Abstract
The presence of bacteria within the pocket epithelium and underlying connective tissue in gingival biopsies from patients with periodontitis has been reported using various methods, including electron microscopy, immunohistochemistry or immunofluorescence using bacteria-specific antibodies, and fluorescent in situ hybridization (FISH) using a fluorescence-labeled oligonucleotide probe. Nevertheless, these methods are not widely used due to technical limitation or difficulties. Here a method to localize bacteria within paraffin-embedded tissues using DIG-labeled DNA probes has been introduced. The paraffin-embedded tissues are the most common form of biopsy tissues available from pathology banks. Bacteria can be detected either in a species-specific or universal manner. Bacterial signals are detected as either discrete forms (coccus, rod, fusiform, and hairy form) of bacteria or dispersed forms. The technique allows other histological information to be obtained: the epithelia, connective tissue, inflammatory infiltrates, and blood vessels are well distinguished. This method can be used to study the role of bacteria in various diseases, such as periodontitis, cancers, and inflammatory immune diseases.
Keywords: Immunology, Issue 99, periodontology, oral microbiology, in situ hybridization, 16S rRNA, bacteria, paraffin-embedded tissue, species-specific, universal
Introduction
Bacteria play a role in the etiology of various oral diseases such as periodontitis, pulpitis, pericoronitis, cellulitis, and osteomyelitis. In order to understand the role of bacteria in the pathogenesis of disease and to monitor the effect of treatments, localization of bacteria within the tissue is important. The presence of bacteria within gingival tissue from patients with periodontitis has been shown using various methods, including electron microscopy1,2, immunohistochemistry and immunofluorescence using bacteria-specific antibodies3-7, and fluorescent in situ hybridization (FISH)8 using a fluorescence-labeled oligonucleotide probe targeting 16S rRNA. Nevertheless, these methods are not widely used due to technical limitation or difficulties. Compared with antibodies, probes targeting 16S rRNA are easy to produce and achieve species-specificity. FISH has proven to be an excellent tool for the visualization of bacteria in their natural environments such as plaque biofilm. However, application of FISH to tissue samples is limited due to autofluorescence of various tissue components. For example, the strong autofluorescence of red blood cells often hampers the application of fluorescence technology to inflamed tissues when they involve bleeding9.
In order to localize bacteria within the inflamed gingival tissues, therefore, an in situ hybridization method using a digoxigenin (DIG)-labeled DNA probe has been developed and successfully applied10,11. Here a detailed protocol for the localization of bacteria within paraffin-embedded tissues using P. gingivalis-specific and universal eubacterial probes has been described. It is particularly focused on standardization of the method so that similar results can be reproduced in other laboratories. This protocol allows localization of bacteria within their histological context and the results are highly reproducible. The described protocol can be used to localize bacteria either in a species-specific or universal manner in various tissues. The universal probe is especially useful to detect bacteria in polymicrobial diseases and to study a potential role of bacteria in diseases where the role of specific bacteria is not known.
Protocol
1. Probe Preparation
- PCR amplification of the probe
- For the P. gingivalis–specific probe, amplify a 343-bp DNA fragment of P. gingivalis 16S rRNA by PCR using the genomic DNA of P. gingivalis and the following primers: 5'- TGC AAC TTG CCT TAC AGA GG-3' and 5'- ACT CGT ATC GCC CGT TAT TC-3'10. Perform amplifications with the following cycle conditions: 35 cycles at 95 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 1 min 20 sec followed by a 5 min extension at 72 °C10,11.
- Amplify the eubacterial probe using a 70-bp DNA fragment (5'-CAGGTGCTGCATGGCTGTCGTCAGCTCGTGTTGTGAAATGTTGGGTTAAG TCCCGCAACGAGCGCAACCC-3') synthesized as an oligonucleotide and the following primers: 5'-CAG GTR CTG CMT GGY-3' and 5'-AGG GTT GCG CTC GTT-3'11. Perform amplifications with the following cycle conditions: 40 cycles at 9 4°C for 30 sec, 60 °C for 30 sec and 72 °C for 1 min 20 s followed by a 5 min extension at 72 °C11,12.
- Precipitate the PCR products (≥500 µl) by adding 3 M sodium acetate (pH 5.2) and 100% ethanol (at a 10:1:2 ratio) and incubating at -20 °C for 2 hr.
- Centrifuge the mixture at 14,000 x g for 15 min, resuspend the pellet in 100 µl of TE buffer, and measure the DNA concentration by optical density at 260 nm.
- DIG-labeling using a DIG DNA labeling and detection kit
- Mix 3 µg of the probe with water to a final volume of 15 µl.
- Denature the DNA at 95 °C for 10 min and quickly chill on ice.
- Add 2 µl of 10x hexanucleotide mix, 2 µl of dNTP-labeling mix, and 1 µl of Klenow enzyme to 15 µl of the denatured DNA.
- Mix and incubate at 37 °C for 24 hr to 48 hr and stop reaction by heating at 65 °C.
- Check the sensitivity of the DIG-labeled probe using a DIG DNA labeling and detection kit
- Manually load 1 µl of serial dilutions of the positive control probe provided in the kit and 1 ng of the DIG-labeled probe on the nylon membrane and dry for 5 min at RT.
- Briefly soak the membrane in 2x SSC buffer (0.3 M NaCl, 30 mM sodium citrate, pH 7.0) and incubate the membrane at 80 °C for 1 hr to immobilize the DNA.
- Briefly soak the membrane in maleic acid buffer (MAB, 0.1 M maleic acid, 0.15 M NaCl, pH 7.5) and incubate with anti-DIG alkaline phosphatase (AP)-conjugated antibody diluted (1:5,000) in 6 ml of blocking solution provided in the kit at RT for 30 min.
- Wash the membrane with MABT (MAB containing 1% Tween 20) and apply 40 µl of the NBT/BCIP solution mixed with 2 ml detection buffer (0.1 M Tris-HCl, 0.1 M NaCl, 0.05 M MgCl2, pH 9.5) onto the membrane for 1 min. If the sensitivity of the labeled probe is not satisfying, repeat steps from 1.1.3 to 1.3.4.
- Measure the DNA concentration of the labeled probe (OD260) and adjust the concentration to 100 ng µl-1.
- Check the specificity of the DIG-labeled probe
- Denature genomic DNA samples (25 ng 3 µl-1 per each sample) from various bacterial species, including the bacterium targeted by the specific probe, at 95 °C for 10 min and quickly chill on ice.
- Manually load the denatured DNA on the nylon membrane and dry for 20 min at RT.
- Briefly soak the membrane in 2x SSC buffer and incubate the membrane at 80 °C for 2 hr to immobilize the DNA.
- Block the membrane with a blocking solution at RT for 1 hr.
- Dilute the probe at 1 ng µl-1 in hybridization buffer, denature the diluted probes, and apply onto the membrane.
- Incubate the membrane at 45 °C for 1 hr.
- Wash the membrane three times with 2x SSC buffer.
- Briefly soak the membrane in MAB.
- Incubate with anti-DIG AP-conjugated antibody diluted (1:2,000) in 6 ml of blocking solution at RT for 1 hr.
- Wash the membrane with MABT and apply 40 µl of the NBT/BCIP solution mixed with 2 ml detection buffer.
- When the probe does not achieve the expected specificity, increase the hybridization temperature until the specificity is observed.
2. In Situ Hybridization
Note: To avoid drying of the specimens and reagents, perform all incubations in a humidified chamber lined with wet paper towels.
- De-paraffinization and rehydration of tissue sections
- Prepare 4 µm thick tissue sections from paraffin-embedded blocks. Formalin, paraformaldehyde, and zinc-based fixative are compatible with this protocol.
- Prepare all reagents using DEPC-treated water.
- Heat slides in a dry oven at 60 °C for 30 min and incubate slides at RT for 30 min.
- Immerse slides in 100% xylene for 5 min three times; use fresh xylene each time. Note: Do this de-waxing procedure in a chemical fume hood.
- Immerse slides in 100% ethanol for 5 min twice, using fresh ethanol each time, and rehydrate specimens in serial 90%, 80% and 70% ethanol solutions for 5 min each.
- Immerse slides in DEPC-treated water for 1 min and wash slides in DEPC-treated PBS (pH 7.4) for 6 min.
- Pretreatments of tissue sections
- Immerse slides in 0.1 N hydrochloric acid for 20 min and wash slides in DEPC-treated water for 30 sec.
- Draw a hydrophobic barrier surrounding specimens using a wax pen. Note: The size of the hydrophobic barrier depends on the size of specimens. Therefore, the volumes of reagents used in the following steps vary depending on the size of specimens but have to fill the hydrophobic barrier (50 µl for small specimens and 400 µl for large specimens).
- Treat specimens with 50 to 400 µl of proteinase K (1 to 10 µg ml-1 in DEPC-treated PBS) in a dry oven at 37 °C for 30 min and wash slides in DEPC-treated 1x PBS for 1 min.
- Soak slides in 4% paraformaldehyde in DEPC-treated PBS for 10 min, then wash slides in DEPC-treated PBS for 1 min.
- Soak slides in 0.1 M triethanolamine-HCl (pH 8.0) containing 0.5% acetic anhydride for 20 min and wash slides in DEPC-treated water for 30 sec.
- Hybridization with probe and washing
- Immerse slides in 2x SSC buffer for 20 min.
- Dilute the probe at 1 ng µl-1 in hybridization buffer (4x SSC, 50% [vol/vol] formamide, 1x Denhardt’s solution, 10% [wt/vol] dextran sulfate, 0.1% [wt/vol] sodium dodecyl sulfate, 0.4 mg ml-1 salmon sperm DNA). For negative controls, mix the labeled probe with a 10-fold excess amount of non-labeled probe.
- Denature the diluted probes and apply 50 to 400 µl onto tissue sections. Place a cover slip on the slide and seal with nail polish.
- Heat slides in PCR machine at 90 °C for 10 min and incubate slides in a humidified chamber O/N at 45 °C or optimal temperature determined at step 1.4.11.
- Cool the slides at 4 °C for 30 min, remove cover slips, and immerse slides in a serial SSC buffer as followings: 4x SSC for 10 min, pre-warmed (45 °C) 2x SSC for 20 min, 2x SSC for 10 min, and 0.2x SSC for 10 min.
- Detection with anti-DIG AP-conjugated antibody
- Wash slides in MABT for 5 min.
- Block with 50 to 400 µl of blocking solution with 1% Tween 20 for 20 min.
- Add 50 to 400 µl of anti-DIG-AP antibody diluted in blocking solution (1:1,000) anld incubate at 37 °C for 90 min.
- Wash slides in MABT for 10 min.
- Immerse slides in detection buffer containing 1% Tween 20 for 5 min.
- Treat with 50 to 400 µ of 1 mM levamisole (in detection buffer containing 1% Tween 20) for 5 min to inactivate endogenous alkaline phosphatase.
- Distribute 50 to 400 µl of the premixed NBT/BCIP solution (diluted in detection buffer at 1:100) onto each specimen and incubate slides in a humidified chamber at RT for 2 to 3 hr until the developed signal is satisfactory.
- Rinse slides with de-ionized water to stop visualization and apply 50 to 400 µl of 0.05% [wt/vol] methyl green as a counter stain at 37 °C for 10 min.
- Rinse slides with de-ionized water, dehydrate in 100% ethanol, and immerse in xylene.
- Apply 80 µl of mounting solution onto each slide and cover with cover slips.
Representative Results
Figure 1 shows dot blotting of DIG-labeled probes compared with the positive control probe provided in the kit to determine their sensitivity. The 343 bp P. gingivalis-specific probe is 25 times more sensitive than the 70 bp eubacterial probe. Figure 2 shows in situ hybridization of gingival tissues obtained from patients with chronic periodontitis for detection of P. gingivalis and eubacteria. The bacterial signals, shown in violet, were detected within the epithelia, the lamina propria, and biofilm located outside the tissue. However, the distribution of P. gingivalis and eubacteria within the gingival tissue was different. At high magnification (1,000X), various shapes (coccus, rod, fusiform, and hairy form) of bacteria were visible using the eubacterial probe, while only rod shaped bacteria were detected by the P. gingivalis-specific probe. The dispersed forms of bacterial signals were also observed.
Hajishengallis et al. demonstrated the essential role of commensal bacteria in the development of P. gingivalis-induced experimental periodontitis in mice14. The application of dextran sulfate sodium (DSS), a tight junction (TJ)-disrupting chemical, onto gingival mucosa induced periodontitis and alveolar bone loss in mice13. The eubacterial probe successfully detected bacteria within the gingival tissues from the DSS group that were not detected by the P. gingivalis-specific probe (Figure 3).
The eubacterial probe is useful for the study of the potential involvement of bacteria in other diseases. Since the International Agency for Research on Cancer designated Helicobacter pylori as a grade I carcinogen for gastric cancer15, potential involvement of bacteria in the development of other cancers has been widely studied. When sections of lung biopsies diagnosed with squamous cell carcinoma were in situ hybridized with the eubacterial probe, bacteria were detected both within the tumor cells and among the surrounding stromal/infiltrating cells (Figure 4). In situ hybridization using the eubacterial probe was also applied to explore the presence of bacteria in the lesions of oral lichen planus, an inflammatory immune disease with unknown etiology. Interestingly, bacterial signals were detected not only within the epithelia but also in the lamina propria where band-like infiltration was observed. Under high magnification, the bacterial signals were detected in the nuclei of many cells (Figure 5).
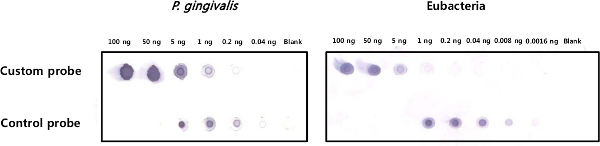 Figure 1. Sensitivity test of the DIG-labeled probes. Serially diluted custom-labeled probes and a positive control probe provided in the kit were dot blotted with anti-DIG-AP-conjugated antibody. After adding the substrate, the blot with the P. gingivalis-specific probe was developed for 30 sec, while the blot with the eubacterial probe was developed for 1 min. Please click here to view a larger version of this figure.
Figure 1. Sensitivity test of the DIG-labeled probes. Serially diluted custom-labeled probes and a positive control probe provided in the kit were dot blotted with anti-DIG-AP-conjugated antibody. After adding the substrate, the blot with the P. gingivalis-specific probe was developed for 30 sec, while the blot with the eubacterial probe was developed for 1 min. Please click here to view a larger version of this figure.
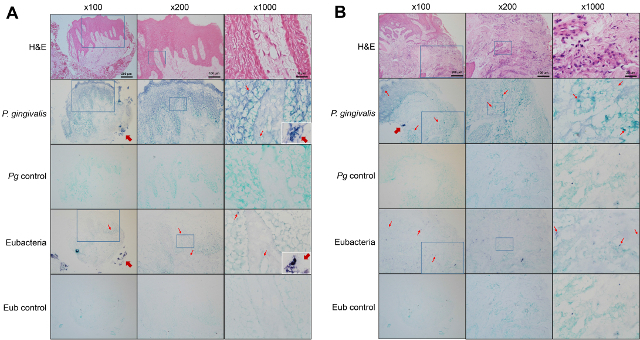 Figure 2. Detection of bacteria within gingival tissues obtained from patients with chronic periodontitis. Sections of gingival biopsies from the healthy site (A) and the diseased site (B) of a patient with periodontitis were subjected to hematoxylin-eosin (H&E) staining or in situ hybridization with either the P. gingivalis-specific probe or the eubacterial probe. Negative controls were hybridized with the probe mixed with 10-fold excess unlabeled probe. The magnified area is indicated with a square box. Representative positive signals, shown in violet, are marked with thin arrows. Thick arrows indicate plaque biofilm outside the tissue. The plaque biofilm examined at 1,000X magnification are shown in insets. Please click here to view a larger version of this figure.
Figure 2. Detection of bacteria within gingival tissues obtained from patients with chronic periodontitis. Sections of gingival biopsies from the healthy site (A) and the diseased site (B) of a patient with periodontitis were subjected to hematoxylin-eosin (H&E) staining or in situ hybridization with either the P. gingivalis-specific probe or the eubacterial probe. Negative controls were hybridized with the probe mixed with 10-fold excess unlabeled probe. The magnified area is indicated with a square box. Representative positive signals, shown in violet, are marked with thin arrows. Thick arrows indicate plaque biofilm outside the tissue. The plaque biofilm examined at 1,000X magnification are shown in insets. Please click here to view a larger version of this figure.
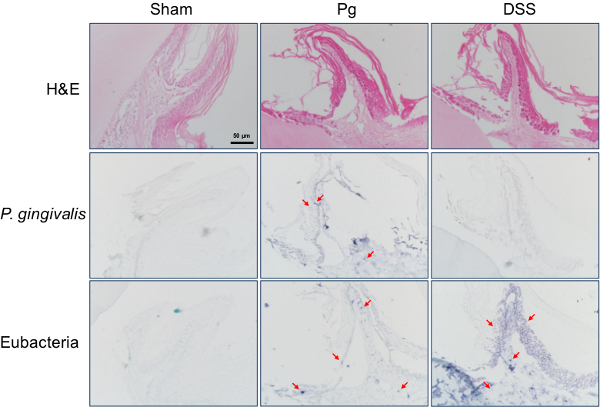 Figure 3. Detection of oral commensal bacteria within the gingival tissue of mouse. Experimental periodontitis was induced in BALB/c mice by oral inoculation of P. gingivalis (Pg) or application of DSS onto gingival mucosa. Gingival sections from mice were subjected to H&E staining or in situ hybridization with either the P. gingivalis-specific probe or the eubacterial probe. Representative positive signals, shown in violet, are marked with arrows. Original magnification is x400. Please click here to view a larger version of this figure.
Figure 3. Detection of oral commensal bacteria within the gingival tissue of mouse. Experimental periodontitis was induced in BALB/c mice by oral inoculation of P. gingivalis (Pg) or application of DSS onto gingival mucosa. Gingival sections from mice were subjected to H&E staining or in situ hybridization with either the P. gingivalis-specific probe or the eubacterial probe. Representative positive signals, shown in violet, are marked with arrows. Original magnification is x400. Please click here to view a larger version of this figure.
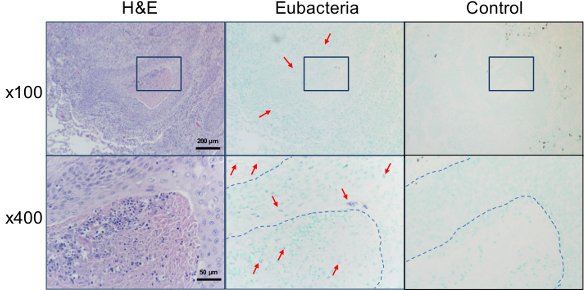 Figure 4. Detection of bacteria within cancer lesion. The sections of lung biopsies from patients diagnosed with squamous cell carcinoma were stained with H&E or in situ hybridized with either the universal eubacterial probe or the negative control probe. The magnified area is indicated with a square box. Representative positive signals, shown in violet, are marked with arrows. Border of tumor cells are indicated with dotted lines. Please click here to view a larger version of this figure.
Figure 4. Detection of bacteria within cancer lesion. The sections of lung biopsies from patients diagnosed with squamous cell carcinoma were stained with H&E or in situ hybridized with either the universal eubacterial probe or the negative control probe. The magnified area is indicated with a square box. Representative positive signals, shown in violet, are marked with arrows. Border of tumor cells are indicated with dotted lines. Please click here to view a larger version of this figure.
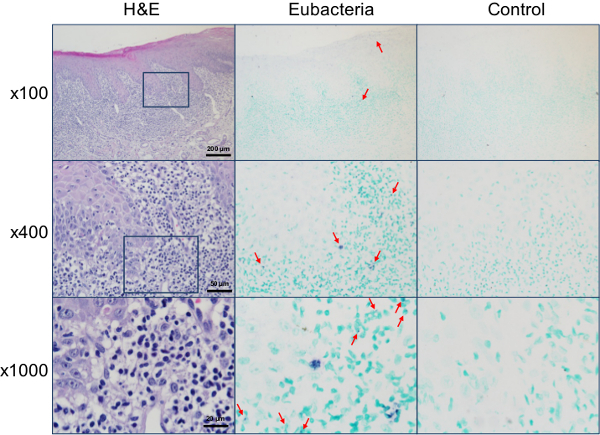 Figure 5. Detection of bacteria within the lesions of oral lichen planus. The sections of biopsy from buccal mucosa diagnosed as oral lichen planus were stained with H&E or in situ hybridized with either the universal eubacterial probe or the negative control probe. The magnified areas are indicated with a square box. Representative positive signals, shown in violet, are marked with arrows. Please click here to view a larger version of this figure.
Figure 5. Detection of bacteria within the lesions of oral lichen planus. The sections of biopsy from buccal mucosa diagnosed as oral lichen planus were stained with H&E or in situ hybridized with either the universal eubacterial probe or the negative control probe. The magnified areas are indicated with a square box. Representative positive signals, shown in violet, are marked with arrows. Please click here to view a larger version of this figure.
Discussion
Here a protocol to localize bacteria within paraffin-embedded tissues using a DIG-labeled DNA probe has been described. The probe targets the DNA or RNA molecules of bacterial 16S rRNA gene, and the 16S rRNA-targeting probes can be designed as either species-specific or universal. The specific hybridization of the P. gingivalis-specific probe to P. gingivalis but not to other oral bacteria has been previously shown10. In contrast, the eubacterial probe hybridized to all bacterial genomic DNA tested (17 different species), although hybridization efficiency varied12. The number of T-nucleotides within the probe determines the efficiency of DIG-labeling. Production of a DIG-labeled probe with high sensitivity is the most critical step for the successful implementation of the described protocol. Treatment of specimens with hydrochloric acid improves the signal-to-noise ratio through extraction of proteins and partial hydrolysis of target sequences. An increase in the concentration of proteinase K increases target accessibility of probes but reduces tissue preservation. Therefore, it is recommended to test several concentrations of proteinase K, depending on the types of tissue. The acetylation step decreases background binding of the negatively charged probe to the proteins of the tissue by acetylating the positively charged amino groups. After a series of these preparation steps, the tissue sections tear easily, thus, have to be handled with caution. Background is minimal compared with FISH. However, the signals from tissues with high endogenous alkaline phosphatase activity such as bones and mucus glands should be critically interpreted.
PCR-amplified DNA probes are easy to produce and stable compared to RNA probes. Immunohistochemical detection of hybridized probes allows identification of histological structures through comparison with H&E stained sections: the epithelia, connective tissue, inflammatory infiltrates, and blood vessels are well distinguished. One of the limitations of the described protocol is the inability to apply multiple probes at the same time. Additionally, the exclusion of the signals from bacterial contaminants on the surface of sections is limited.
This protocol can be adapted to detect other bacterial transcripts within tissue sections16. In addition, this protocol can be extended to double staining with immunohistochemical detection of a host cell marker.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This study was supported by a grant (2013R1A1A3005669) from the National Research Foundation of Korea and a grant (HI13C0016) of the Korean Health Technology R&D Project, Ministry of Health & Welfare.
References
- Takarada H, Cattoni M, Sugimoto A, Rose GG. Ultrastructural studies of human gingiva. II. The lower part of the pocket epithelium in chronic periodontitis. J. Periodontol. 1974;45(3):155–169. doi: 10.1902/jop.1974.45.3.155. [DOI] [PubMed] [Google Scholar]
- Allenspach-Petrzilka GE, Guggenheim B. Bacterial invasion of the periodontium; an important factor in the pathogenesis of periodontitis. J. Clin. Periodontol. 1983;10(6):609–617. doi: 10.1111/j.1600-051x.1983.tb01299.x. [DOI] [PubMed] [Google Scholar]
- Saglie FR, et al. The presence of bacteria in the oral epithelium in periodontal disease. II. Immunohistochemical identification of bacteria. J. Periodontol. 1986;57(8):492–500. doi: 10.1902/jop.1986.57.8.492. [DOI] [PubMed] [Google Scholar]
- Christersson LA, Albini B, Zambon JJ, Wikesjö UM, Genco RJ. Tissue localization of Actinobacillus actinomycetemcomitans. in human periodontitis. I. Light, immunofluorescence and electron microscopic studies. J. Periodontol. 1987;58(8):529–539. doi: 10.1902/jop.1987.58.8.529. [DOI] [PubMed] [Google Scholar]
- Saglie FR, Pertuiset J, Rezende MT, Nestor M, Marfany A, Cheng J. In situ. correlative immuno-identification of mononuclear infiltrates and invasive bacteria in diseased gingiva. J. Periodontol. 1988;59(10):688–696. doi: 10.1902/jop.1988.59.10.688. [DOI] [PubMed] [Google Scholar]
- Rautemaa R, et al. Intracellular localization of Porphyromonas gingivalis. thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 2004;10(5):298–305. doi: 10.1111/j.1601-0825.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- Marttila E, et al. Intracellular localization of Treponema denticola. chymotrypsin-like proteinase in chronic periodontitis. J. Oral Microbiol. 2014;6 doi: 10.3402/jom.v6.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AV, da Silva CM, Haffajee A, Colombo AP. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ. hybridization. J. Periodontal Res. 2007;42(3):236–243. doi: 10.1111/j.1600-0765.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- Abrams K, Caton J, Polson A. Histologic comparisons of interproximal gingival tissues related to the presence or absence of bleeding. J. Periodontol. 1984;55(11):629–632. doi: 10.1902/jop.1984.55.11.629. [DOI] [PubMed] [Google Scholar]
- Kim YC, et al. Presence of Porphyromonas gingivalis. and plasma cell dominance in gingival tissues with periodontitis. Oral Dis. 2010;16(4):375–381. doi: 10.1111/j.1601-0825.2009.01649.x. [DOI] [PubMed] [Google Scholar]
- Choi YS, et al. Porphyromonas gingivalis. and dextran sodium sulfate induce periodontitis through the disruption of physical barriers. Eur. J. Inflammation. 2013;10:419–431. [Google Scholar]
- Choi YS, Kim YC, Ji S, Choi Y. Increased bacterial invasion and differential expression of tight junction proteins, growth factors, and growth factor receptors in periodontal lesions. J. Periodontol. 2014;85(8):e313–e322. doi: 10.1902/jop.2014.130740. [DOI] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55(3):541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, et al. A Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and the complement. Cell Host Microbe. 2011;10(5):497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axon A. Gastric cancer and Helicobacter pylori. Aliment. Pharmacol. Ther. 2002;16(suppl. 4):83–88. doi: 10.1046/j.1365-2036.16.s4.14.x. [DOI] [PubMed] [Google Scholar]
- Fenhalls G, et al. In situ. detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect. Immun. 2002;70(11):6330–6338. doi: 10.1128/IAI.70.11.6330-6338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


