Abstract
BACKGROUND
Large epidemiologic studies support the role of dyslipidemia in preeclampsia; however, the etiology of preeclampsia or whether dyslipidemia plays a causal role remains unclear. We examined the association between the genetic predisposition to dyslipidemia and risk of preeclampsia using validated genetic markers of dyslipidemia.
METHODS
Preeclampsia cases (n = 164) and normotensive controls (n = 110) were selected from live birth certificates to nulliparous Iowa women during the period August 2002 to May 2005. Disease status was verified by medical chart review. Genetic predisposition to dyslipidemia was estimated by 4 genetic risk scores (GRS) (total cholesterol (TC), LDL cholesterol (LDL-C), HDL cholesterol (HDL-C), and triglycerides) on the basis of established loci for blood lipids. Logistic regression analyses were used to evaluate the relationships between each of the 4 genotype scores and preeclampsia. Replication analyses were performed in an independent, US population of preeclampsia cases (n = 516) and controls (n = 1,097) of European ancestry.
RESULTS
The GRS related to higher levels of TC, LDL-C, and triglycerides demonstrated no association with the risk of preeclampsia in either the Iowa or replication population. The GRS related to lower HDL-C was marginally associated with an increased risk for preeclampsia (odds ratio (OR) = 1.03, 95% confidence interval (CI) = 0.99–1.07; P = 0.10). In the independent replication population, the association with the HDL-C GRS was also marginally significant (OR = 1.03, 95% CI: 1.00–1.06; P = 0.04).
CONCLUSIONS
Our data suggest a potential effect between the genetic predisposition to dyslipidemic levels of HDL-C and an increased risk of preeclampsia, and, as such, suggest that dyslipidemia may be a component along the causal pathway to preeclampsia.
Keywords: blood pressure, dyslipidemia, genetic epidemiology, genetic risk score, hypertension, preeclampsia.
Preeclampsia is a multisystem disorder of pregnancy characterized by de novo hypertension and proteinuria with onset after 20 weeks gestation.1 In the absence of proteinuria, de novo hypertension with new onset of any of the following will also qualify a woman for a diagnosis of preeclampsia: thrombocytopenia, renal insufficiency, impaired liver function, pulmonary edema, or cerebral/visual disturbances.2 Complicating approximately 3% of all deliveries in the United States,3,4 preeclampsia is a leading cause of maternal and infant morbidity and mortality worldwide.5,6 Without intervention, the mother is at substantial risk for seizures (eclampsia), renal and liver failure, stroke, and death.4 In the infant, preeclampsia increases the risk for intrauterine growth restriction, prematurity, and perinatal death.5 Despite considerable research, there are no clinically useful screening tools for identifying women in whom preeclampsia will develop1; the only “cure” is delivery.5
Evidence of the heritability of preeclampsia is apparent. Family studies have demonstrated that preeclampsia is more common among daughters and sisters of previously preeclamptic women.7–12 However, the underlying genetics are complex, and the extent to which genes and pathways contribute to preeclampsia remains unknown.13 Numerous candidate gene studies and a few genome-wide association studies (GWAS) have been performed to identify genetic markers of preeclampsia.13 A recent meta-analysis reported 7 variants in 6 genes that were significantly associated with preeclampsia. Some of these variants are also independent genetic risk factors for dyslipidemia and are involved in lipid metabolism.13 One identified genetic variant, rs268 of the LPL gene, is also associated with reduced lipoprotein lipase activity and dyslipidemia.14 Because dyslipidemia can play a role in endothelial cell dysfunction,15 a known mechanism in the pathophysiology of preeclampsia, evaluating the genetic components of lipid metabolism, and of dyslipidemia specifically, may help to elucidate the pathophysiology of preeclampsia.
A recent meta-analysis of 46 lipid GWAS comprising more than 100,000 individuals of European descent established a comprehensive genetic profile of 95 loci for various blood lipids.16 In 2013, this meta-analysis was updated with an additional 62 loci.17 In the current study, we calculated 4 genetic risk scores (GRS) on the basis of the well-established single nucleotide polymorphisms (SNPs) for total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglycerides as proxies for the genetic predisposition to dyslipidemia. We then evaluated the effects of these dyslipidemia GRS on the risk of preeclampsia. To our knowledge, this is the first study to assess the relationship between a GRS for dyslipidemia and risk for preeclampsia.
METHODS
SOPHIA
Study population and phenotype definition.
The Study of Pregnancy Hypertension in Iowa, SOPHIA, is a population-based case-control study designed to examine the roles of maternal-fetal human leukocyte antigen sharing and sexual history with the baby’s father in preeclampsia. A detailed description of this study has been described elsewhere.18–20 Briefly, 3,078 primaparous women who resided in one of 42 Iowa counties and delivered a live, singleton birth from August 2002 to May 2005 were identified from electronic birth certificates. Final case–control status was determined based on computer-assisted telephone interviews and review of prenatal and hospital records.
Based on information abstracted from the telephone interviews and medical charts, 274 preeclampsia cases and 190 normotensive controls were ascertained. DNA samples belonging to Caucasian women who consented to future genetic studies with sufficient DNA were genotyped and analyzed in a GWAS (n = 177 preeclampsia cases and n = 116 normotensive controls).20
Preeclampsia was defined according to National Heart, Lung, and Blood Institute guidelines: (i) sustained de novo hypertension (systolic blood pressure (BP) ≥140 mmHg or diastolic BP ≥90 mmHg) on at least 2 occasions at least 6h apart after the 20th week of pregnancy, and (ii) accompanying proteinuria defined as a urine protein concentration ≥300mg/l (equivalent to a 24-h urine sample containing >300mg of protein; a urine sample concentration of ≥30mg/dl urinary protein or dipstick protein test value of 1+ or greater based on 2 or more samples collected at least 4h apart; 1 or more urinary dipstick value of 2+ near the end of pregnancy; or 1 or more catheterized dipstick value of 1+ during delivery hospitalization).21 Only women who had no evidence of hypertension or proteinuria during their pregnancy were included as normotensive controls.
GWAS genotyping and imputation.
The genotyping methods have been described in detail elsewhere.20 Briefly, buccal cell DNA was extracted from cytobrush samples using Puregene DNA tissue kits (Gentra Systems, Minneapolis, MN) following the manufacturer’s protocol with minor modifications.22 After extraction, DNA samples were assessed for quality. Genotyping of the buccal cell DNA was performed at the Rockefeller University Genomics Resource Center using Affymetrix Genome-wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA). Sample quality was assessed using the Dynamic Model algorithm, and genotyping calls were generated using the Birdseed algorithm available in Genotyping Console 4.0 (Affymetrix). We used MaCH23 to impute SNPs on chromosome 1–22 using the 379 samples of European ancestry from the 1000 Genomes Project24 as the referent panel. Haplotypes for the SOPHIA subjects were estimated using MaCH, and imputation was performed using minimac.25 After accounting for imputation quality, we included 164 preeclampsia cases and 110 normotensive controls in our final analysis.
Exposure assessment—GRS calculation.
To estimate the genetic predisposition to dyslipidemia, 4 lipid (TC, triglycerides, LDL-C, and HDL-C) GRS were calculated based on well-established SNPs for blood lipids reported by 2 recent meta-analyses of GWAS.16,17 These GRS represent the genetic contribution for increased levels of TC, triglycerides, and LDL-C, as well as decreased levels of HDL-C. Only SNPs with genotyped data or high imputation quality scores (MACH r 2 ≥ 0.8) were included; 17 SNPs were excluded due to poor genotyping quality in the SOPHIA population. A total of 68, 40, 50, and 69 SNPs for TC, triglycerides, LDL-C, and HDL-C, respectively, were included in the GRS analysis (Supplementary Table 1). We assumed that each SNP acted independently and in an additive manner.
The genotype scores were calculated using a weighted method in a manner similar to that described by other researchers.26–29 Each SNP was weighted by its relative effect size (β coefficient) obtained from the reported meta-analysis data.16,30 The genotype scores were then calculated by multiplying each β coefficient by the number of corresponding risk alleles and then summing the products. This calculation resulted in a TC genotype score out of 198.65, an LDL-C score out of 120.88, an HDL-C score out of 72.17, and a triglyceride score out of 263.67, twice the sum of the β coefficients. To make the genotype scores easier to interpret, these values were divided by 198.65, 120.88, 72.17, and 263.67 and multiplied by 136, 100, 138, and 80 (the total number of the risk alleles), respectively. This allowed us to interpret the β coefficients as per-1 risk allele increase in the GRS.
Replication population
Study population.
A total of 516 preeclampsia cases and 449 normotensive controls of European ancestry were selected from Institutional Review Board-approved hospital- and internet-based collections at 5 US sites and matched to population controls (Supplementary Table 2).
Case status was verified by review of medical records at each site. Preeclampsia was defined as new onset of hypertension after 20 weeks of gestation with systolic BP ≥140 or diastolic BP ≥90 on 2 occasions at least 6h apart in the presence of proteinuria of 300mg/dl on a 24-h collection or at least +1 on a dipstick. In Boston and University of Southern California samples, case definition included superimposed preeclampsia (onset of hypertension after 20 weeks and new onset proteinuria in women with chronic hypertension). Pregnant women without a history of chronic hypertension and with no evidence of hypertensive disorders of pregnancy were defined as normotensive controls.
Cardiovascular disease array genotyping.
DNA was extracted from blood, saliva, or buccal samples at each site using standard protocols. Samples were genotyped on a cardiovascular gene-centric 50 K SNP array v1.0.31 SNPs were clustered into genotypes using the Illumina Beadstudio software and subjected to quality control filters at the sample and SNP level. For this study, samples of European ancestry were identified by principal component (PC) analysis, including HapMap3 European (CEU), African (YRI), and Asian (JPT+CHB) panels as reference standards.32 To increase power, cardiovascular disease array genotypes from 648 additional EA population-based women from the Atherosclerosis Risk in Communities study from the CARe study33 were individually matched to cases using nearest-neighbor genetic matching at each site. Samples were excluded for individual call rates <90%, duplicate discordance, heterozygosity >3 SD from the mean, any of the first 3 PCs >3 SD from the mean, and excess relatedness (pi-hat >0.125). SNPs were removed for call rates <95%, departure from Hardy-Weinberg equilibrium with P < 10−6 in controls, differential missingness between cases and controls (P < 0.05), or batch effects (P < 10−6).
GRS calculation.
GRS were calculated as described for the SOPHIA population with some modifications. Since the cardiovascular disease gene-centric array was designed before recent lipid GWAS discoveries, only a limited number of index SNPs for TC, HDL-C, LDL-C, and triglycerides were directly genotyped. Thus, in order to improve power to detect association of a lipid GRS with preeclampsia, we included a proxy SNP with the strongest correlation (pairwise r 2 > 0.8) to the untyped index SNP in the 1KG CEU sample. Using this approach, 27 loci for TC, 19 loci for LDL-C, 23 loci for HDL-C, and 16 loci for triglycerides were available for assessment. A weighted GRS for each trait was calculated for each individual. If a genotype in the score was missing for a particular individual, then the expected value was imputed based on the sample allele frequency. Division by twice the sum of the β coefficients (118.46 for TC, 65.79 for LDL-C, 37.25 for HDL-C, and 150.11 for triglyceride) and multiplication by the number of risk alleles (54, 38, 46, and 32) allowed us to interpret the β coefficients as per-1 risk allele increase in the GRS.
Statistical analysis
Chi-square and t-tests were run to compare proportions and means between case and control groups. Four GRS were modeled—one for each of the lipid types. Logistic regression was used to estimate the odds ratios (ORs) for preeclampsia risk. Multivariate logistic regression was used in order to evaluate and adjust for potential confounders. A priori, only variables that produced at least a 10% change in the OR for preeclampsia would be retained in the final models. The covariates examined included body mass index (BMI) (continuous), maternal age at delivery (continuous), education, preconception smoking (yes/no), smoking during pregnancy (nonsmoker, 1st trimester only, and 2nd/3rd trimester), physical activity (metabolic equivalents per week; continuous), paternal seminal fluid exposure,19 and human leukocyte antigen sharing. Ultimately, none of the evaluated covariates met the threshold for retention in any of the models. However, due to its significant difference between preeclamptic and normotensive women and biologic relevance to the associations tested, bivariate models for the SOPHIA population are also presented with an adjustment for prepregnancy BMI. Finally, assessment of the relationship between each SNP and the risk of preeclampsia was evaluated with univariate logistic regression using PLINK. Adequacy of model fit was examined using the Hosmer and Lemeshow goodness of fit test.
In the replication study, multivariate logistic regression analysis was used to test for association of each lipid GRS with preeclampsia, adjusting for 10 PCs and collection site. BMI and other relevant covariates were not available for the replication population.
RESULTS
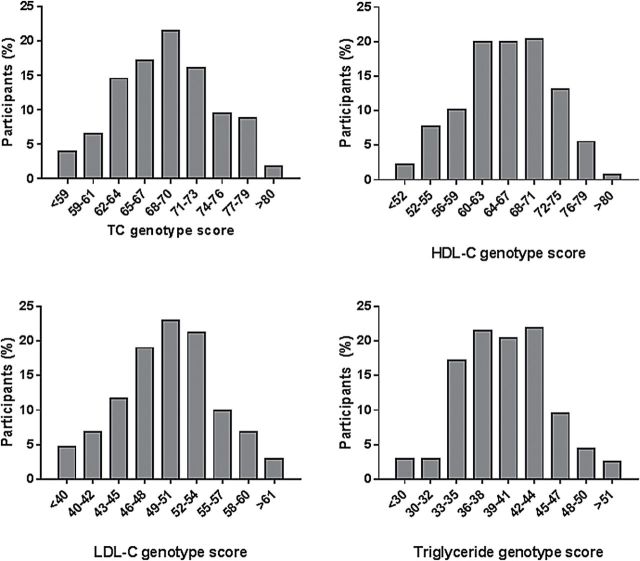
Characteristics of the study participants are shown in Table 1. Women who developed preeclampsia had a significantly higher prepregnancy BMI, engaged in significantly less leisure time physical activity, and were less likely to smoke than the normotensive control women. Preeclamptic women and women who remained normotensive during pregnancy did not differ with respect to age at delivery, level of education, or mean GRS for TC, LDL-C, HDL-C, or triglycerides. In addition, all 4 genotype scores were normally distributed among study participants (Figure 1).
Table 1.
Characteristics of study participants by case status, Study of Pregnancy-induced Hypertension in Iowa, 2004–2005
| Variable | Preeclampsia | Normotensive controls | P |
|---|---|---|---|
| N | 164 | 110 | |
| Age at birth (years) | 26.2 (4.9) | 26.4 (5.2) | 0.85 |
| Prepregnancy BMI (kg/m2) | 27.3 (5.9) | 23.9 (4.9) | <0.0001 |
| Smoked during pregnancy, n (%) | |||
| Nonsmoker | 129 (78.7) | 86 (78.2) | 0.02 |
| First trimester only | 22 (13.4) | 6 (5.5) | |
| Second/third trimester | 13 (7.9) | 18 (16.4) | |
| Education, n (%) | |||
| High school diploma or less | 28 (17.1) | 20 (18.2) | 0.27 |
| Some college | 66 (40.2) | 34 (30.9) | |
| College graduate | 70 (42.7) | 56 (50.9) | |
| Physical activity (metabolic equivalent hours/week) | 6.2 (10.3) | 11.2 (17.8) | 0.001 |
| Total cholesterol genotype score | 68.8 (6.3) | 69.0 (5.3) | 0.81 |
| LDL cholesterol genotype score | 50.2 (6.0) | 49.8 (5.6) | 0.57 |
| HDL cholesterol genotype score | 66.2 (6.6) | 64.9 (7.4) | 0.12 |
| Triglyceride genotype score | 40.5 (5.1) | 40.3 (4.8) | 0.80 |
Data are means (SD) or percent, unless otherwise indicated; t-tests used to test difference between cases and controls for continuous variables. Chi-square tests were performed to test differences between cases and controls for categorical variables. Abbreviations: BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Figure 1. Lipid genotype scores in pre-eclamptic and normotensive women from the Study of Pregnancy Hypertension in Iowa. The histograms represent the percentage of participants whose genotype score falls within a particular category. Abbreviations: TC, total cholesterol; LDL‐C, low density lipoprotein cholesterol; HDL‐C, high density lipoprotein cholesterol.
Table 2 presents the results from the analyses of the TC, LDL-C, HDL-C, and triglyceride genotype scores and the associated risk of preeclampsia. No significant associations were observed between the GRS for increased levels of TC, LDL-C, or triglycerides and the risk for preeclampsia: TC (OR = 1.00, 95% confidence interval (CI) = 0.96, 1.04), LDL-C (OR = 1.01, 95% CI = 0.97, 1.06), or triglycerides (OR = 1.01, 95% CI = 0.96, 1.06). Additionally, the risk of preeclampsia among women with GRS in the second, third, and fourth quartiles did not significantly differ from the risk among women with GRS in the first quartile (P for trend = 0.89, TC = 0.35, LDL-C = 0.52, triglycerides). However, a marginally significant association was observed between the GRS associated with lower levels of HDL-C and preeclampsia risk (OR = 1.03, 95% CI = 0.99, 1.07; P = 0.10), though the test for trend across the quartiles was not significant (P for trend = 0.46). Results were consistent with multivariate adjustment for prepregnancy BMI. Each of the GRS explained <1% of the deviance in preeclampsia outcomes whereas BMI, smoking, and physical activity explained 11% of the deviance in preeclampsia outcome.
Table 2.
Association between the lipid genotype scores and risk for preeclampsia, Study of Pregnancy-induced Hypertension in Iowa, 2004–2005
| Continuous | Quartile | P for trend | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| Total cholesterol | ||||||
| n (case/control subjects) | 43/28 | 40/28 | 43/27 | 38/27 | ||
| Median (range) | 62.5 (49.6–65.0) | 67.1 (65.0–69.4) | 71.0 (69.4–73.1) | 76.2 (73.2–81.1) | ||
| OR (95% CI) | ||||||
| Unadjusted | 1.00 (0.96, 1.04) | 1.00 (Ref) | 0.93 (0.47, 1.83) | 1.04 (0.53, 2.04) | 0.92 (0.46, 1.82) | 0.89 |
| BMI adjusted | 1.01 (0.96, 1.05) | 1.00 (Ref) | 1.11 (0.54, 2.27) | 1.23 (0.60, 2.52) | 1.09 (0.53, 2.24) | 0.75 |
| LDL cholesterol | ||||||
| n (case/control subjects) | 43/28 | 27/29 | 46/26 | 48/27 | ||
| Median (range) | 43.7 (30.6–46.4) | 48.0 (46.5–49.5) | 51.2 (49.6–53.8) | 56.8 (53.8–65.4) | ||
| OR (95% CI) | ||||||
| Unadjusted | 1.01 (0.97, 1.06) | 1.00 (Ref) | 0.61 (0.30, 1.23) | 1.15 (0.59, 2.27) | 1.16 (0.59, 2.26) | 0.35 |
| BMI adjusted | 1.03 (0.98, 1.07) | 1.00 (Ref) | 0.69 (0.32, 1.45) | 1.39 (0.68, 2.84) | 1.39 (0.69, 2.81) | 0.15 |
| HDL cholesterol | ||||||
| n (case/control subjects) | 39/27 | 29/28 | 53/27 | 43/28 | ||
| Median (range) | 57.0 (47.6–60.6) | 63.0 (60.6–64.9) | 67.8 (64.9–70.6) | 73.6 (70.6–81.5) | ||
| OR (95% CI) | ||||||
| Unadjusted | 1.03 (0.99, 1.07) | 1.00 (Ref) | 0.72 (0.35, 1.47) | 1.36 (0.69, 2.67) | 1.06 (0.54, 2.11) | 0.46 |
| BMI adjusted | 1.03 (0.99, 1.07) | 1.00 (Ref) | 0.73 (0.34, 1.55) | 1.47 (0.72, 2.98) | 1.16 (0.57, 2.37) | 0.33 |
| Triglycerides | ||||||
| n (case/control subjects) | 47/28 | 22/27 | 46/27 | 49/28 | ||
| Median (range) | 35.1 (27.8–37.1) | 38.6 (37.2–39.4) | 41.7 (39.5–43.2) | 45.6 (43.2–53.7) | ||
| OR (95% CI) | ||||||
| Unadjusted | 1.01 (0.96, 1.06) | 1.00 (Ref) | 0.49 (0.23, 1.01) | 1.02 (0.52, 1.98) | 1.04 (0.54, 2.02) | 0.52 |
| BMI adjusted | 1.00 (0.95, 1.05) | 1.00 (Ref) | 0.42 (0.19, 0.90) | 0.88 (0.44, 1.76) | 0.90 (0.45, 1.79) | 0.84 |
P value for test for trend from Cochrane–Armitage test for trend. Abbreviations: BMI, body mass index; CI, confidence interval; HDL, high-density lipoprotein; LDL, low-density lipoprotein; OR, odds ratio.
To ensure that an individual SNP was not driving the results, logistic regression analysis was conducted to evaluate the relationship between each SNP and preeclampsia risk. While none of the SNPs assessed reached statistical significance after the Bonferroni correction (P < 0.0003), 1 SNP (rs964184) did achieve a P value of 0.0088 (OR = 1.90 for every addition of a G allele); as such, sensitivity analyses were performed excluding rs964184 from all 4 GRS. Results were consistent with those of the main analyses (data not shown).
Replication analysis
Table 3 presents the results from the replication analyses of the GRS for TC, LDL-C, HDL-C, and triglyceride genotype scores and the associated risk of preeclampsia. The GRS results were similar to those from the original SOPHIA analysis: TC (OR = 0.98, 95% CI = 0.95 1.01), LDL-C (OR = 0.98, 95% CI = 0.95, 1.02), or triglycerides (OR = 1.00, 95% CI = 0.96, 1.05). Effect estimates from the GRS analysis of HDL-C were not only similar to those from the SOPHIA analysis, but were statistically significant (OR = 1.03, 95% CI = 1.00, 1.06). The risk of preeclampsia among women with GRS in the second, third, and fourth quartiles did not significantly differ from the risk among women with GRS in the first quartile (P for trend = 0.26, TC = 0.16, LDL-C = 0.12, HDL-C = 0.84, triglycerides). Meta-analysis of the HDL-C results between SOPHIA and the replication population also demonstrated a significant association between the HDL-C GRS and preeclampsia risk (OR = 1.03, 95% CI = 1.01, 1.05).
Table 3.
Association between the lipid genotype scores and risk for preeclampsia, preeclampsia replication population
| Continuous | Quartile | P for trend | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| Total cholesterol | ||||||
| n (case/control subjects) | 136/267 | 133/270 | 122/280 | 125/280 | ||
| Median (range) | 29.5 (20.4–31.2) | 32.5 (31.2–33.6) | 34.6 (33.6–35.9) | 37.6 (35.9–43.5) | ||
| OR (95% CI) | ||||||
| PCA adjusted | 0.98 (0.95, 1.01) | 1.00 (Ref) | 1.01 (0.74, 1.38) | 0.91 (0.67, 1.25) | 0.85 (0.63, 1.16) | 0.26 |
| LDL cholesterol | ||||||
| n (case/control subjects) | 130/273 | 140/263 | 131/272 | 115/289 | ||
| Median (range) | 15.8 (7.5–17.5) | 18.8 (17.5–19.9) | 21.0 (19.9–22.4) | 23.9 (22.4–29.4) | ||
| OR (95% CI) | ||||||
| PCA adjusted | 0.98 (0.95, 1.02) | 1.00 (Ref) | 1.02 (0.75, 1.40) | 1.07 (0.78, 1.46) | 0.81 (0.59, 1.10) | 0.16 |
| HDL cholesterol | ||||||
| n (case/control subjects) | 125/278 | 122/281 | 127/276 | 142/262 | ||
| Median (range) | 19.4 (10.7–21.8) | 23.2 (21.8–24.6) | 25.9 (24.6–27.1) | 28.7 (27.2–34.0) | ||
| OR (95% CI) | ||||||
| PCA adjusted | 1.03 (1.00, 1.06) | 1.00 (Ref) | 0.98 (0.72, 1.35) | 1.06 (0.77, 1.46) | 1.31 (0.95, 1.81) | 0.12 |
| Triglycerides | ||||||
| n (case/control subjects) | 133/270 | 133/270 | 113/290 | 137/267 | ||
| Median (range) | 18.0 (11.0–19.2) | 20.1 (19.2–21.0) | 21.8 (21.0–22.8) | 24.0 (22.8–29.6) | ||
| OR (95% CI) | ||||||
| PCA adjusted | 1.00 (0.96, 1.05) | 1.00 (Ref) | 1.00 (0.73, 1.36) | 0.76 (0.55, 1.05) | 1.00 (0.73, 1.37) | 0.84 |
P value for test for trend from Cochrane–Armitage test for trend. Abbreviations: CI, confidence interval; HDL, high-density lipoprotein; LDL, low-density lipoprotein; OR, odds ratio; PCA, principal components analysis for population stratification.
Logistic regression was performed in the replication population to evaluate for potential associations between individual SNPs and preeclampsia risk; 3 SNPs were identified (rs11024739, P < 0.0001; rs103294, P = 0.0009; and rs6073952, P = 0.007). As such, sensitivity analyses were performed for each applicable GRS without these loci. Results were consistent with those from the main analyses (data not shown).
Because the replication population included only a subset of the SNPs included in the SOPHIA analysis (Supplementary Table 3), a sensitivity analysis was performed in the SOPHIA dataset evaluating only those SNPs utilized in the replication set. Results were similar to the original SOPHIA results.
DISCUSSION
Several GWAS and many candidate gene studies have been performed to identify underlying genetic risk factors for preeclampsia; however, results have been inconsistent. One meta-analysis found a validated association between the LPL gene and risk for preeclampsia, potentially implicating dyslipidemia in the etiology of preeclampsia. Despite the emerging evidence of a strong role of dyslipidemia in the etiology of preeclampsia, no studies to our knowledge have examined genetic predisposition to dyslipidemia and preeclampsia. Our study is the first, to examine the relationships between well-established GRS for circulating lipid levels and preeclampsia. We did not find a statistically significant association between the GRS for elevated levels of LDL-C, TC, and triglycerides and preeclampsia risk in either the main study population or in the replication population. We did, however, identify a statistically significant relationship between the genetic predisposition for decreased levels of HDL-C and increased risk for preeclampsia.
It is important to note that the loci included in each of the risk scores only explain ~12% of the variation in plasma lipid levels.17,26 Therefore, as additional variants, particularly rare variants, are discovered, the association between the GRS for circulating lipid levels and risk of preeclampsia will need to be reevaluated. As lipid measures during pregnancy were not collected in SOPHIA, we could not directly measure the association between the GRS and lipid levels in our population of women; however, large GWAS have demonstrated the validity of the GRS used for this analysis based on their ability to predict circulating levels of lipids.
Epidemiologic studies have shown a strong association between dyslipidemia and preeclampsia; however, most studies examined levels in the third trimester of pregnancy and the ability of lipid levels to predict preeclampsia remains unclear.34 Women who experience preeclampsia are more likely to develop dyslipidemia later-in-life suggesting a potential shared etiology for both preeclampsia and dyslipidemia. The most common patterns of dyslipidemia experienced during preeclamptic pregnancies are elevated triglycerides and reduced HDL-C levels.34–36 Levels of circulating lipids indicative of dyslipidemia have been implicated in the pathogenesis of endothelial dysfunction in atherosclerosis.37 Because one of the classic hallmarks of preeclampsia is endothelial dysfunction, dyslipidemia is a plausible candidate in the pathogenesis of preeclampsia.15 Our data suggest that lower levels of HDL-C may be a component or an effect modifier along the causal pathway to preeclampsia. This finding is intriguing as HDL-C has antithrombotic properties, the ability to inhibit inflammation and oxidation, and is reported to enhance endothelial repair. However, it should be noted that the observed effects explain an extremely small (<1%) percentage of risk for preeclampsia. In comparison, anthropometric measures such as BMI and lifestyle risk factors such as smoking and low physical activity explains just over 11% of the preeclampsia risk in our population. As such, the clinical utility of this GRS to identify women at risk for preeclampsia is low compared to other well-known risk factors.
A major strength of our study includes the strict case definition of preeclampsia confirmed by BP measures and urinary protein levels recorded in the medical charts in our initial study population. We also used a population-based, nonhypertensive control group that underwent the same strict protocol of medical chart abstraction to verify normotensive status. In addition, we had high-quality genotype data from a large replication cohort to confirm our findings.
In conclusion, we found that genetic predisposition for decreased levels of HDL-C, estimated by a GRS, were associated with increased risk for preeclampsia. In contrast, genetic predisposition for elevated levels of TC, triglycerides, and LDL-C was not associated with the risk of preeclampsia in our study. Future research should focus on additional loci, particularly rare variants, associated with circulating lipid levels measured during pregnancy.
SUPPLEMENTARY MATERIAL
Supplementary materials are available at American Journal of Hypertension (http://ajh.oxfordjournals.org).
FUNDING
The SOPHIA study was supported by the National Institute of Child Health and Human Development , National Institutes of Health (R01 HD32579), and the Verto Institute. The replication study was funded in part by a Gertie Marx Grant from the Society for Obstetric Anesthesia and Perinatology.
DISCLOSURE
The authors declared no conflict of interest.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge contribution of samples for IBC array genotyping by preeclampsia investigators from Boston (B. Bateman, S.A. Karumanchi), Iowa (J. Murray), University of Southern California (M.L. Wilson), and Yale (E. Norwitz), as well as contribution of genotype data from the preeclampsia CARe IBC study (ARIC).
REFERENCES
- 1. Solomon CG, Seely EW. Preeclampsia – searching for the cause. N Engl J Med 2004; 350:641–642. [DOI] [PubMed] [Google Scholar]
- 2. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet Gynecol 2013; 122:1122–1131. [DOI] [PubMed] [Google Scholar]
- 3. Roberts JM, Cooper DW. Pathogenesis and genetics of pre-eclampsia. Lancet 2001; 357:53–56. [DOI] [PubMed] [Google Scholar]
- 4. Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol 2009; 33:130–137. [DOI] [PubMed] [Google Scholar]
- 5. Hubel CA. Oxidative stress in the pathogenesis of preeclampsia. Proc Soc Exp Biol Med 1999; 222:222–235. [DOI] [PubMed] [Google Scholar]
- 6. Ghulmiyyah L, Sibai B. Maternal mortality from preeclampsia/eclampsia. Semin Perinatol 2012; 36:56–59. [DOI] [PubMed] [Google Scholar]
- 7. Sutherland A, Cooper DW, Howie PW, Liston WA, MacGillivray I. The indicence of severe pre-eclampsia amongst mothers and mothers-in-law of pre-eclamptics and controls. Br J Obstet Gynaecol 1981; 88:785–791. [DOI] [PubMed] [Google Scholar]
- 8. Chesley LC, Cooper DW. Genetics of hypertension in pregnancy: possible single gene control of pre-eclampsia and eclampsia in the descendants of eclamptic women. Br J Obstet Gynaecol 1986; 93:898–908. [DOI] [PubMed] [Google Scholar]
- 9. Arngrimsson R, Björnsson S, Geirsson RT, Björnsson H, Walker JJ, Snaedal G. Genetic and familial predisposition to eclampsia and pre-eclampsia in a defined population. Br J Obstet Gynaecol 1990; 97:762–769. [DOI] [PubMed] [Google Scholar]
- 10. Cincotta RB, Brennecke SP. Family history of pre-eclampsia as a predictor for pre-eclampsia in primigravidas. Int J Gynaecol Obstet 1998; 60:23–27. [DOI] [PubMed] [Google Scholar]
- 11. Nilsson E, Salonen Ros H, Cnattingius S, Lichtenstein P. The importance of genetic and environmental effects for pre-eclampsia and gestational hypertension: a family study. BJOG 2004; 111:200–206. [DOI] [PubMed] [Google Scholar]
- 12. Esplin MS, Fausett MB, Fraser A, Kerber R, Mineau G, Carrillo J, Varner MW. Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med 2001; 344:867–872. [DOI] [PubMed] [Google Scholar]
- 13. Buurma AJ, Turner RJ, Driessen JH, Mooyaart AL, Schoones JW, Bruijn JA, Bloemenkamp KW, Dekkers OM, Baelde HJ. Genetic variants in pre-eclampsia: a meta-analysis. Human Reprod Update 2013; 19:289–303. [DOI] [PubMed] [Google Scholar]
- 14. Fisher RM, Humphries SE, Talmud PJ. Common variation in the lipoprotein lipase gene: effects on plasma lipids and risk of atherosclerosis. Atherosclerosis 1997; 135:145–159. [DOI] [PubMed] [Google Scholar]
- 15. Taylor R, Roberts J. Endothelial Cell Dysfunction, 3rd edn Elsevier: San Diego, 2009. [Google Scholar]
- 16. Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010; 466:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013; 45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Triche EW, Harland KK, Field EH, Rubenstein LM, Saftlas AF. Maternal-fetal HLA sharing and preeclampsia: variation in effects by seminal fluid exposure in a case-control study of nulliparous women in Iowa. J Reprod Immunol 2014; 101–102:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saftlas AF, Rubenstein L, Prater K, Harland KK, Field E, Triche EW. Cumulative exposure to paternal seminal fluid prior to conception and subsequent risk of preeclampsia. J Reprod Immunol 2014; 101-102:104–110. [DOI] [PubMed] [Google Scholar]
- 20. Zhao L, Triche EW, Walsh KM, Bracken MB, Saftlas AF, Hoh J, Dewan AT. Genome-wide association study identifies a maternal copy-number deletion in PSG11 enriched among preeclampsia patients. BMC Pregnancy Childbirth 2012; 12:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberts JM, Pearson GD, Cutler JA, Lindheimer MD; National Heart Lung and Blood Institute. Summary of the NHLBI Working Group on Research on Hypertension During Pregnancy. Hypertens Pregnancy 2003; 22:109–127. [DOI] [PubMed] [Google Scholar]
- 22. Saftlas AF, Waldschmidt M, Logsden-Sackett N, Triche E, Field E. Optimizing buccal cell DNA yields in mothers and infants for human leukocyte antigen genotyping. Am J Epidemiol 2004; 160:77–84. [DOI] [PubMed] [Google Scholar]
- 23. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34:816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA; 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44:955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qi Q, Liang L, Doria A, Hu FB, Qi L. Genetic predisposition to dyslipidemia and type 2 diabetes risk in two prospective cohorts. Diabetes 2012; 61:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qi Q, Forman JP, Jensen MK, Flint A, Curhan GC, Rimm EB, Hu FB, Qi L. Genetic predisposition to high blood pressure associates with cardiovascular complications among patients with type 2 diabetes: two independent studies. Diabetes 2012; 61:3026–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cornelis MC, Qi L, Zhang C, Kraft P, Manson J, Cai T, Hunter DJ, Hu FB. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med 2009; 150:541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y, Qi Q, Workalemahu T, Hu FB, Qi L. Birth weight, genetic susceptibility, and adulthood risk of type 2 diabetes. Diabetes Care 2012; 35:2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, Barnard J, Baumert J, Beitelshees AL, Bhangale TR, Chen YD, Gaunt TR, Gong Y, Hopewell JC, Johnson T, Kleber ME, Langaee TY, Li M, Li YR, Liu K, McDonough CW, Meijs MF, Middelberg RP, Musunuru K, Nelson CP, O’Connell JR, Padmanabhan S, Pankow JS, Pankratz N, Rafelt S, Rajagopalan R, Romaine SP, Schork NJ, Shaffer J, Shen H, Smith EN, Tischfield SE, van der Most PJ, van Vliet-Ostaptchouk JV, Verweij N, Volcik KA, Zhang L, Bailey KR, Bailey KM, Bauer F, Boer JM, Braund PS, Burt A, Burton PR, Buxbaum SG, Chen W, Cooper-Dehoff RM, Cupples LA, deJong JS, Delles C, Duggan D, Fornage M, Furlong CE, Glazer N, Gums JG, Hastie C, Holmes MV, Illig T, Kirkland SA, Kivimaki M, Klein R, Klein BE, Kooperberg C, Kottke-Marchant K, Kumari M, LaCroix AZ, Mallela L, Murugesan G, Ordovas J, Ouwehand WH, Post WS, Saxena R, Scharnagl H, Schreiner PJ, Shah T, Shields DC, Shimbo D, Srinivasan SR, Stolk RP, Swerdlow DI, Taylor HA, Jr, Topol EJ, Toskala E, van Pelt JL, van Setten J, Yusuf S, Whittaker JC, Zwinderman AH, Anand SS, Balmforth AJ, Berenson GS, Bezzina CR, Boehm BO, Boerwinkle E, Casas JP, Caulfield MJ, Clarke R, Connell JM, Cruickshanks KJ, Davidson KW, Day IN, de Bakker PI, Doevendans PA, Dominiczak AF, Hall AS, Hartman CA, Hengstenberg C, Hillege HL, Hofker MH, Humphries SE, Jarvik GP, Johnson JA, Kaess BM, Kathiresan S, Koenig W, Lawlor DA, März W, Melander O, Mitchell BD, Montgomery GW, Munroe PB, Murray SS, Newhouse SJ, Onland-Moret NC, Poulter N, Psaty B, Redline S, Rich SS, Rotter JI, Schunkert H, Sever P, Shuldiner AR, Silverstein RL, Stanton A, Thorand B, Trip MD, Tsai MY, van der Harst P, van der Schoot E, van der Schouw YT, Verschuren WM, Watkins H, Wilde AA, Wolffenbuttel BH, Whitfield JB, Hovingh GK, Ballantyne CM, Wijmenga C, Reilly MP, Martin NG, Wilson JG, Rader DJ, Samani NJ, Reiner AP, Hegele RA, Kastelein JJ, Hingorani AD, Talmud PJ, Hakonarson H, Elbers CC, Keating BJ, Drenos F; LifeLines Cohort Study. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet 2012; 91:823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One 2008; 3:e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG, Keating BJ, Yang Q, Chen MH, Lapchyk N, Crenshaw A, Ziaugra L, Rachupka A, Benjamin EJ, Cupples LA, Fornage M, Fox ER, Heckbert SR, Hirschhorn JN, Newton-Cheh C, Nizzari MM, Paltoo DN, Papanicolaou GJ, Patel SR, Psaty BM, Rader DJ, Redline S, Rich SS, Rotter JI, Taylor HA, Jr, Tracy RP, Vasan RS, Wilson JG, Kathiresan S, Fabsitz RR, Boerwinkle E, Gabriel SB. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet 2010; 3:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spracklen CN, Smith CJ, Saftlas AF, Robinson JG, Ryckman KK. Maternal hyperlipidemia and the risk of preeclampsia: a meta-analysis. Am J Epidemiol 2014; 180:346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallos ID, Sivakumar K, Kilby MD, Coomarasamy A, Thangaratinam S, Vatish M. Pre-eclampsia is associated with, and preceded by, hypertriglyceridaemia: a meta-analysis. BJOG 2013; 120:1321–1332. [DOI] [PubMed] [Google Scholar]
- 36. Gohil JT, Patel PK, Gupta P. Estimation of lipid profile in subjects of preeclampsia. J Obstet Gynaecol India 2011; 61:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown MC, Best KE, Pearce MS, Waugh J, Robson SC, Bell R. Cardiovascular disease risk in women with pre-eclampsia: systematic review and meta-analysis. Eur J Epidemiol 2013; 28:1–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.