahl L, Ojcius DM, Viala J, et al. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cell Microbiol. 2006;8:1047–57. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013;39:432–41. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]