Abstract
Higher eukaryotic organisms cannot live without oxygen; yet, paradoxically, oxygen can be harmful to them. The oxygen molecule is chemically relatively inert because it has two unpaired electrons located in different pi * anti-bonding orbitals. These two electrons have parallel spins, meaning they rotate in the same direction about their own axes. This is why the oxygen molecule is not very reactive. Activation of oxygen may occur by two different mechanisms; either through reduction via one electron at a time (monovalent reduction), or through the absorption of sufficient energy to reverse the spin of one of the unpaired electrons. This results in the production of reactive oxidative species (ROS). There are a number of ways in which the human body eliminates ROS in its physiological state. If ROS production exceeds the repair capacity, oxidative stress results and damages different molecules. There are many different methods by which oxidative stress can be measured. This manuscript focuses on one of the methods named cell gel electrophoresis, also known as “comet assay” which allows measurement of DNA breaks. If all factors known to cause DNA damage, other than oxidative stress are kept constant, the amount of DNA damage measured by comet assay is a good parameter of oxidative stress. The principle is simple and relies upon the fact that DNA molecules are negatively charged. An intact DNA molecule has such a large size that it does not migrate during electrophoresis. DNA breaks, however, if present result in smaller fragments which move in the electrical field towards the anode. Smaller fragments migrate faster. As the fragments have different sizes the final result of the electrophoresis is not a distinct line but rather a continuum with the shape of a comet. The system allows a quantification of the resulting “comet” and thus of the DNA breaks in the cell.
Keywords: Molecular Biology, Issue 99, Comet assay analysis, Single-cell gel electrophoresis, DNA breaks, Oxidative stress
Introduction
Comet assay was first developed by Swedish scientists Ostling and Johanson1. DNA is a negatively charged molecule. During electrophoresis, smaller, damaged DNA fragments travel faster toward a positively charged anode2. The smaller the DNA fragments, the faster their migration toward the anode to form a typical “comet” with a “head” composed of intact, undamaged DNA and a “tail” composed of damaged DNA fragments3. Damaged DNA can be repaired by different mechanisms in the body4, which keep the generation of DNA breaks fairly balanced5. If, however, the balance is in favour of DNA breaks , the breaks can accumulate and will ultimately contribute to the development of a disease.
Our DNA suffers approximately 0.000165% damage at a given time6. Single stranded DNA breaks refer to a defect on one of the two strands of the DNA double helix whereas double-stranded DNA breaks are caused when both strands of the DNA double helix are damaged. There are various factors which may enhance the amount of DNA breaks at a given time such as the age of the cell, cigarette smoke, various drugs or oxidative stress7-9. If all factors leading to enhanced DNA breaks are kept constant, then the quantification of DNA breaks by means of comet assay is a good indirect parameter of oxidative stress.
The method of comet assay is used increasingly today in medicine including ophthalmology. One reason for this is that oxidative stress is more and more recognized as a pathogenetic mechanism for various diseases8-11 and comet assay is one method with which oxidative stress can indirectly be quantified.
Protocol
Studies on comet assay performed in the University of Basel, Department of Ophthalmology were approved by the Local Ethical Committee of Basel
1. Preparation of Reagents
Prepare Dulbecco's Phosphate Buffered Saline (PBS) (Ca++, Mg++ free) solution by adding PBS (1 L packet) and 990 ml of dH2O to the media. Adjust the pH to 7.4. Store at RT.
Prepare lysing solution: To prepare 1,000 ml add 2.5 M NaCl (146.1 g), 100 mM EDTA (37.2 g) and 10 mM Trizma base (1.2 g) in dH2O.
Prepare electrophoresis buffer (300 mM NaOH/1 mM EDTA) by adding 30 ml of 10 M NaOH and 7.5 ml of 0.2M EDTA, q.s. to 1,500 ml dH2O and mix well. The total volume depends on the gel box capacity. Prior to use, measure the pH of the buffer to ensure a pH of >13.
Prepare neutralization buffer containing 0.4 M Tris (48.5 gm added to ~800 ml dH2O, adjust pH to 7.5), concentrated (>10 M HCl) in 1,000 ml with dH2O. Store the solution at RT.
Prepare staining solution. To Ethidium Bromide (EtBr; 10x Stock, 20 µg/ml) add 10 mg of EtBr in 50 ml dH2O and store at RT. For 1x Stock - mix 1 ml with 9 ml dH2O. CAUTIOUS! Handle EtBr with adequate precaution as it is a known carcinogen.
2. Isolation of Leukocytes
Obtain human blood samples (20 ml) with anti-coagulant such as heparin using venous puncture.
- Separate the lymphocytes using a density gradient cell separator such as Histopaque.
- Briefly, dilute blood in 1:1 ratio with PBS or RPMI (without FBS) and layer over 600 µl cell separator liquid.
- Centrifuge 1,800 x g for 20 min at 4 °C. Aspirate the ‘buffy’ coat into 3-5 ml of PBS/ RPMI and centrifuged at 300 x g for 10 min to pellet the lymphocytes.
- Retrieve lymphocytes from just above boundary between PBS (RPMI) and cell separator liquid, using a pipettor. Remove the leukocyte bands from the interface between plasma and the cell separator liquid layers of each tube and collect into one 50 ml tube.
Bring the total volume to 50 ml with cold Dulbecco’s Modified Eagle Medium (DMEM). Wash the cell suspension three times with DMEM at 300 x g for 10 min.
Count the total numbers of cells using a Haemocytometer. Suspend the cells in PBS and aliquote into microcentrifuge tubes at 107 cells/tube.
Pipet 0.4 ml of cells into a 5 ml plastic disposable tube. Add 1.2 ml 1% low-gelling-temperature agarose at 40 °C. Mix and rapidly and pipet 1.2 ml of cell suspension onto the agarose-covered surface of a pre-coated slide. Avoid producing bubbles.
Allow the agarose to gel for about 2 min. Be consistent with the time and temperature used for gelling, and ensure that agarose is fully set before submerging in lysis solution. After fully set, submerge into lysing solution at ~4 ºC.
3. Lysis and Electrophoresis
NOTE: The procedure described is for electrophoresis under pH >13 alkaline conditions.
After at least 2 hr at ~4 ºC, gently remove slides from the lysing Solution. Place slides side by side on the horizontal gel box near one end, sliding them as close together as possible.
Fill the buffer reservoirs with freshly made pH >13 electrophoresis buffer until the liquid level completely covers the slides (avoid bubbles over the agarose).
Let slides sit in the alkaline buffer for 20 min to allow for unwinding of the DNA and the expression of alkali-labile damage.
Turn on power supply to 25 volts (~0.75 V/cm) and adjust the current to 300 milliamperes by raising or lowering the buffer level. Electrophorese the slides for 30 min.
Turn off the power. Gently lift the slides from the buffer and place on a drain tray. Drop wise coat the slides with neutralization Buffer. Allow the slides to sit for at least 5 min. Drain slides and repeat two more times.
Stain the slides with 80 µl 1x Ethidium Bromide (EtBr) and leave for 5 min and then dip in chilled distilled water to remove excess stain. Then place a coverslip over the slide and store them immediately. Store slides for a month at RT in dry conditions. NOTE: Other DNA stains (e.g., SYBR-green) may also be used. Caution! wear protective gloves as most dyes are mutagenic.
4. Quantification of DNA Breaks
To visualize the DNA damage, observe the EtBr-stained DNA slide under a 40X objective fluorescent microscope.
Assess DNA damage quantitatively in the cells by measuring the length of DNA migration and the percentage of migrated DNA (Figure 1A). Quantify DNA damage (Figure 1B) by the Tail- moment and Olive-moment. Definitions of Tail-Moment and Olive –Moment have previously been given8-10. NOTE: In our experience it's enough to stick by one of the parameters. The cells are photographed by setting the automated tool that scans over the microscope slide and photographs cells. Quantification of DNA breaks can be done by image analysis software packages. There are several different software packages that are custom made to capture the image of the cells and quantify DNA damage. We recommend using the same software package throughout the experiment. The lab technician identifies the intact cell or comet to be scored on the computer and clicks with the mouse on it. The comet or intact cell is then automatically scored.
Representative Results
Although the method of comet assay renders reproducible results, it can be influenced by a variety of factors. One such factor is whether or not the leukocytes are cryopreserved prior to lysis and gel-electrophoresis or whether this step is performed on freshly prepared leukocytes. Figure 2 shows that in Group 2, where leukocytes were cryopreserved prior to lysis and gel electrophoresis ss- DNA breaks were significantly higher, than in Group 1 where lysis and gel -electrophoresis was done on freshly prepared cells.
The assay can also be used to indirectly assess systemic oxidative stress. Table 1 shows the descriptive statistics for Tail- moment and Olive- moment in smokers and healthy subjects. The data reveals that smoking half a pack of cigarettes daily more than doubles ssDNA breaks in the circulating leukocytes9.

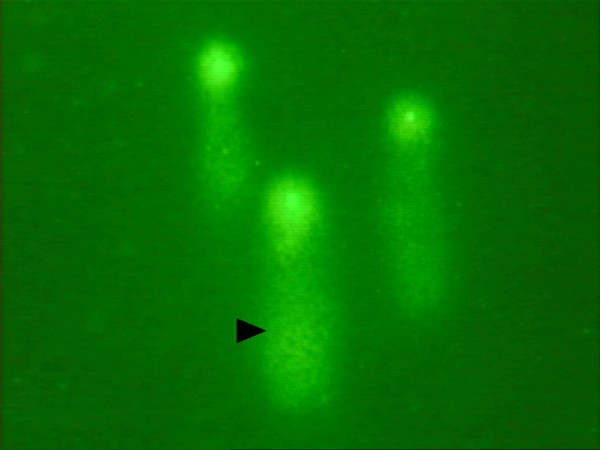 Figure 1. (A) Depicts the microscope used during comet assay analysis. (B) Δ depicts a typical “comet” with a bright head and tail (damaged smaller DNA fragments). Please click here to view a larger version of this figure.
Figure 1. (A) Depicts the microscope used during comet assay analysis. (B) Δ depicts a typical “comet” with a bright head and tail (damaged smaller DNA fragments). Please click here to view a larger version of this figure.
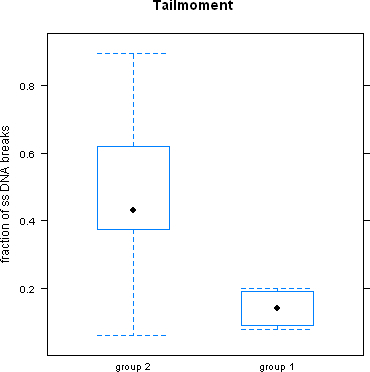 Figure 2.
ssDNA breaks were significantly higher when leukocytes were cryopreserved (Group 2) than when freshly prepared leukocytes were used prior to lysis and gel-electrophoresis.
Figure 2.
ssDNA breaks were significantly higher when leukocytes were cryopreserved (Group 2) than when freshly prepared leukocytes were used prior to lysis and gel-electrophoresis.
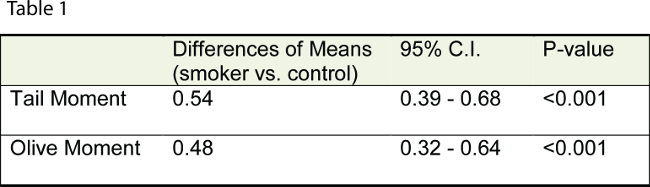 Table 1. Smokers had more than double the number of ssDNA breaks in comparison to age and sex matched healthy non-smokers.
Table 1. Smokers had more than double the number of ssDNA breaks in comparison to age and sex matched healthy non-smokers.
Discussion
Swedish scientists Ostling and Johanson were the first in their field to use comet assay to quantify DNA damage in cells1. The neutral conditions that the two scientists used, however, only allowed the detection of double-stranded DNA breaks. Singh et al. later adapted the assay for use under alkaline conditions, which produced a sensitive version that could assess double and single-stranded DNA breaks and detect alkali-labile sites12. Since its initial development, the assay has been modified at various steps to make it suitable for assessing types of DNA damage in different cell types13-15.
As with all techniques, paying rigorous attention to technical details is important to obtain accurate results16. Based on our experience, best results are obtained if every methodological step—from solution preparation to comet quantification—is performed by expert laboratory technicians. The use of fresh and correctly prepared media, adequate pipetting techniques, exact timing and (as previously mentioned in the results section) the use of freshly prepared leukocytes are essential in order for lysis and gel electrophoresis to obtain accurate results17.
All the individual steps in this assay are equally important for obtaining reliable results.16 In general best results are obtained if every single methodological step from solution preparation till comet quantification is performed by expert lab technicians.Important seem the use of fresh and correctly prepared media, adequate pipetting techniques, exact timing and as already referred to in the results section the use of freshly prepared leukocytes for lysis and gel-electrophoresis to obtain adequate results from the assay17.
As do other methodologies, the comet assay technique has its advantages and drawbacks. Being a sensitive method, the assay is vulnerable to factors (e.g., UV light) which could augment DNA breaks and thus affect results. Any factor that may enhance oxidative stress, except that which is being researched, should be avoided. Stressors can increase level of oxidative stress not only in leukocytes but also in all other blood mediums and lymphocytes. As mentioned earlier there are many different and more direct ways of measuring oxidative stress18,19. The comet assay is one of many methods which has certain advantages. These include its relative low cost, the small number of cells required (<10,000 cells) and thus the small samples of blood required per patient, the relative short period of time required to quantify DNA damage in cells (approximately 3 days), its sensitivity and its wide-spread applicability to asses DNA damage in different cell-types. In the past we choose to work with leukocytes since we had experience with this cell-type and it was applicable for the particular study planned. Another advantage of the comet assay is that it can be used to detect different types and levels of DNA damage and thus can be applied in various other areas of studies besides oxidative stress such as DNA repair studies, supplementation trials or genotoxicity studies.
Oxidative stress is now recognized as playing a critical role in the pathogenesis of multiple diseases. The comet assay method, while one of many ways of measuring oxidative stress18,19, is relatively simple, versatile and inexpensive. When mastered, this method can be used in all areas of medicine in which oxidative stress plays role 8-10,20,21.
Disclosures
None.
Acknowledgments
We would like to thank the LHW Stiftung, in Triesen Liechtenstein, for financially supporting our research on oxidative stress.
References
- Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123(1):291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- Phillips JL, Singh NP, Lai H. Electromagnetic fields and DNA damage. Pathophysiology. 2009;16(2-3):79–88. doi: 10.1016/j.pathophys.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Dhawan A, Bajpayee M, Parmar D. Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol. 2009;25(1):5–32. doi: 10.1007/s10565-008-9072-z. [DOI] [PubMed] [Google Scholar]
- Aiub CA, Pinto LF, Felzenszwalb I. DNA-repair genes and vitamin E in the prevention of N-nitrosodiethylamine mutagenicity. Cell Biol Toxicol. 2009;25(4):393–402. doi: 10.1007/s10565-008-9093-7. [DOI] [PubMed] [Google Scholar]
- Bartsch H, Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch Surg. 2006;391(5):499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- Martin LJ. DNA damage and repair: relevance to mechanisms of neurodegeneration. J Neuropathol Exp Neurol. 2008;67(5):377–387. doi: 10.1097/NEN.0b013e31816ff780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy CK, Benzie IF, Cho P. UV-mediated DNA strand breaks in corneal epithelial cells assessed using the comet assay procedure. Photochem Photobiol. 2005;81(3):493–497. doi: 10.1562/2004-10-20-RA-347. [DOI] [PubMed] [Google Scholar]
- Mozaffarieh M, et al. Comet assay analysis of single-stranded DNA breaks in circulating leukocytes of glaucoma patients. Mol Vis. 2008;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- Mozaffarieh M, Konieczka K, Hauenstein D, Schoetzau A, Flammer J. Half a pack of cigarettes a day more than doubles DNA breaks in circulating leukocytes. Tob Induc Dis. 2010;8(14) doi: 10.1186/1617-9625-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarieh M, Schotzau A, Josifova T, Flammer J. The effect of ranibizumab versus photodynamic therapy on DNA damage in patients with exudative macular degeneration. Mol Vis. 2009;15:1194–1199. [PMC free article] [PubMed] [Google Scholar]
- Pertynska-Marczewska M, Merhi Z. Relationship of Advanced Glycation End Products With Cardiovascular Disease in Menopausal Women. Reprod Sci. 2014. [DOI] [PMC free article] [PubMed]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells) Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Gaivão I, Sierra LM. Drosophila comet assay: insights, uses, and future perspectives. Front Genet. 2014;5:304. doi: 10.3389/fgene.2014.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. DNA damage in lens epithelial cells and peripheral lymphocytes from age-related cataract patients. Ophthalmic Res. 2014;51(3):124–128. doi: 10.1159/000356399. [DOI] [PubMed] [Google Scholar]
- Huet SA, Vasseur L, Camus S, Chesné C, Fessard V. Performance of comet and micronucleus assays in metabolic competent HepaRG cells to predict in vivo genotoxicity. Toxicol Sci. 2014;138(2):300–309. doi: 10.1093/toxsci/kfu004. [DOI] [PubMed] [Google Scholar]
- Danson S, Ranson M, Denneny O, Cummings J, Ward TH. Validation of the comet-X assay as a pharmacodynamic assay for measuring DNA cross-linking produced by the novel anticancer agent RH1 during a phase I clinical trial. Cancer chemotherapy and pharmacology. 2007;60:851–861. doi: 10.1007/s00280-007-0432-9. [DOI] [PubMed] [Google Scholar]
- Moller P, Moller L, Godschalk RW, Jones GD. Assessment and reduction of comet assay variation in relation to DNA damage: studies from the European Comet Assay Validation Group. Mutagenesis. 2010;25:109–111. doi: 10.1093/mutage/gep067. [DOI] [PubMed] [Google Scholar]
- Chan SW, Chevalier S, Aprikian A, Chen JZ. Simultaneous quantification of mitochondrial DNA damage and copy number in circulating blood: a sensitive approach to systemic oxidative stress. BioMed Res Int. 2013. pp. 157–547. [DOI] [PMC free article] [PubMed]
- Sanchez-Quesada JL, Perez A. Modified lipoproteins as biomarkers of cardiovascular risk in diabetes mellitus. Endocrinol Nutr. 2013;60:518–528. doi: 10.1016/j.endonu.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Gandhi G, Kaur G, Nisar UA. Cross-sectional case control study on genetic damage in individuals residing in the vicinity of a mobile phone base station. Electromagn Biol Med. 2014;9:1–11. doi: 10.3109/15368378.2014.933349. [DOI] [PubMed] [Google Scholar]
- Cabarkapa A, et al. Protective effect of dry olive leaf extract in adrenaline induced DNA damage evaluated using in vitro comet assay with human peripheral leukocytes. Toxicol In Vitro. 2014;28(3):451–456. doi: 10.1016/j.tiv.2013.12.014. [DOI] [PubMed] [Google Scholar]


