In a household outbreak of extended-spectrum β-lactamase–producing ST131 Escherichia coli, molecular analysis of fecal samples from all household members showed that the index ST131 strain was the predominant and most extensively shared E. coli strain, and a persistent intestinal colonizer.
Keywords: E. coli, ST131, ESBL
Abstract
Background. Reasons for the successful global dissemination of multidrug-resistant Escherichia coli sequence type 131 (ST131) are undefined, but may include enhanced transmissibility or ability to colonize the intestine compared with other strains.
Methods. We identified a household in which 2 young children had urinary tract infection (UTI) caused by an extended-spectrum β-lactamase (ESBL)–producing, multidrug-resistant ST131 E. coli strain. We assessed the prevalence of ST131 intestinal colonization among the 7 household members (6 humans, 1 dog). Fecal samples, collected 3 times over a 19-week period, were cultured selectively for E. coli. Isolates were characterized using clone-specific polymerase chain reaction to detect ST131 and its ESBL-associated H30Rx subclone, pulsed-field gel electrophoresis, extended virulence genotyping, and antimicrobial susceptibility testing.
Results. In total, 8 different E. coli pulsotypes (strains) were identified. The index patient's urine isolate represented ST131-H30Rx strain 903. This was the most widely shared and persistent strain in the household, colonizing 5 individuals at each sampling. In contrast, the 7 non-ST131 strains were each found in only 1 or 2 household members at a time, with variable persistence. The ST131 strain was the only strain with both extensive virulence and antimicrobial resistance profiles.
Conclusions. An ESBL-producing ST131-H30Rx strain caused UTI in 2 siblings, plus asymptomatic intestinal colonization in multiple other household members, and was the household's most extensively detected and persistent fecal E. coli strain. Efficient transmission and intestinal colonization may contribute to the epidemiologic success of the H30Rx subclone of E. coli ST131.
Extraintestinal pathogenic Escherichia coli (ExPEC) strains colonize the human intestinal tract but have enhanced capacity to cause extraintestinal infections, including bacteremia, pneumonia, urinary tract infection (UTI), bone and joint infections, intra-abdominal infections, and meningitis [1]. In recent years there has been rapid global emergence and spread of antimicrobial-resistant ExPEC strains, driven largely by the clonal expansion of a single sequence type (ST), ST131 [2]. Reasons for ST131's widespread dissemination are unclear but may include within-household transmission, which has been documented for other ExPEC strains [3, 4]. To date, few studies have assessed uninfected household contacts of patients with ST131 infection.
CASE REPORT
The index patient was a 40-day-old female infant born by cesarean delivery at 32 5/7 weeks' gestation. Following birth, she stayed in the neonatal intensive care unit (ICU) for 2 days and was discharged 5 days later. At 40 days of life, she presented with poor feeding, sleepiness, and dark, malodorous urine. She was lethargic, mottled, tachycardic, and hypothermic. The infant was admitted to the hospital and treated empirically with intravenous ampicillin and cefotaxime.
Complete blood count, a blood culture, and cerebrospinal fluid analysis were unremarkable. Urine contained large amounts of blood and gram-negative bacilli, and grew >100 000 colony-forming units (CFU)/mL of extended-spectrum β-lactamase (ESBL)–producing E. coli, resistant to multiple antibiotics (Table 1). Renal ultrasonography and voiding cystourethrography were normal. She received a 10-day course of cefepime, with clinical cure.
Table 1.
Antimicrobial Susceptibility of Escherichia coli Urine Isolatesa From the Index Patient and Her Sibling
| Antimicrobial | MIC in mg/L, S/I/R Interpretation |
||||
|---|---|---|---|---|---|
| Index Patient Episode |
Sister |
||||
| Initial | Second (3 wk After Initial) | Third (6 wk After Initial) | Fourth (9 wk After Initial) | (11 wk After Index Initial) | |
| Ampicillin | >16 R | >16 R | >16 R | >16 R | >16 R |
| Ampicillin-sulbactam | 16/8 I | 16/8 I | 16/8 I | 16/8 I | 16/8 I |
| Piperacillin-tazobactam | ≤16/4 S | ≤16/4 S | ≤16/4 S | ≤16/4 S | ≤16/4 S |
| Cefazolin | >16 R | >16 R | >16 R | >16 R | >16 R |
| Cefdinir | >2 R | >2 R | >2 R | >2 R | >2 R |
| Cefepime | 8 S | >16 R | 8 S | 8 S | 8 S |
| Ceftriaxone | >32 R | >32 R | >32 R | >32 R | >32 R |
| Cephalothin | >16 R | >16 R | >16 R | >16 R | >16 R |
| Ertapenem | ≤0.25 S | ≤0.25 S | ≤0.25 S | ≤0.25 S | ≤0.25 S |
| Meropenem | ≤1 S | ≤1 S | ≤1 S | ≤1 S | ≤1 S |
| Ciprofloxacin | >2 R | >2 R | >2 R | >2 R | >2 R |
| Levofloxacin | >4 R | >4 R | >4 R | >4 R | >4 R |
| Amikacin | ≤8 S | ≤8 S | ≤8 S | ≤8 S | ≤8 S |
| Gentamicin | >8 R | >8 R | >8 R | >8 R | >8 R |
| Tobramycin | 8 I | 8 I | 8 I | 8 I | 8 I |
| Nitrofurantoin | ≤32 S | ≤32 S | ≤32 S | ≤32 S | ≤32 S |
| TMP-SMX | >2/38 R | ≤0.5/9.5 S | >2/38 R | >2/38 R | >2/38 R |
| Fosfomycin | 128 I | ≤64 S | ≤64 S | ≤64 S | ≤64 S |
Abbreviations: I, intermediate; MIC, minimum inhibitory concentration; R, resistant; S, susceptible; TMP-SMX, trimethoprim-sulfamethoxazole.
a All specimens were obtained by sterile in-and-out catheterization. Susceptibility testing was performed by the clinical microbiology laboratory using agar dilution.
Ten days after completing this first antibiotic course, the infant returned with malodorous urine and emesis. Urine again contained blood, leukocytes, and gram-negative bacilli. She was rehospitalized and given empiric cefepime. A urine culture grew >100 000 CFU/mL of E. coli with a susceptibility pattern resembling that of the previous isolate (Table 1), although the trimethoprim-sulfamethoxazole (TMP-SMX) result was now susceptible (minimum inhibitory concentration [MIC] ≤0.5/9.5 mg/L), and the cefepime MIC had increased to 16 mg/L (resistant), prompting antibiotic change to meropenem. Blood cultures were negative and repeat renal ultrasonography was normal. After 3 days of meropenem therapy, which achieved clinical cure, she was discharged with 7 more days of therapy with oral TMP-SMX.
One week later, the infant again returned with malodorous urine and crying with urination. Urine culture grew >100 000 CFU/mL of E. coli with the same susceptibility pattern as the initial urine isolate (Table 1). She was rehospitalized and received meropenem for 14 days, which again achieved clinical cure. Voiding cystourethrography as well as ultrasonography of the kidneys and spinal cord were normal. An on-treatment surveillance urine culture was negative.
Nine days after completion of this third antibiotic course, a surveillance urine culture grew >100 000 CFU/mL of E. coli with the same susceptibility profile as the urine isolates from the first and third episodes. Although the infant was asymptomatic, she received a fourth antibiotic course (14 days of nitrofurantoin), followed by continuous daily nitrofurantoin prophylaxis. A dimercaptosuccinic acid renal scan and limited immunodeficiency evaluation (quantitative immunoglobulin levels, complete blood count with peripheral smear, and human immunodeficiency virus testing) were normal. The infant continued on prophylactic nitrofurantoin for 6 months after her last UTI episode. To date, she has been off prophylactic antibiotics for 7 months and has not had a recurrence.
Family history revealed that 2.5 months after the index patient's initial presentation, her 2-year-old sister was diagnosed with UTI caused by an E. coli strain with a susceptibility profile resembling the index patient's isolates (Table 1). This sibling was treated with a 10-day course of nitrofurantoin and has not had a recurrence of UTI. The index patient's 2 other siblings had remained healthy. Her father was a janitor in an academic medical center ICU. Her mother, currently a homemaker, had worked previously as a labor and delivery ward nurse. Notably, the neonatal ICU to which the index patient was admitted had experienced no infections with ESBL-producing gram-negative organisms.
Because 2 young children within the same household developed multidrug-resistant E. coli UTI despite lacking traditional risk factors for antimicrobial-resistant infections, we hypothesized that intestinal colonization and transmission of antimicrobial-resistant E. coli was occurring among household members. We therefore evaluated all household members for intestinal colonization with antibiotic-resistant E. coli over a 19-week period and determined the clonal background, resistance profiles, and virulence factor profiles of all colonizing E. coli isolates, for comparison with the index patient's third clinical UTI isolate.
METHODS
Clinical Information
Clinical and laboratory data were obtained by retrospective review of the index patient's electronic medical record. Antimicrobial susceptibility testing of the clinical urine isolates from the index patient and her sister was performed by the Mayo Clinic clinical microbiology laboratory using agar dilution [5].
Fecal Sample Collection
After obtaining informed consent, stool samples were collected from all household members, including the index patient, 2 parents, 3 older siblings, and 1 pet dog. Fecal samples were cultured on Tergitol-7 agar plates, with and without ciprofloxacin supplementation (4 mg/L), to recover total and ciprofloxacin-resistant gram-negative bacilli. Indole-positive, citrate-negative colonies with a consistent colonial morphology were identified presumptively as E. coli. To assess for persistence of colonization, human household members underwent repeat fecal sampling at 10 and 19 weeks.
Characterization of E. coli Isolates
From each fecal specimen, 10 colonies of presumptive E. coli were selected for DNA isolation, molecular analysis, and susceptibility testing. The urine isolate from the index patient's third UTI episode was retrieved from the hospital clinical microbiology laboratory for similar analysis. The major phylogenetic group (A, B1, B2, C, D, E, F) was determined using the updated Clermont method [6]. Isolates were further characterized using established polymerase chain reaction (PCR) assays to detect ST131 E. coli and its H30-Rx subclone [7, 8], ST95 [9], ST73 [10], and blaCTX-M (group 1) and blaCTX-M-15 [11]; XbaI pulsed-field gel electrophoresis (PFGE) to resolve unique pulsotypes (strains) [12]; and extended virulence genotyping for 52 ExPEC-associated virulence genes [13] (Table 2). All PCR testing was done in duplicate using relevant positive and negative controls. Susceptibility to 26 antibiotics (Table 3) was assessed by disk diffusion using Clinical and Laboratory Standards Institute–specified procedures and interpretive criteria [5].
Table 2.
Virulence Genes Among Escherichia coli Strains From Fecal Samples of 6 Household Members as Obtained on 3 Different Samplings Over 19 Weeks
| Virulence Geneb,c | Presence or Absence of Virulence Genea |
|||||||
|---|---|---|---|---|---|---|---|---|
| ST131 |
Non-ST131 |
|||||||
| 903 (n = 15) | 1908 (n = 3) | 583 (n = 3) | 1701 (n = 6) | 1835 (n = 3) | 1836 (n = 2) | 1837 (n = 4) | 1924 (n = 2) | |
| Adhesins | ||||||||
| papAHCG | + | − | − | + | − | + | − | − |
| papEF | + | − | + | + | − | + | − | − |
| papG II | + | − | − | + | − | − | − | − |
| papG III | − | − | − | − | − | + | − | − |
| sfa/focDE | − | − | − | − | − | + | − | − |
| focG | − | − | − | − | − | + | − | − |
| afa/draBC | − | − | + | − | − | − | − | − |
| iha | + | − | + | − | − | − | − | − |
| fimH | + | + | + | + | + | + | + | + |
| hra | + | + | − | − | − | + | − | − |
| Toxins | ||||||||
| hlyD | + | − | − | − | − | + | − | − |
| cnf1 | − | − | − | − | − | + | − | − |
| sat | + | + | + | + | + | + | + | + |
| pic | − | − | − | − | − | + | − | − |
| vat | − | − | − | + | − | + | − | − |
| Siderophores | ||||||||
| iroN | − | − | − | − | − | + | − | − |
| fyuA | + | + | + | + | − | + | − | − |
| ireA | − | − | − | + | − | + | − | − |
| iutA | + | + | + | − | − | − | − | − |
| Protectins | ||||||||
| kpsM II | + | − | + | + | − | + | − | − |
| K1 | − | − | + | + | − | − | − | − |
| K5 | + | − | − | − | − | − | − | − |
| traT | + | + | − | + | + | − | − | − |
| Miscellaneous | ||||||||
| usp | + | − | − | + | − | + | − | − |
| ompT | − | − | − | + | − | + | + | − |
| H7 fliC | − | − | − | + | − | − | − | − |
| malX | + | − | − | + | − | + | − | − |
| clbB, clbN | − | − | − | + | − | − | − | − |
Abbreviation: ST, sequence type.
a Plus sign, gene present in ≥50% of isolates of strain. Minus sign, gene present in <50% of isolates of strain.
b Genes listed are those detected in ≥1 isolate. Definitions: papAHCG, papEF, papG II, and pap G III (P fimbriae structural subunits, assembly, and tip adhesion variants), sfa/focDE (S and F1C fimbriae), focG (F1C fimbriae adhesion), afa/draBC (Dr-binding adhesins), iha (adhesin-siderophore receptor), fimH (type 1 fimbriae adhesin), hra (heat-resistant agglutinin), hlyD (alpha hemolysin), cnf1 (cytotoxic necrotizing factor), sat (secreted auto-transporter toxin), pic (protein associated with intestinal colonization), vat (vacuolating toxin), iroN (salmochelin receptor), fyuA (yersiniabactin receptor), ireA (siderophore receptor), iutA (aerobactin receptor), kpsM ll (group 2 capsules), K1 (group 2 capsule variant), K5 (group 2 capsule variant), traT (serum resistance-associated), usp (uropathogenic-specific protein), ompT (outer membrane protease), H7 fliC (flagellar variant), malX (pathogenicity island marker), clbB and clbN (colibactin polyketide synthesis).
c Genes sought but not detected: papG allele I (variant P adhesin), sfaS (S fimbriae adhesin), afaE8 (variant afimbrial adhesin), bmaE (M fimbriae), gafD (G fimbriae), F17 (adhesin variant), clpG (adhesin variant), hlyF (variant hemolysin), cdtB (cytolethal distending toxin), tsh (temperature-sensitive hemagglutinin), astA (toxin of enteroaggregative Escherichia coli), kpsMT III (group 3 capsule), K15 (capsule variant), K2/K100 (capsule variant), rfc (O4 lipopolysaccharide synthesis), cvaC (microcin V), ibeA (invasion of brain endothelium), and iss (increased serum survival).
Table 3.
Antibiotic Resistance Patterns Among Fecal Escherichia coli Strains From 6 Household Members at 3 Samplings Over a 19-Week Period
| Resistance Phenotypea,b | Prevalence of Resistancea, No. of Isolates (Column %) |
|||||||
|---|---|---|---|---|---|---|---|---|
| ST131 | Non-ST131 |
|||||||
| 903 (n = 15) | 1908 (n = 3) | 583 (n = 3) | 1701 (n = 6) | 1835 (n = 3) | 1836 (n = 2) | 1837 (n = 4) | 1924 (n = 2) | |
| Ampicillin | 15 (100) | 3 (100) | 0 (0) | 1 (17) | 1 (33) | 0 (0) | 0 (0) | 1 (50) |
| Piperacillin | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Piperacillin-tazobactam | 1 (7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ampicillin-sulbactam | 14 (93) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Cefepime | 14 (93) | 2 (67) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ceftriaxone | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Cefazolin | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ceftazidime | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Aztreonam | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Gentamicin | 13 (87) | 1 (33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Streptomycin | 15 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Tetracycline | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Levofloxacin | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Nalidixic acid | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| TMP-SMX | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Chloramphenicol | 0 (0) | 2 (67) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| ESBL producer | 15 (100) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Abbreviations: ESBL, extended-spectrum β-lactamase; ST, sequence type; TMP-SMX, trimethoprim-sulfamethoxazole.
a Intermediate results were considered resistant. Antibiotic susceptibility was assessed using disk diffusion.
b Drugs shown are those to which at least 1 isolate was resistant. No resistance was detected to ertapenem, meropenem, cefoxitin, amikacin, or nitrofurantoin.
Resistance and Virulence Scores
Antibiotic resistance scores were the total number of antibiotics with an intermediate or resistant result. Virulence scores were the total number of virulence genes detected, adjusted for multiple detection of the pap (P fimbriae), sfa/foc (S and F1C fimbriae), kpsM II (group 2 capsule), and clb (colibactin) operons. Isolates were operationally defined as ExPEC if they contained ≥2 of the following: papAH and/or papC: (counted as 1), sfa/focDE (S and F1C fimbriae), afa/draBC (Dr-binding adhesins), iutA (aerobactin receptor), and kpsM II [14].
Institutional Review Board Approval
This study was approved by the institutional review boards of the Veterans Affairs Medical Center, Minneapolis, and the Mayo Clinic, Rochester, Minnesota.
RESULTS
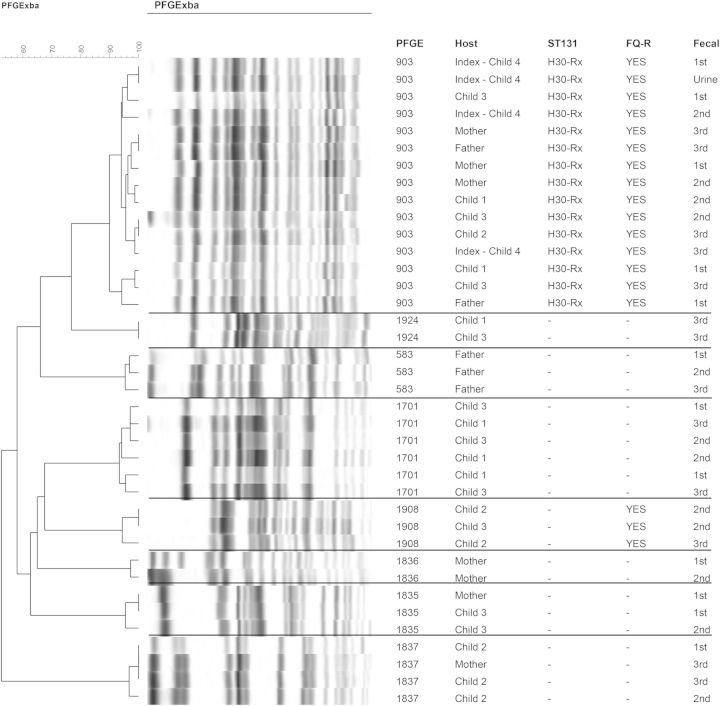
Over a 19-week period (16 January–30 May 2014), 6 human household members each contributed 3 fecal specimens (1 initially, another after approximately 10 weeks, a third after approximately 19 weeks), for 18 total samples. Overall, among the multiple colonies analyzed per sample, 8 distinct strains were identified by PFGE (Figure 1), including 3 from phylogroup A (strains 583, 1837, and 1924), 2 from phylogroup B1 (strains 1835 and 1908), and 3 from phylogroup B2 (strains 903 [ST131], 1701 [ST95], and 1836 [ST73]). All household members except the index patient were colonized with multiple strains at ≥2 samplings each, whereas the index patient had only a single strain, 903, identified at all time points (Figure 2). The mother shared more strains with the children than did the father. No E. coli was found in the single fecal specimen from the pet dog (Figure 2).
Figure 1.
Pulsed-field gel-electrophoresis profiles of Escherichia coli isolates of all 6 human household members. Isolates were from a urine sample of the index patient and fecal samples of all household members from 3 different occasions over a 19-week period. Horizontal lines separate different pulsotypes (ie, strains). Note the index patient's urine isolate near top of dendrogram, interposed among fecal isolates of pulsotype 903. Scale is percentage of similarity according to Dice similarity coefficients. Sibling with urinary tract infection is child 3. Abbreviations: 1st, 2nd, and 3rd: first, second, and third round of fecal sampling; FQ-R, fluoroquinolone resistant; PFGE, pulsotype; ST131, sequence type 131.
Figure 2.
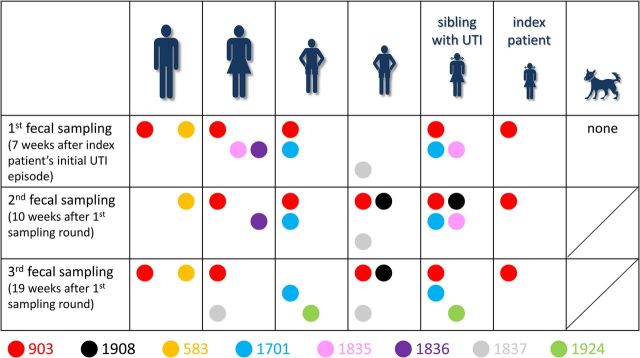
Escherichia coli strains isolated from 6 household members during 3 rounds of fecal sampling over 19 weeks. Household member symbols: large figures, parents; small figures, children; figures in skirts, females; animal, pet dog. Colors indicate different pulsed-field gel electrophoresis types (strains). Up to 10 E. coli colonies (as available) per fecal sample were analyzed; size of circles is independent of the proportional prevalence of the strain in the sample. The first fecal sample of the index patient was collected during a urinary tract infection (UTI), all other samples are not associated with UTI episodes.
Strain 903, which accounted for the index subject's clinical UTI isolate, represented the fluoroquinolone-resistant ESBL-associated H30Rx subclone of ST131. This was the most prevalent and extensively shared strain among household members, occurring overall in 15 of 18 specimens, and at various times in all 6 human household members (Figure 2). Strain 903 was detected at all 3 samplings in 3 subjects (index patient, 1 older sister, and mother), and at 1 or 2 samplings each in the other 3 household members. In contrast, the 7 non-ST131 strains were detected in only 1 (strains 583 and 1836) or 2 individuals (strains 1701, 1835, 1837, 1908, and 1924) at any sampling (Figure 2). Additionally, of these non-ST131 strains, only strains 538, 1701, and 1837 persisted through all 3 samplings, and in only 1 or 2 individuals each.
Antimicrobial Resistance
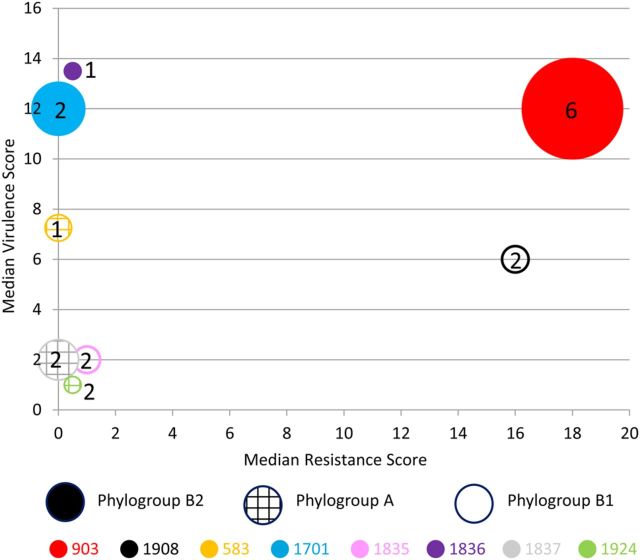
The 8 strains differed greatly from one another according to resistance profiles. Six were susceptible to most antibiotics, whereas 2 (strains 903 and 1908) were extensively antimicrobial-resistant ESBL producers (Table 3); blaCTX-M-15, encoding the CTX-M-15 ESBL, was found in both strains 903 and 1908. The highest resistance scores were associated with strains 903 (median, 18) and 1908 (median, 16). The remaining strains had low median resistance scores of 0–1 (Figure 3).
Figure 3.
Bacterial characteristics and ecological behavior of colonizing Escherichia coli strains. Circles represent the 8 different fecal E. coli strains. Shading pattern within circle indicates phylogroup (hatched, group A; none, group B1; solid, group B2). Size of circle reflects number of fecal samples (of 18 total) from which the strain was isolated (15 for 903; 6 for 1701; 4 for 1837; 3 each for 1908, 583, and 1835; and 2 each for 1836 and 1924). Number within or near the circle indicates the number of human household members (of 6) from whom the strain was isolated.
Virulence Factors
Virulence gene content likewise varied greatly by strain (Table 2). Virulence scores were higher (median value) for strains 903 (12), 1836 (13.5), and 1701 (12), compared with the other strains: 583 (7.25), 1908 (6), 1835 (2), 1837 (2), and 1924 (1) (Figure 3). Of the 8 strains, only ST131 strain 903 had high values for both resistance score and virulence score (Figure 3). Strains 903, 583, 1701, and 1836 qualified molecularly as ExPEC.
DISCUSSION
We describe a household in which an extensively multidrug-resistant, ESBL-producing E. coli strain from ST131-H30Rx caused multiple UTI episodes in 2 siblings and was highly prevalent and persistent (over the 19-week study period) as an intestinal colonizer among all 6 human household members. These findings support that E. coli ST131 is a threatening pathogen, including for children, and also a persistent intestinal colonizer that likely is efficiently transmitted among household members. Moreover, the (co-colonized) parents' healthcare employment histories suggest possible importation of the strain into the household from a healthcare-associated reservoir, as proposed previously in a case of recurrent pediatric ESBL E. coli infection [15].
Although all 3 colonizing phylogroup B2 strains represented STs commonly associated with human extraintestinal infection (ST95, ST73, and ST131), only the ST131 strain caused clinical infection and extensive colonization. ST131 is a newly emerged, pandemic antimicrobial-resistant clonal group [2, 16]. Its ESBL and sepsis-associated subclone, H30Rx, has been proposed as a driving force behind the rapid spread of ST131 [7, 17]. Reasons for the successful worldwide expansion of H30Rx are unknown, but may include enhanced transmissibility or colonization ability, increased virulence, and/or more extensive antimicrobial resistance [2]. The ST131 strain 903 identified here is a relatively uncommon type within ST131 [12] but was encountered previously among clinical isolates from our institution (J. R. Johnson, unpublished data).
The ST131 H30Rx strain was the most widely shared E. coli strain in the household, occurring in 5 of 6 human household members at each sampling, and at 2 or 3 sampling points per individual. In contrast, the non-ST131 strains were present in only 1–2 household members at any sampling, and were less persistent than the ST131 strain. Although this is only a single-household observation, it suggests that ST131 H30Rx has enhanced transmissibility and persistence compared with other E. coli strains. Enhanced persistence might be due to superior intestinal colonization ability compared with other E. coli strains. Relevant to this, in a previous study of mouse intestinal colonization, the test E. coli ST131 strain (which likely represented H30Rx, based on its reported characteristics) was a better colonizer than the commensal E. coli comparator strains [18].
The origin of the multidrug-resistant ST131 isolate in the present index infant patient is unclear. Although it is possible that the infant acquired the strain during her short stay in the neonatal ICU, this unit had experienced no prior infections with ESBL-producing gram-negative organisms. The genetic similarity between the infant's urine isolate and the household members' ST131 colonization isolates suggests that the infant acquired the ST131 isolate postnatally, from a household contact. Because she was born by cesarean delivery, it is unlikely that she acquired the strain from exposure to maternal rectal flora during birth. The index patient's parents may have acquired the strain from their hospital workplace, as exposure to healthcare settings is a risk factor for E. coli ST131, at least with E. coli clinical isolates [19]. Route of transmission was not investigated here and may have been from parent to child, child to child, child to adult, or to multiple household members from a common environmental source [20].
A combination of more extensive resistance and greater virulence capabilities may contribute generally to ST131's ecological success. Here, the ST131 strain was the only strain detected that had both an extensive resistance profile and an extensive virulence gene profile. In other studies of antibiotic-resistant E. coli, ST131 isolates have had higher virulence scores than non-ST131 isolates [8, 21, 22]. However, although it is tempting to speculate that its extensive virulence gene content enables ST131 to outcompete other strains in colonizing the gut and, presumably, the urothelium, animal models have been inconclusive regarding ST131's comparative virulence [18, 23, 24].
This study has limitations, including reliance on retrospective medical record review for the index patient and her sibling, and analysis of a single household. We also were unable to obtain other relevant urine isolates (ie, the index patient's first 2 UTI isolates and asymptomatic bacteriuria isolate, and the sibling's UTI isolate) to confirm that all represented the same ST131 strain 903 and to compare them genetically with the other ST131 study isolates. We are quite confident that this is the case, however, because these urine isolates all exhibited the same distinctive resistance profile (with minor variation) as did ST131 strain 903 (Table 1), and both children had persistent intestinal colonization with this strain. Study strengths include our ability to obtain and extensively molecularly characterize multiple E. coli isolates from all household members over a 5-month period.
In conclusion, this recent household outbreak of multidrug-resistant UTI and urosepsis is, to our knowledge, the only reported instance of pediatric UTI involving a shared ST131 strain, and represents the most extensive within-household sharing of ST131 among human household members documented to date. Our data indicate that the H30Rx subclone of E. coli ST131 includes highly resistant and virulent strains with a superior ability to colonize the human intestinal tract and to spread among household contacts, thereby putting children without traditional risk factors for drug resistance at risk for ST131 infection. Approaches to prevent the acquisition and spread of this ST131 subclone are urgently needed.
Notes
Financial support. This work was supported by a Clinical and Translational Science Award (number KL2 TR000136) from the National Center for Advancing Translational Science (to R. B.); and is also based in part on work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (grant number 1 I01 CX000192 01 to J. R. J.).
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli: “The other bad E. coli.” J Lab Clin Med 2002; 139:155–62. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R, Johnson JR. A new clone sweeps clean: the enigmatic emergence of Escherichia coli ST131. Antimicrob Agents Chemother 2014; 58:4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JR, Clabots C. Sharing of virulent Escherichia coli clones among household members of a woman with acute cystitis. Clin Infect Dis 2006; 43:e101–8. [DOI] [PubMed] [Google Scholar]
- 4.Foxman B, Manning SD, Tallman P, et al. Uropathogenic Escherichia coli are more likely than commensal E. coli to be shared between heterosexual sex partners. Am J Epidemiol 2002; 156:1133–40. [DOI] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21 Wayne, PA: CLSI, 2011. [Google Scholar]
- 6.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 2013; 5:58–65. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JR, Tchesnokova V, Johnston B, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 2013; 207:919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colpan A, Johnston B, Porter S, et al. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 2013; 57:1256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidet P, Metais A, Mahjoub-Messai F, et al. Detection and identification by PCR of a highly virulent phylogenetic subgroup among extraintestinal pathogenic Escherichia coli B2 strains. Appl Environ Microbiol 2007; 73:2373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont O, Christenson JK, Daubie AS, Gordon DM, Denamur E. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J Microbiol Methods 2014; 101:24–7. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JR, Urban C, Weissman SJ, et al. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-beta-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother 2012; 56:2364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Nicolas-Chanoine MH, DebRoy C, et al. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg Infect Dis 2012; 18:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000; 181:261–72. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JR, Murray AC, Gajewski A, et al. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 2003; 47:2161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woods KL, Johnson JR, Padkowsky S, et al. Community-associated Escherichia coli harboring CTX-M beta-lactamases from urine cultures from pediatric patients. Antimicrob Agents Chemother 2012; 56:2209–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 2014; 27:543–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price LB, Johnson JR, Aziz M, et al. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 2013; 4:e00377–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vimont S, Boyd A, Bleibtreu A, et al. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 2012; 7:e46547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol 2013; 34:361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fornasini M, Reves RR, Murray BE, Morrow AL, Pickering LK. Trimethoprim-resistant Escherichia coli in households of children attending day care centers. J Infect Dis 1992; 166:326–30. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 2010; 51:286–94. [DOI] [PubMed] [Google Scholar]
- 22.Olesen B, Hansen DS, Nilsson F, et al. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol 2013; 51:1779–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavigne JP, Vergunst AC, Goret L, et al. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 2012; 7:e34294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun 2012; 80:1554–62. [DOI] [PMC free article] [PubMed] [Google Scholar]