Abstract
Premise of the study:
Plann automates the process of annotating a plastome sequence in GenBank format for either downstream processing or for GenBank submission by annotating a new plastome based on a similar, well-annotated plastome.
Methods and Results:
Plann is a Perl script to be executed on the command line. Plann compares a new plastome sequence to the features annotated in a reference plastome and then shifts the intervals of any matching features to the locations in the new plastome. Plann’s output can be used in the National Center for Biotechnology Information’s tbl2asn to create a Sequin file for GenBank submission.
Conclusions:
Unlike Web-based annotation packages, Plann is a locally executable script that will accurately annotate a plastome sequence to a locally specified reference plastome. Because it executes from the command line, it is ready to use in other software pipelines and can be easily rerun as a draft plastome is improved.
Keywords: chloroplast, GenBank, genome annotation, plastome, Sequin
As next-generation sequencing becomes the primary source of molecular variation for phylogenetic analyses, sequencing and assembling whole plastomes is more feasible than ever. Recent papers (Kane et al., 2012; Bock et al., 2013; Ripma et al., 2014) have highlighted the ease of genome skimming techniques for assembling the relatively high-coverage plastome and chondriome (mitochondrial genome) from lower-level genomic DNA short-read sequencing. De novo assembly programs can easily assemble large contiguous sequences of plastome; these sequences can be aligned to the generally syntenic and conserved gene order of the plastome, which makes whole-plastome alignments ideal for phylogenomic and phylogeographic studies (Cronn et al., 2008; Parks et al., 2009, 2012; Straub et al., 2012; Njuguna et al., 2013). Once a plastome is assembled, however, it still needs to be annotated. Generally, new plastomes are annotated in reference to known plastid genes and features, using available online tools such as DOGMA (Wyman et al., 2004) or CpGAVAS (Liu et al., 2012). These tools are excellent for annotating and visualizing a single plastome sequence from a single taxon, but can become tedious for annotation of plastomes for which a well-annotated sequenced relative already exists. In this situation, an interactive interface that repeatedly queries a broad range of annotated plastomes is needlessly complex.
Here we introduce Plann (Plastome Annotator), a command-line tool to automate the annotation of newly assembled plastomes based on a single well-annotated related reference plastome. Because it runs locally on the user’s machine (or on a Unix server), it can be easily run independently or pipelined into existing workflows. Its inputs are minimal: it requires only the new plastome sequence and the GenBank-formatted file of the related annotated plastome to match. Its output can be immediately used in the National Center for Biotechnology Information’s (NCBI) command-line tool tbl2asn to create a GenBank-ready submission. Features in the reference genome that were not matched in the new plastome are included as alignments in the report so that more attention can be paid to possible misassemblies in those regions. Plastome sequences are often used for deep phylogenetics in plants because of their highly conserved gene order and low mutation rate. In closely related taxa, the coding sequence of the genes is nearly identical, with few insertions or deletions; the variation is mostly present in the intergenic spacers. Because of these features, annotating the plastome using the template of a closely related pre-annotated plastome is a straightforward process.
METHODS AND RESULTS
The reference gene sequences to be used in the search can be obtained from a GenBank record that is already validated and has annotations describing the names and qualifiers for all of the genes in the plastome. Plann searches for sequences in the newly assembled plastome similar to those known genes and transforms those matched genes to their corresponding genomic locations. Because it only searches one sequence against one sequence, it is a very fast process: it should only take a few seconds to run.
Plann consists of Perl scripts contained in a GitHub repository (https://github.com/daisieh/plann/releases/tag/v1.1) and licensed under a BSD open-source license. It uses two freely available command-line tools from NCBI: BLASTN and tbl2asn. The graphical user interface (GUI) application Sequin, also available from NCBI, can be used to generate the template file required by tbl2asn and to validate the output of tbl2asn. It has been tested on Unix and Unix-like platforms, including Linux and Mac OS X. To use Plann, make sure that both BLASTN and tbl2asn are directly available as executables from the command line, then execute the script plann.pl. The input files required are the new plastome sequence (in FASTA format), the reference plastome (in GenBank format), and the Sequin template file. The output is a Sequin file, ready for submission to NCBI, and a text report with the genes that were not aligned. These problematic genes can be manually edited in a text editor and added back to the Sequin .tbl file, which can then be rerun through tbl2asn. If it turns out that the sequence is incorrect, it can be edited and then rerun through Plann again.
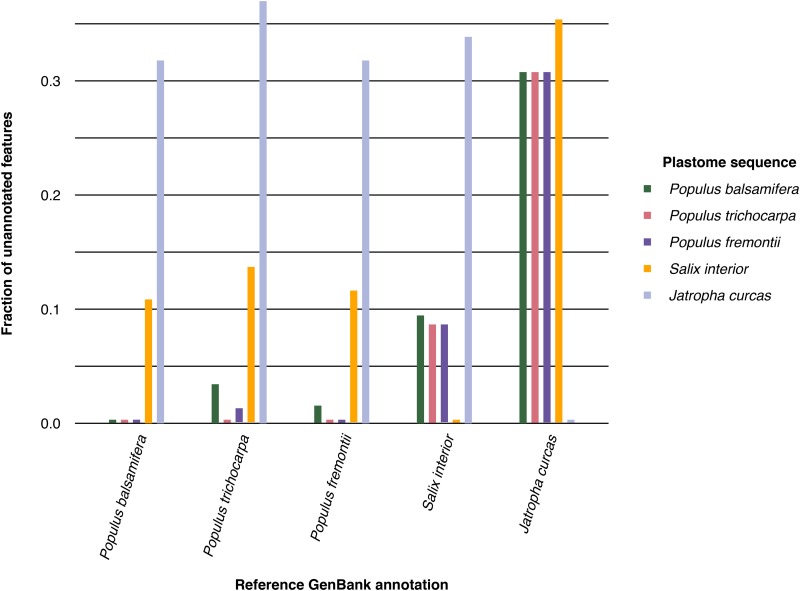
To validate the annotations produced by Plann, we reciprocally annotated plastomes of taxa at varying phylogenetic distances. The reference GenBank records used were NC_024735.1 (Populus balsamifera), NC_009143.1 (P. trichocarpa), NC_024734.1 (P. fremontii), NC_024681.1 (Salix interior), and NC_012224.1 (Jatropha curcas). This analysis can be found in the repository at test/analysis.sh. The comparative results are presented in Fig. 1. Nearly all features were successfully annotated within a genus. Even for a distantly related pair of taxa, Plann was able to identify nearly 70% of the features present in the annotation.
Fig. 1.
For each GenBank record along the x-axis, the number of genes annotated by Plann for the raw plastome sequence is represented by a bar. Note that the self-to-self annotation is always equivalent to the number of genes present in the original GenBank record.
CONCLUSIONS
Now that whole-genome shotgun sequencing is inexpensive and widely available, many plant systematists will have access to sufficiently deep sequencing to assemble whole plastome sequences to use in phylogenetic and phylogeographic studies. In these types of studies, it is not uncommon to generate new whole plastome sequences for many individuals. Annotating many plastomes from scratch using an interactive Web application can be both time-consuming and tedious, and the annotations used may not be as detailed as the desired reference plastome annotation. A potential use case for Plann would be a phylogeographic study in which multiple (from a few to perhaps several hundred) complete plastome assemblies are obtained for a single species (or closely related species) for which a good fully annotated plastome sequence already exists.
It should be emphasized that Plann is a tool for expeditiously generating annotations based on an existing reference; therefore, its annotation can only be as good as the reference annotation used. If genes are missing or poorly annotated in the reference GenBank file, the Plann output will also be missing genes and be poorly annotated. Genes that have many mutations in coding regions (relative to the reference plastome) may also be missed by Plann, although sequence variation in the intergenic spacers will not affect Plann at all.
Plann is unlike the widely used Web-based applications DOGMA and CpGAVAS in that Plann is not interactive and is designed to run quickly, with minimal user interaction, from the command line. Using Plann, the 10 plastome sequences in the validation test were annotated in less than five minutes, compared with an hour per plastome for CpGAVAS or many hours (depending on user speed) for DOGMA. Automated command-line scripts such as Plann will make plastome annotation a quick and simple part of a workflow instead of a tedious and time-consuming afterthought for publication.
LITERATURE CITED
- Bock D. G., Kane N. C., Ebert D. P., Rieseberg L. H. 2013. Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: Neither from Jerusalem nor an artichoke. New Phytologist 201: 1021–1030. [DOI] [PubMed] [Google Scholar]
- Cronn R., Liston A., Parks M., Gernandt D. S., Shen R., Mockler T. 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Research 36: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane N., Sveinsson S., Dempewolf H., Yang J. Y., Zhang D., Engels J. M. M., Cronk Q. 2012. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. American Journal of Botany 99: 320–329. [DOI] [PubMed] [Google Scholar]
- Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics 13: 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njuguna W., Liston A., Cronn R., Ashman T. L., Bassil N. 2013. Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Molecular Phylogenetics and Evolution 66: 17–29. [DOI] [PubMed] [Google Scholar]
- Parks M., Cronn R., Liston A. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M., Cronn R., Liston A. 2012. Separating the wheat from the chaff: Mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae). BMC Evolutionary Biology 12: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripma L. A., Simpson M. G., Hasenstab-Lehman K. 2014. Geneious! Simplified genome skimming methods for phylogenetic systematic studies: A case study in Oreocarya (Boraginaceae). Applications in Plant Sciences 2: 1400062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S. C. K., Parks M., Weitemier K., Fishbein M., Cronn R. C., Liston A. 2012. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. American Journal of Botany 99: 349–364. [DOI] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K., Boore J. L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]