Abstract
Immune tolerance in pregnancy requires that the immune system of the mother undergoes distinctive changes in order to accept and nurture the developing fetus. This tolerance is initiated during coitus, established during fecundation and implantation, and maintained throughout pregnancy. Active cellular and molecular mediators of maternal-fetal tolerance are enriched at the site of contact between fetal and maternal tissues, known as the maternal-fetal interface, which includes the placenta and the uterine and decidual tissues. This interface is comprised of stromal cells and infiltrating leukocytes, and their abundance and phenotypic characteristics change over the course of pregnancy. Infiltrating leukocytes at the maternal-fetal interface include neutrophils, macrophages, dendritic cells, mast cells, T cells, B cells, NK cells, and NKT cells that together create the local micro-environment that sustains pregnancy. An imbalance among these cells or any inappropriate alteration in their phenotypes is considered a mechanism of disease in pregnancy. Therefore, the study of leukocytes that infiltrate the maternal-fetal interface is essential in order to elucidate the immune mechanisms that lead to pregnancy-related complications. Described herein is a protocol that uses a combination of gentle mechanical dissociation followed by a robust enzymatic disaggregation with a proteolytic and collagenolytic enzymatic cocktail to isolate the infiltrating leukocytes from the murine tissues at the maternal-fetal interface. This protocol allows for the isolation of high numbers of viable leukocytes (>70%) with sufficiently conserved antigenic and functional properties. Isolated leukocytes can then be analyzed by several techniques, including immunophenotyping, cell sorting, imaging, immunoblotting, mRNA expression, cell culture, and in vitro functional assays such as mixed leukocyte reactions, proliferation, or cytotoxicity assays.
Keywords: Immunology, Issue 99, Decidua, Dissociation, Isolation, Leukocytes, Myometrium, Placenta, Pregnancy, Uterus
Introduction
Immune tolerance in pregnancy is a period when distinctive changes occur within the immune system of the mother. These changes allow the mother to tolerate the fetus, a semi-allogenic graft1. The fetus expresses paternal major histocompatibility complex (MHC) antigens2, and fetal cells have been found in the maternal circulation3; however, the fetus is not rejected4,5. This enigma is not fully understood.
The most recent hypothesis states that maternal-fetal tolerance is created during coitus and fecundation6,7 and maintained to sustain a full-term pregnancy8-10. A breakdown of this maternal-fetal tolerance is considered a mechanism of disease during early and late stages of pregnancy10-16. Maternal-fetal tolerance involves the participation of various leukocyte sub-populations, including T cells (regulatory T cells, Th1 cells, Th2 cells, and Th17 cells), macrophages, neutrophils, mast cells, NK cells, and NKT cells, dendritic cells, and B cells, that change in density and localization throughout pregnancy15,17-19. Maternal-fetal tolerance is enriched at the maternal-fetal interface20 - the anatomical site where the immune system of the mother interacts with the fetal antigens20,21.
The maternal-fetal interface is created during placentation when the fetal extravillous trophoblast cells invade the uterine mucosa22-24. On the fetal side of this interface, the membranes surrounding the fetus create a specialized epithelial surface within the placenta, and the syncytiotrophoblast cells control the nutrient exchange through their direct contact with maternal blood22. On the maternal side of the interface, the decidua recruits a heterogeneous pool of leukocytes that in mice account for 30% to 50% of all decidual cells. In addition to their participation in maternal immune tolerance, these cells are key contributors to different processes during pregnancy, e.g., the protection of the reproductive tract from infections, fecundation, embryo implantation7,25, decidual angiogenesis26, vascular remodeling24,27, trophoblast invasion28, placental development24,25, and, ultimately, labor and delivery15,17. Therefore, the study of the leukocytes involved in maternal-fetal tolerance is essential to elucidating the pathogenesis of pregnancy-related complications.
While the use of immunohistochemistry and immunofluorescence has generated data for the direct visualization and localization of uterine, decidual, or placental leukocytes29,30, flow cytometry analysis has further revealed specific subsets of leukocytes in each of these tissues31,32. Additionally, flow cytometry has been used to determine the density and proportion of maternal-fetal interface leukocytes33 and expression levels of extracellular and intracellular proteins8-10,34. Flow cytometric analysis of leukocytes at the maternal-fetal interface requires a single-cell suspension. In order to isolate infiltrating leukocytes from the decidual, uterine, and placental tissues, two methods of tissue dissociation have been used: mechanical and enzymatic. Both methods allow the separation of infiltrated leukocytes from the extracellular matrix (ECM) of these tissues. Enzymatic tissue dissociation is superior to mechanical tissue dissociation as it allows a higher yield of leukocytes with less shear-force-associated damage35. Consequently, mechanical tissue dissociation requires pooling tissues36, which may increase the variability and heterogeneity of the samples. Yet, mechanical dissociation may be the choice when the antigen of interest can be altered by enzymatic dissociation or when the functionality of the cells of interest need to be preserved (e.g., cytotoxicity of NK cells)35.
The use of proteolysis with specific enzymes to degrade the ECM eliminates the low yields observed with mechanical dissociation. Several studies have reported the use of trypsin32, collagenase37, DNase31, dispase38, and commercial cocktails of various enzymes32,39. However, the nature and concentration of different enzymes and the duration of digestion must be meticulously defined and validated in order to ensure maintenance of the integrity of the cell surface antigenic epitopes required for immunophenotyping. The various surface structures are differentially susceptible to destruction by different enzymes, with some enzymes, such as trypsin, being notorious for stripping leukocyte surface epitopes recognized by many monoclonal antibodies.
Introduced herein is a method using a proteolytic and collagenolytic enzymatic cocktail, called Accutase. This enzymatic solution is gentle enough while still efficient in dissociating murine tissues at the maternal-fetal interface, and does not require the addition of other dissociating reagents or serum to terminate the dissociation reaction. Moreover, it is ready to use as supplied and, although the time of dissociation needs to be validated, it is more robust than the above-noted enzymes40,41.
The utilization of a combination of both types of tissue disaggregation improves the quality and amount of cells obtained; thus, several studies have implemented the combined use of mechanical and enzymatic dissociation with satisfactory results31,32,37. The protocol described herein was established and validated in our laboratory; it uses a combination of a gentle mechanical dissociation followed by a robust enzymatic disaggregation. This protocol allows the isolation and further study of the infiltrating leukocytes in murine tissues at the maternal-fetal interface (uterus, decidua, and placenta). The following protocol also maintains the integrity of cell surface markers and yields enough viable cells for downstream applications as demonstrated by flow cytometric analysis. Finally, this protocol maintains the consistency of cell preparation for the analysis and comparison of different murine tissues composing the maternal-fetal interface.
Protocol
Before working with the samples mentioned in this protocol, animal ethical approval must be given by the Local Research Ethics Committee and the Institutional Review Boards. When working with animal blood, cells, or hazardous agents as mentioned in this protocol, the proper biosafety and laboratory safety actions must be followed.
1. Mouse Handling and Tissue Collection
Prepare a sterile workstation and obtain sterile tools for tissue collection. These tools will include large and small surgical scissors, forceps, and fine-tip tweezers. Include small Petri dishes labeled with the appropriate tissue names, filled with 1x phosphate-buffered saline (PBS) solution (3 - 5 ml) equilibrated to RT (20 - 22 °C). Also include a large Petri dish, unlabeled, filled with 1x PBS solution (10 - 15 ml).
Euthanize a pregnant mouse (10.5 to 19.0 days post-coitum (dpc) or prior to delivery) using carbon dioxide (CO2). To ensure that the mouse is euthanized, use the cervical dislocation technique. Note: Prior to 10.5 dpc, it is difficult to manually separate the decidual tissues from the uterine tissues. If the isolation of decidual leukocytes is not required, leukocytes from the uterine tissues of non-pregnant mice or those from mice at <10.5 dpc can also be performed by following this protocol.
Using a pair of sterile surgical scissors, completely remove the lower abdominal portion of the skin and muscular tissue. With forceps, move aside all other organs until the uterus and ovaries are visible. With the uterus still attached, use the forceps to move it outside of the peritoneal cavity (Figure 1A).
Locate the ovaries distal to the oviduct and uterotubal junction of each uterine horn. Using the small surgical scissors, make an incision at the uterotubal junction and separate the uterine horns from the mesentery containing the uterine vessels; then, excise at the cervix to detach the uterine horns from the peritoneal cavity (Figure 1B).
With small surgical scissors, make an incision between the cervix and the lower segment of the uterine horns. Leave attached a portion of the cervix during this process (Figure 1C) to avoid opening the uterine horn.
Immerse the uterine horns (including the cervix) in a large Petri dish filled with 1x PBS solution (10 - 15 ml) to hydrate the implantation sites.
While still immersed in 1x PBS solution, trim any fat from the uterine horns with the small surgical scissors. Keep the tissues immersed in 1x PBS solution throughout the dissection.
Holding the uterine horns in place with the forceps, remove one of the implantation sites from the uterus with a transverse cut through the inter-implantation region, using the small surgical scissors (Figure 1D). The implantation site contains the fetus, surrounded by the chorioallantoic membrane, and the uterine, decidual, and placental tissues.
Using the forceps, hold the site in place and make a small incision in the uterine wall adjacent to the placental and decidual tissues (Figure 1E). Trim around the perimeter of the placenta/decidua until these become separated from both the uterine wall and the chorioallantoic membrane that includes the fetus (Figure 1F).
Remove the placenta and attached decidual tissues and place in 1x PBS solution (3 - 5 ml; see step 1.12).
Place the fetus surrounded by the chorioallantoic membrane attached to the uterine tissues in a Petri dish filled with 1x PBS solution (3 - 5 ml). The hydration of these tissues will facilitate the separation of the chorioallantoic membrane from the uterine wall (see step 1.14).
Using a pair of fine-tip tweezers, gently peel the decidual tissue (white-grey layer) from the placental surface. Perform this step carefully to maintain the integrity of the placental and decidual tissues (Figure 1G).
Place the placenta and decidual tissues in two separately labeled Petri dishes filled with 1x PBS solution (3 - 5 ml each).
Gently remove the uterine wall from the chorioallantoic membrane with fine-tip tweezers. These are the uterine tissues.
Place the uterine tissues in a Petri dish filled with 1x PBS solution (3 - 5 ml).
Repeat steps 1.8 through 1.15 until all implantation sites have been processed.
Collect several uterine, decidual, and placental tissues to improve the yield of cells during enzymatic digestion. The amount of tissues depends on the litter size; however, leukocyte isolation requires >2 pieces of tissue. Note: For more detailed information regarding the dissection of the implantation sites on different gestational days, view the pictures found in Chapter Two of The Guide to Investigation of Mouse Pregnancy42.
2. Uterine, Decidual, and Placental Tissue Dissociation
Label several sterile 2 ml conical tubes with the appropriate tissue name, and place a vial with 1,000 µl of enzymatic solution on ice.
From the Petri dish filled with 1x PBS solution, transfer two pieces (100 - 150 mg) of the uterine, decidual, or placental tissues to a labeled 2 ml conical tube.
Add 500 µl of cold enzymatic solution into each 2 ml conical tube.
With fine sterile scissors, begin to dissociate the tissue immersed in the enzymatic solution until it is finely minced. Over time, the suspension will develop a milky appearance. This step should not exceed more than 2 min to prevent over-manipulation of the sample.
Once the tissue has been dissociated, add an extra 500 µl of cold enzymatic solution to the 2 ml conical tube.
Place the 2 ml conical tube on ice.
Repeat steps 2.2 through 2.6 with each individual tissue until all tissues have been processed.
Transfer all 2 ml conical tubes with homogenized tissue (minced tissue + enzymatic solution) samples from the ice into an incubator at 37 ºC and perform a gentle orbital agitation (80 rpm). Incubate for 30 - 35 min. Make sure the temperature of the incubator is stabilized before starting the incubation time.
Following incubation, remove the 2 ml conical tubes with dissociated tissues from the incubator and place them on ice to prevent any further digestion.
Label sterile 50 ml conical tubes according to tissue type. Remove the lids and replace them with a sterile cell strainer (100 µm pore size).
Pour the dissociated tissues from the 2 ml conical tubes through the cell strainer using ~20 ml of 1x PBS solution. The fragments of tissue will remain in the cell strainer and the cell suspension will pass through it.
Wash the dissociated tissues in the cell strainer 2 - 3 times with 20 ml of 1x PBS solution using a transfer pipette.
Rinse the empty 2 ml conical tubes with 1x PBS solution (1 ml) and pour the contents through the cell strainers.
Remove the cell strainers from the 50 ml conical tubes and replace the lids. Centrifuge the tubes at 1,250 x g for 10 min at 4 ºC.
Carefully, aspirate the supernatant without disturbing the cell pellet. The cell pellet contains the isolated leukocytes and stromal cells.
Re-suspend the cell pellet in 1 ml of RPMI culture medium without fetal bovine serum (FBS). Mix gently. Do not use vortex.
Add 500 µl of neat FBS to a 5 ml polystyrene plastic tube and slowly overlay the cell suspension.
Centrifuge for 10 min at 1,100 x g without the brake at RT to pellet viable cells while cellular debris is retained at the PBS/FBS interface.
Aspirate the supernatant without touching the cell pellet. The cell pellet contains mostly viable cells.
- Optionally: To concentrate the mononuclear cells, use a 20% saturated polysaccharide solution, as described below.
- Resuspend the cell pellet in 1,000 µl of RPMI culture medium without FBS.
- Add 1 ml of 20% saturated polysaccharide solution to a 5 ml polystyrene plastic tube and slowly overlay the cell suspension on top of this solution, according to the manufacturer’s instructions. While layering the sample, avoid mixing the 20% saturated polysaccharide solution with the cell suspension.
- Centrifuge with a swing-out rotor at 500 x g for 30 min at RT without brake. It is very important to turn off the brake to avoid mixing the cells and losing the gradient. Mononuclear cells will be found in the interface between the 20% saturated polysaccharide solution and the 1x PBS solution.
- Aspirate and discard the supernatant. The cell pellet contains mostly mononuclear cells. Note: To perform viability staining and intracellular staining, see step 3. To perform immunophenotyping, see step 4. To perform viability staining and extracellular staining only, see step 5. To perform a cell culture of isolated leukocytes, see step 6. To perform magnetic cell sorting, see step 7.
3. Viability Staining for Fixable Cells
Resuspend the cell pellet in 1,100 µl of 1x PBS solution. Mix gently using a micro-pipette.
Transfer 100 µl of the cell suspension into a 5 ml polystyrene plastic tube. Add 400 µl of 1x PBS solution to the 5 ml polystyrene tube. This tube is a control for tissue auto-fluorescence. Store at 4 ºC until step 3.5.
Transfer the remaining 1,000 µl of the cell suspension to a new 5 ml polystyrene plastic tube. Add 1 µl of the viability dye and gently homogenize. Incubate for 30 min in darkness at 4 ºC.
Following incubation, add 1,000 µl of 1x PBS solution. Mix gently by hand.
Centrifuge both 5 ml polystyrene plastic tubes (steps 3.2 and 3.3) at 1250 x g for 10 min at 4 ºC. Aspirate and discard the supernatant. Note: The cell pellet (step 3.3) will be used in step 4. The cell pellet (step 3.2) will serve as a control for tissue auto-fluorescence.
4. Immunophenotyping
Resuspend the cell pellet in 50 µl of anti-mouse CD16/CD32 (diluted 1:100 in FACS buffer [0.1% BSA, 0.05% sodium azide, 1x PBS solution, ph 7.4]). Mix the cell suspension gently. Do not use the vortex.
Incubate the cell suspension in the 5 ml polystyrene tube in darkness at 4 °C for 10 min.
Following incubation, add 50 µl of each anti-leukocyte monoclonal antibody reacting with extracellular cell surface markers or isotype-matched control antibody (antibody dilutions must be validated). For example, to analyze leukocyte sub-populations, use anti-mouse antibodies reacting with the following leukocyte-cell surface markers: CD45, Ly6G, F4/80, CD11c, CD49b, B220, CD3, CD4, and CD8 (Table 1).
Once antibodies against extracellular markers have been added to the 5 ml polystyrene plastic tube, mix gently. Incubate in darkness for 30 min at 4 °C.
During this incubation, prepare and pre-warm the fixation buffer solution (diluted with deionized water, 1:5) to 37 ºC, if required. Keep the buffer at 37 ºC in darkness until its use is necessary.
After 30 min incubation, add 500 µl of FACS buffer in the 5 ml polystyrene plastic tube and gently mix. This step is needed to wash the cells and remove any excess of unbound antibody.
Centrifuge the samples at 1,250 x g at 4 °C for 10 min. Aspirate and discard the supernatant. Note: If intracellular staining is not required, see step 5. If intracellular staining is required, perform permeabilization and intracellular staining here, and then proceed with the fixation (step 4.8).
Add 500 µl of the pre-warmed fixation buffer solution to the cell pellet and incubate in darkness for 10 min at 37 ºC.
Dispense 500 µl of FACS buffer into the 5 ml polystyrene plastic tube. Mix gently.
Centrifuge the samples at 1,250 x g at 4 °C for 10 min. Aspirate and discard the supernatant.
Add 500 µl of FACS buffer to each sample. These samples can now be analyzed by flow cytometry. Acquire the samples immediately for best results.
5. Viability Staining for Unfixable Cells
After extracellular immunophenotyping, resuspend the cell pellet in 500 µl of FACS buffer.
Add 1 µl of 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI, 200 µg/ml) just before acquiring the sample.
Visualize the samples within 1 - 2 min after adding DAPI using a flow cytometer.
6. Cell Culture
Prepare a workstation under a sterile fume hood. Pre-warm the RPMI culture medium to 37 °C using a water bath.
Resuspend the cell pellet in 1000 µl of FACS buffer for cell counting.
Count the number of viable cells using an automatic cell counter or hemocytometer and 0.4% trypan blue solution, following the manufacturer's instructions. Record the total live cell count.
Centrifuge the cells at 1,250 x g at 4 °C for 10 min to remove the FACS buffer.
Resuspend the cell pellet in 500 µl of pre-warmed RPMI culture medium by gently pipetting 2 to 3 times.
Plate the desired number of cells in a sterile 24-well plate and incubate at 37 °C. The incubation time will be determined by the research question.
7. Magnetic Cell Sorting
Set up a clean workstation with the following materials: MS columns, magnetic cell separator, 15 ml conical tubes, 30 µm pre-separation filters, and a multi-stand.
Resuspend the cell pellet in 500 µl of MACS buffer (0.5% BSA, 2mM EDTA, 1x PBS solution, pH 7.2).
Using an automatic cell counter or hemocytometer and 0.4% trypan blue solution, count and record the total live cell count.
Centrifuge the cells at 1,250 x g at 4 °C for 10 min. Aspirate and discard the supernatant. Add another 500 µl of MACS buffer.
Proceed to perform the magnetic separation following the manufacturer’s recommendations.
Determine purity by flow cytometry or microscopy.
Representative Results
The dissection of murine tissues from the maternal-fetal interface is shown in Figure 1; this procedure includes opening the peritoneal cavity (Figure 1A,B), uterine horns (Figure 1C) including the implantation sites (Figure 1D), and the collection of the uterine tissues (Figure 1E), placenta (Figure 1F), and decidual tissues (Figure 1G) at 16.5 dpc. Figure 2 shows the morphology of isolated macrophages (F4/80+) collected from the decidual and uterine tissues at 16.5 dpc using magnetic cell sorting. Isolated macrophages maintain the ability to release cytokines (data not shown). The yield of viable cells isolated from the decidua, uterus, and placenta is shown in Figure 3, and cell viability is greater than 70% in all tissues. Figure 4 shows the gating strategy for analyzing polymorphonuclear and mononuclear leukocytes within the singlets and viability gates, including T cells (CD45+CD3+), neutrophils (CD45+Ly6G+), macrophages (CD45+F4/80+), dendritic cells (CD45+CD11c+), and NK cells (CD45+CD49b+) at 16.5 dpc. A high proportion of macrophages co-express CD11c. Figure 5 shows neutrophils, macrophages, and T cells in the decidual, uterine, and placental tissues at 16.5 dpc. Figure 6 shows the gating strategy for analyzing lymphocytes within the singlets and viability gates, including T cells (CD45+CD3+) and B cells (CD45+B220+) at 16.5 dpc. T cells include CD4+ and CD8+ T cells. Murine tissues at the maternal-fetal interface also include CD3+CD4-CD8- (gamma-delta T cells) in high proportions. Figure 7 shows T cells and B cells in the decidual, uterine, and placental tissues at 16.5 dpc.
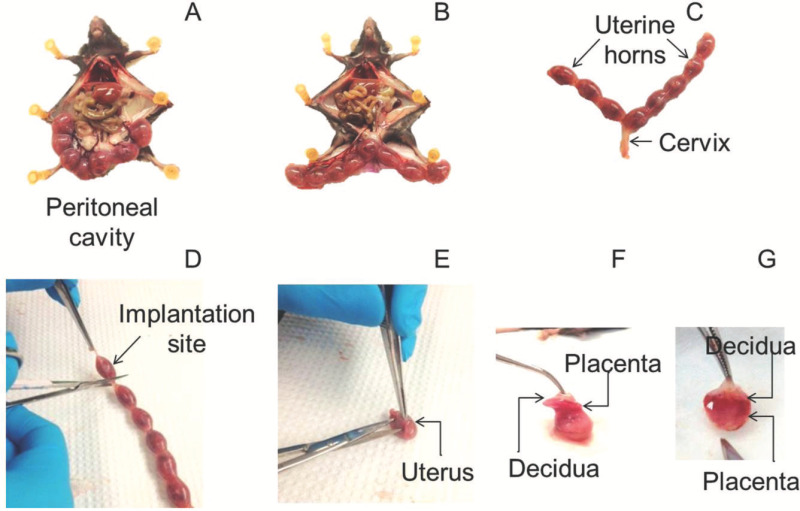 Figure 1. Tissue dissection. (A) Uterine horns at 16.5 dpc in a B6 mouse. The uterine horns are attached to the mesentery and draining vessels. (B) Uterine horns dissected from the mesentery and still attached to the cervix. (C) Uterine horns including the implantation sites and cervix. (D) Dissection of an implantation site. (E) Dissection of the uterine tissues from the implantation site. (F) Separation of the placenta and decidua from the implantation site. (G) Detachment of the decidua (white-gray layer) from the placenta. Please click here to view a larger version of this figure.
Figure 1. Tissue dissection. (A) Uterine horns at 16.5 dpc in a B6 mouse. The uterine horns are attached to the mesentery and draining vessels. (B) Uterine horns dissected from the mesentery and still attached to the cervix. (C) Uterine horns including the implantation sites and cervix. (D) Dissection of an implantation site. (E) Dissection of the uterine tissues from the implantation site. (F) Separation of the placenta and decidua from the implantation site. (G) Detachment of the decidua (white-gray layer) from the placenta. Please click here to view a larger version of this figure.
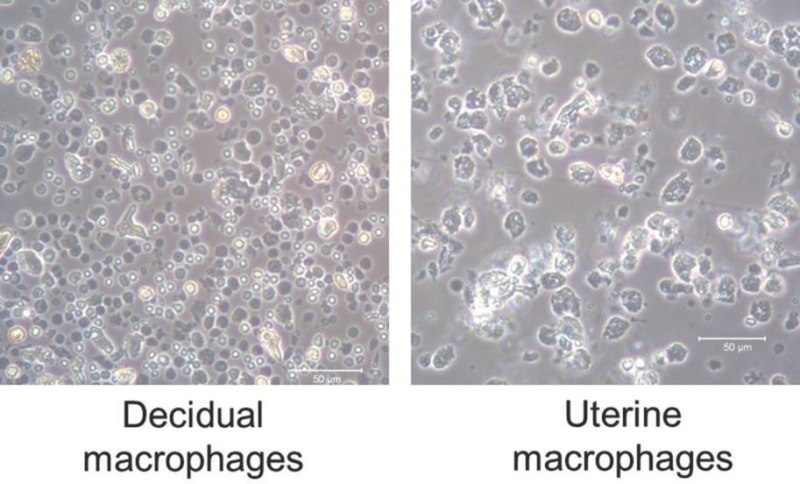 Figure 2. Macrophages isolated from decidual and uterine tissues. Macrophages (F4/80+ cells) isolated from the decidual and uterine tissues at 16.5 dpc using magnetic cell sorting. 20X magnification. Please click here to view a larger version of this figure.
Figure 2. Macrophages isolated from decidual and uterine tissues. Macrophages (F4/80+ cells) isolated from the decidual and uterine tissues at 16.5 dpc using magnetic cell sorting. 20X magnification. Please click here to view a larger version of this figure.
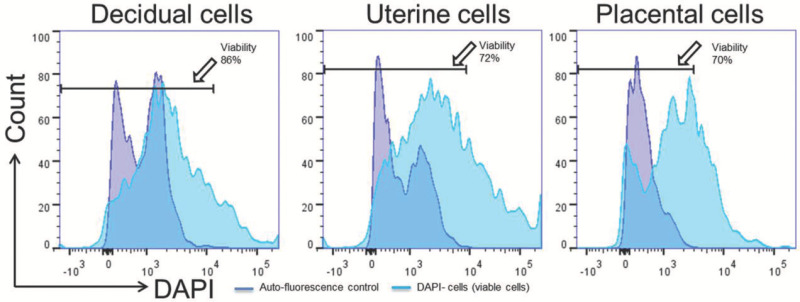 Figure 3. Viability of isolated cells. Viable cells (DAPI- cells represented with arrows) isolated from decidual, uterine, and placental tissues. Please click here to view a larger version of this figure.
Figure 3. Viability of isolated cells. Viable cells (DAPI- cells represented with arrows) isolated from decidual, uterine, and placental tissues. Please click here to view a larger version of this figure.
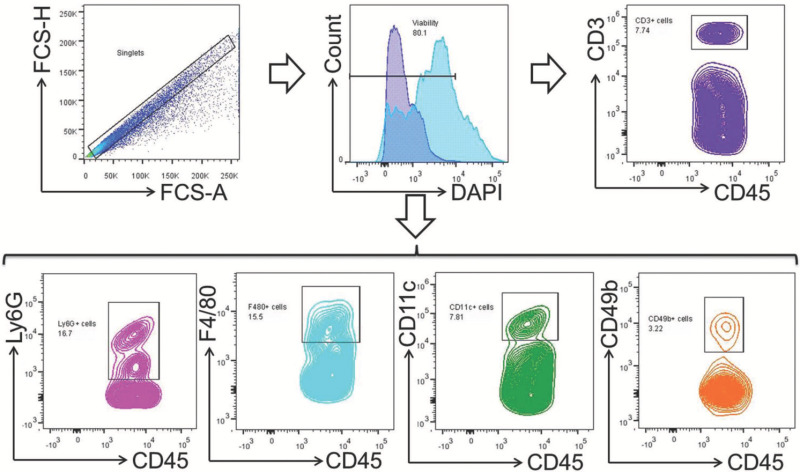 Figure 4. Gating strategy for polymorphonuclear and mononuclear leukocytes. Total leukocyte population was gated within the singlets and viability gates. T cells (CD45+CD3+), macrophages (CD45+F4/80+), neutrophils (CD45+Ly6G+), dendritic cells (CD45+CD11c+), and NK cells (CD45+CD49b+) were gated within the total-leukocyte gate (CD45+). Please click here to view a larger version of this figure.
Figure 4. Gating strategy for polymorphonuclear and mononuclear leukocytes. Total leukocyte population was gated within the singlets and viability gates. T cells (CD45+CD3+), macrophages (CD45+F4/80+), neutrophils (CD45+Ly6G+), dendritic cells (CD45+CD11c+), and NK cells (CD45+CD49b+) were gated within the total-leukocyte gate (CD45+). Please click here to view a larger version of this figure.
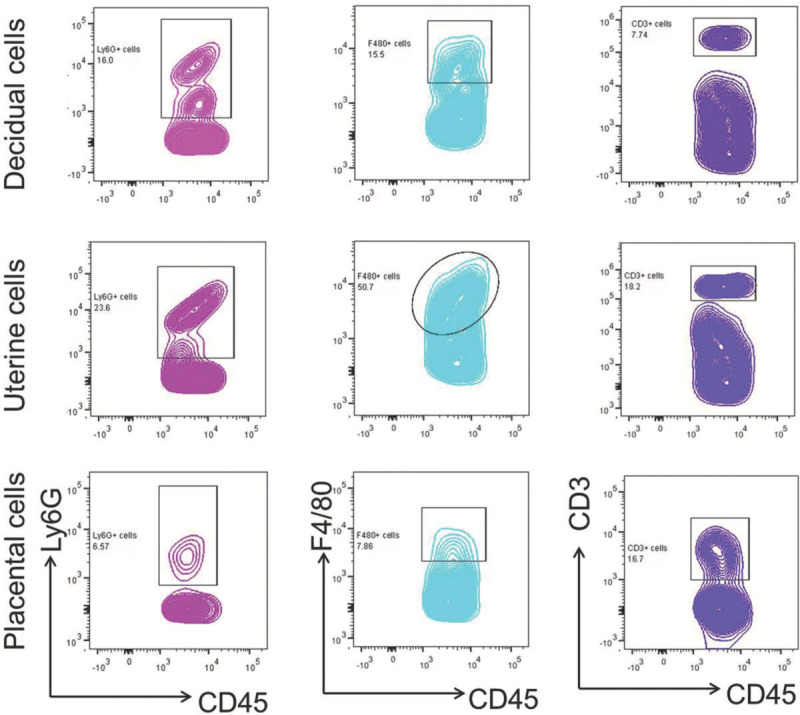 Figure 5. Neutrophils, macrophages, and T cells in murine tissues at the maternal-fetal interface. Neutrophils (CD45+Ly6G+), macrophages (CD45+F4/80+), and T cells (CD45+CD3+) were gated within the viability and total-leukocyte (CD45+) gates in isolated decidual, uterine, and placental cells. Please click here to view a larger version of this figure.
Figure 5. Neutrophils, macrophages, and T cells in murine tissues at the maternal-fetal interface. Neutrophils (CD45+Ly6G+), macrophages (CD45+F4/80+), and T cells (CD45+CD3+) were gated within the viability and total-leukocyte (CD45+) gates in isolated decidual, uterine, and placental cells. Please click here to view a larger version of this figure.
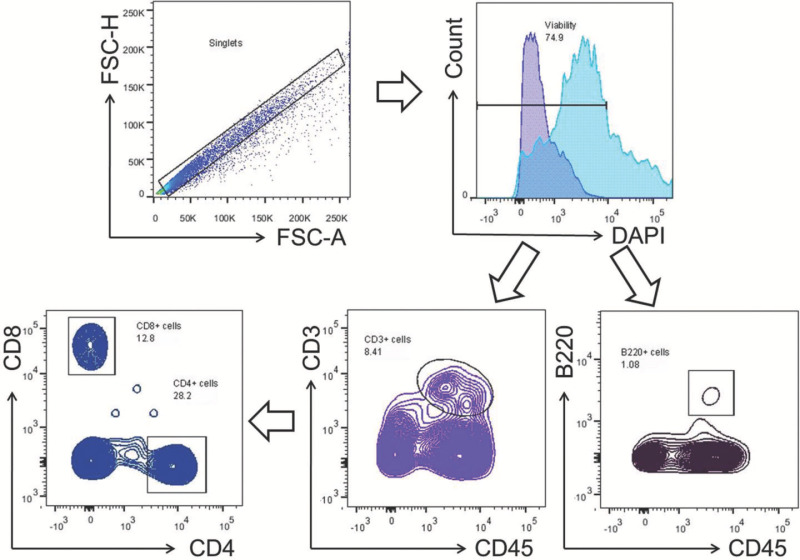 Figure 6. Gating strategy for lymphocytes. Mononuclear cells were gated within the singlets and viability gates. T cells (CD3+) and B cells (B220+) were gated within the viability gate. CD4+ and CD8+ T cells were gated within the T-cell gate (CD3+). Please click here to view a larger version of this figure.
Figure 6. Gating strategy for lymphocytes. Mononuclear cells were gated within the singlets and viability gates. T cells (CD3+) and B cells (B220+) were gated within the viability gate. CD4+ and CD8+ T cells were gated within the T-cell gate (CD3+). Please click here to view a larger version of this figure.
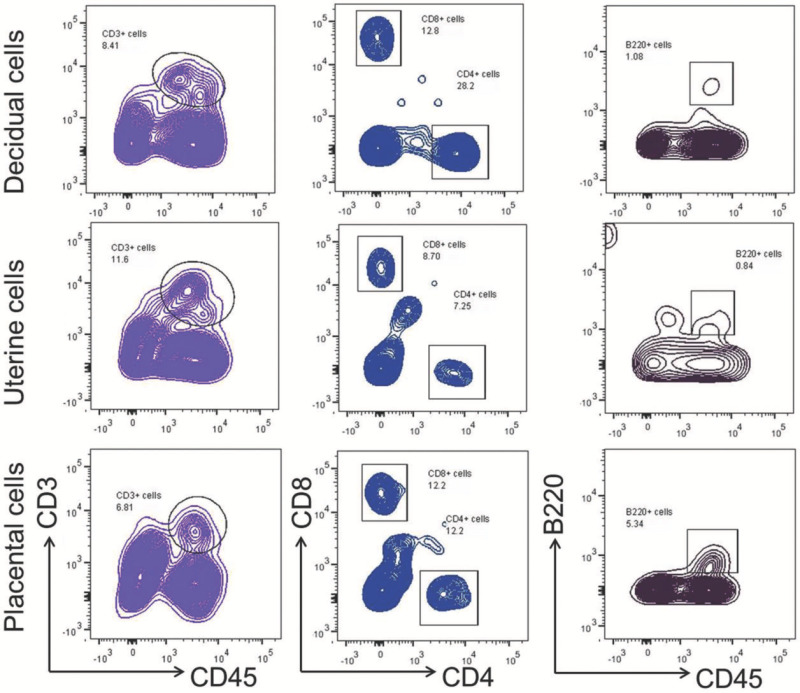 Figure 7. Lymphocyte sub-populations in murine tissues at the maternal-fetal interface. T cells (CD3+) and B cells (B220+) were gated within the viability gate in isolated decidual, uterine, and placental cells. CD4+ and CD8+ T cells were gated within the T-cell gate (CD3+). Please click here to view a larger version of this figure.
Figure 7. Lymphocyte sub-populations in murine tissues at the maternal-fetal interface. T cells (CD3+) and B cells (B220+) were gated within the viability gate in isolated decidual, uterine, and placental cells. CD4+ and CD8+ T cells were gated within the T-cell gate (CD3+). Please click here to view a larger version of this figure.
| Cell marker | Fluorochrome | Clone | Company | Catalog Number |
| LIVE/DEAD | DAPI | - | Life Technologies | L23105 |
| CD45 | V450 | 30-F11 | BD Biosciences | 560501 |
| CD3 | FITC | 145-2C11 | BD Biosciences | 553062 |
| CD4 | APC | RM4-5 | BD Biosciences | 553051 |
| CD8 | PE-CF594 | 53-6.7 | BD Biosciences | 562283 |
| B220 | APC-Cy7 | RA3-6B2 | BD Biosciences | 552094 |
| F4/80 | PE | BM8 | eBiosciences | 12-4801-82 |
| Ly6G | APC-Cy7 | 1A8 | BD Biosciences | 560600 |
| CD49b | APC | DX5 | BD Biosciences | 560628 |
| CD11c | PE-Cy7 | HL3 | BD Biosciences | 558079 |
Table 1. List of antibodies utilized for leukocyte subset immunophenotyping
Discussion
The collection of consistent data that records the abundance and phenotypic characteristics of infiltrating leukocytes at the maternal-fetal interface is essential to understanding the pathogenesis of pregnancy-related complications. Several techniques have been described that facilitate the isolation of infiltrating leukocytes from the murine tissues at the maternal-fetal interface throughout pregnancy31,38,39,43-46. However, each technique is different, uses different enzymes or enzyme combinations, requires different dissociation times, does not specify quantities of tissue, and, most importantly, does not always specify the viability of the isolated cells. The protocol described herein allows the isolation of infiltrating leukocytes from the murine tissues at the maternal-fetal interface with high viability, and provides detailed information about the commercial reagents, buffer preparation, tissue quantities, and incubation times validated in the laboratory.
One of the most critical steps of the leukocyte isolation process is the tissue dissociation; this step involves mechanical homogenization and/or enzymatic reactions that can alter the integrity of extracellular proteins used in phenotypic characterization47. The protocol described herein offers a novel approach that combines the use of gentle mechanical and enzymatic tissue dissociation techniques to preserve the integrity of extracellular markers in the leukocytes isolated from the placenta and the decidual and uterine tissues of mice.
Single enzymes and combinations of different enzymes have been used to isolate infiltrating leukocytes from the murine tissues at the maternal-fetal interface31,32,39,44. In many cases, these enzymes that are prepared to specific concentrations by hand in the laboratory may be subject to human error. Here, instead, a ready-to-use purified collagenase/neutral protease cocktail, Accutase, has been implemented in the laboratory; as a commercially available enzyme preparation, it has been shown to provide reliable results in cell culture48. This enzymatic solution is known to effectively detach macrophages from the culture plates without scraping and, most importantly, without losing surface antigens48. This prepared enzyme has also been used to process the digestion of human and animal nervous system tissues, resulting in viable isolated cells that remain sustainable for long periods, which allows their subsequent culture47. Moreover, this enzymatic solution preserves CD24 antigenicity in isolated cells from central nervous system tissues47. When compared to Liberase-1, another cocktail of collagenase and neutral protease, neither Accutase nor Liberase-1 generate free DNA aggregates; however, Accutase is gentler than Liberase-1 during tissue dissociation47. The collagenase/neutral protease cocktail used in this protocol has also demonstrated superiority to trypsin in the preservation of CD44, a cancer stem cell surface marker49. This laboratory’s studies have consistently noted that this enzymatic solution preserves mouse leukocyte surface antigens. Indeed, informative differences have been found in the expression of extracellular markers in macrophages (CD11b+F4/80+ cells), neutrophils (CD11b+Ly6G+ cells), NKT cells (CD3+CD49b+ cells), T cells (CD3+ cells) and B cells (B220+CD19+ cells), including CD4, CD8, CD69, CD25, CD40L, PD1, CD44, CD62L, and CTLA4, and in cytokine release. Therefore, the method described herein is optimal for immunophenotyping of infiltrating leukocytes at the maternal-fetal interface in mice, as shown in the representative results.
One important advantage of this method is the simultaneous determination of several extracellular and intracellular antigens within the viability gate. The dye most used to determine cell viability is propidium iodide (PI); however, its use is limited since it can be used only in combination with FITC. This is because PI cannot be adequately distinguished from most other fluorochromes excited by blue and red lasers50. A solution is to use DAPI50 or any other dye that is excited by UV lasers but not by blue and red lasers commonly used for immunophenotyping. This protocol allows immunophenotyping of viable cells as it includes the use of DAPI50 for unfixable cells or the use of viability dye for fixable cells.
A second important advantage of the protocol described herein is that it allows the isolation of leukocytes with a high yield of viable cells. The representative data shows that 70% to 89% of the isolated cells are viable. This is of great importance as this protocol has allowed the study of the functional properties of the isolated cells from murine tissues at the maternal-fetal interface. For example, cultures of the decidual and uterine macrophages and the study of cytokine release under stimulation have been performed using this method.
To achieve successful results, the application of the described protocol is important when considering the following factors: 1) tissue collection must be performed within 5 to 10 min after opening the peritoneal cavity, and these tissues must be placed on ice to preserve the viability of the isolated cells; 2) mechanical tissue homogenization must be performed using fine-tip scissors and cannot exceed 2 min since longer periods have been demonstrated to reduce the yield of viable cells; 3) the duration of incubation with the enzymatic solution must be less than 1 hr since its activity diminishes after this period of time; 4) the temperature of incubation with the enzymatic solution must be maintained at 37 °C to obtain the optimal activity of this cocktail of enzymes; 5) cell pellet manipulation must be done gently with micropipettes because use of the vortex can damage the integrity of the cells (isolated cells are differentiated and abrupt manipulation can easily reduce their viability); 6) buffer and centrifuge temperatures must be kept at the same temperature as the cell suspension; 7) isolated cells must be processed for immunophenotyping or used immediately as their viability reduces rapidly; and 8) when performing immunophenotyping, samples must be acquired using a flow cytometer immediately for best results.
A limitation of this protocol is the cost of the enzymatic solution, which is more expensive than other enzymes with similar functions, e.g., trypsin, dispase II, and collagenase. However, the advantages that this enzymatic solution displays over and above the described enzymes are superior.
Besides immunophenotyping and cell culture and sorting, the future applications of this protocol are numerous and varied. For example, it is now possible to determine DNA methylation, expression of target genes, in vitro leukocyte functionality (e.g., phagocytosis, cytotoxicity, T-cell proliferation and plasticity assays, etc.), and production of reactive oxygen species in the leukocytes isolated from murine tissues at the maternal fetal interface. Indeed, description of new and rare leukocytes in murine tissues at the maternal-fetal interface can be also performed.
Disclosures
The authors disclose no conflicts of interest.
Acknowledgments
NGL was supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. We gratefully acknowledge Maureen McGerty and Amy E. Furcron (Wayne State University) for their critical reading of the manuscript.
References
- Trowsdale J, Betz AG. Mother's little helpers: mechanisms of maternal-fetal tolerance. Nat Immunol. 2006;7(3):241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- King A, et al. Evidence for the expression of HLAA-C class I mRNA and protein by human first trimester trophoblast. J Immunol. 1996;156(6):2068–2076. [PubMed] [Google Scholar]
- Bonney EA, Matzinger P. The maternal immune system's interaction with circulating fetal cells. J Immunol. 1997;158(1):40–47. [PubMed] [Google Scholar]
- Tafuri A, Alferink J, Moller P, Hammerling GJ, Arnold B. T cell awareness of paternal alloantigens during pregnancy. Science. 1995;270(5236):630–633. doi: 10.1126/science.270.5236.630. [DOI] [PubMed] [Google Scholar]
- Chaouat G, Petitbarat M, Dubanchet S, Rahmati M, Ledee N. Tolerance to the foetal allograft. Am J Reprod Immunol. 2010;63(6):624–636. doi: 10.1111/j.1600-0897.2010.00832.x. [DOI] [PubMed] [Google Scholar]
- Robertson SA, et al. Seminal fluid drives expansion of the CD4+CD25+ T regulatory cell pool and induces tolerance to paternal alloantigens in mice. Biol Reprod. 2009;80(5):1036–1045. doi: 10.1095/biolreprod.108.074658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SA, Moldenhauer LM. Immunological determinants of implantation success. Int J Dev Biol. 2014;58(2-4):205–217. doi: 10.1387/ijdb.140096sr. [DOI] [PubMed] [Google Scholar]
- Aluvihare VR, Kallikourdis M, Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. 2004;5(3):266–271. doi: 10.1038/ni1037. [DOI] [PubMed] [Google Scholar]
- Rowe JH, Ertelt JM, Xin L, Way SS. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature. 2012;490(7418):102–106. doi: 10.1038/nature11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150(1):29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Sakai M, Sasaki Y, Nakashima A, Shiozaki A. Inadequate tolerance induction may induce pre-eclampsia. J Reprod Immunol. 2007;76(1-2):30–39. doi: 10.1016/j.jri.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Lee J, et al. A signature of maternal anti-fetal rejection in spontaneous preterm birth: chronic chorioamnionitis, anti-human leukocyte antigen antibodies, and C4d. PLoS One. 2011;6(2):0016806. doi: 10.1371/journal.pone.0016806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn A, et al. Pregnancy-associated diseases are characterized by the composition of the systemic regulatory T cell (Treg) pool with distinct subsets of Tregs. Clin Exp Immunol. 2012;167(1):84–98. doi: 10.1111/j.1365-2249.2011.04493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lopez N, Laresgoiti-Servitje E. T regulatory cells: regulating both term and preterm labor. Immunol Cell Biol. 2012;90(10):919–920. doi: 10.1038/icb.2012.48. [DOI] [PubMed] [Google Scholar]
- Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. 2014;23(10):46. doi: 10.1038/cmi.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lopez N, Guilbert LJ, Olson DM. Invasion of the leukocytes into the fetal-maternal interface during pregnancy. J Leukoc Biol. 2010;88(4):625–633. doi: 10.1189/jlb.1209796. [DOI] [PubMed] [Google Scholar]
- Timmons B, Akins M, Mahendroo M. Cervical remodeling during pregnancy and parturition. Trends Endocrinol Metab. 2010;21(6):353–361. doi: 10.1016/j.tem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring's health. Nat Med. 2013;19(5):548–556. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- Wambach CM, Patel SN, Kahn DA. Maternal and fetal factors that contribute to the localization of T regulatory cells during pregnancy. Am J Reprod Immunol. 2014;71(5):391–400. doi: 10.1111/aji.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266(5190):1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23(1):3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- Croy BA, et al. Imaging of vascular development in early mouse decidua and its association with leukocytes and trophoblasts. Biol Reprod. 2012;87(5) doi: 10.1095/biolreprod.112.102830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AP, Gerber SA, Croy BA. Uterine natural killer cells pace early development of mouse decidua basalis. Mol Hum Reprod. 2014;20(1):66–76. doi: 10.1093/molehr/gat060. [DOI] [PubMed] [Google Scholar]
- Lima PD, Zhang J, Dunk C, Lye SJ, Anne Croy B. Leukocyte driven-decidual angiogenesis in early pregnancy. Cell Mol Immunol. 2014. [DOI] [PMC free article] [PubMed]
- Robson A, et al. Uterine natural killer cells initiate spiral artery remodeling in human pregnancy. FASEB J. 2012;26(12):4876–4885. doi: 10.1096/fj.12-210310. [DOI] [PubMed] [Google Scholar]
- Lash GE, et al. Regulation of extravillous trophoblast invasion by uterine natural killer cells is dependent on gestational age. Hum Reprod. 2010;25(5):1137–1145. doi: 10.1093/humrep/deq050. [DOI] [PubMed] [Google Scholar]
- Kruse A, Merchant MJ, Hallmann R, Butcher EC. Evidence of specialized leukocyte-vascular homing interactions at the maternal/fetal interface. Eur J Immunol. 1999;29(4):1116–1126. doi: 10.1002/(SICI)1521-4141(199904)29:04<1116::AID-IMMU1116>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Degaki KY, Chen Z, Yamada AT, Croy BA. Delta-like ligand (DLL)1 expression in early mouse decidua and its localization to uterine natural killer cells. PLoS One. 2012;7(12):28. doi: 10.1371/journal.pone.0052037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habbeddine M, Verbeke P, Karaz S, Bobe P, Kanellopoulos-Langevin C. Leukocyte Population Dynamics and Detection of IL-9 as a Major Cytokine at the Mouse Fetal-Maternal Interface. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell A, Erlbacher E. Ch. 53. In: Yamada AT, Croy BA, DeMayo FJ, Adamson SL, editors. The Guide to Investigation of Mouse Pregnancy. Elsevier Academic Press; 2014. pp. 619–635. [Google Scholar]
- Rinaldi SF, Catalano RD, Wade J, Rossi AG, Norman JE. Decidual neutrophil infiltration is not required for preterm birth in a mouse model of infection-induced preterm labor. J Immunol. 2014;192(5):2315–2325. doi: 10.4049/jimmunol.1302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V, et al. Uterine DCs are crucial for decidua formation during embryo implantation in mice. J Clin Invest. 2008;118(12):3954–3965. doi: 10.1172/JCI36682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr EL, Szary A, Parr MB. Measurement of natural killer activity and target cell binding by mouse metrial gland cells isolated by enzymic or mechanical methods. J Reprod Fertil. 1990;88(1):283–294. doi: 10.1530/jrf.0.0880283. [DOI] [PubMed] [Google Scholar]
- Arck PC, et al. Murine T cell determination of pregnancy outcome. Cell Immunol. 1999;196(2):71–79. doi: 10.1006/cimm.1999.1535. [DOI] [PubMed] [Google Scholar]
- Male V, Gardner L, Moffett A. Isolation of cells from the feto-maternal interface. Curr Protoc Immunol. 2012;7(7):1–11. doi: 10.1002/0471142735.im0740s97. [DOI] [PubMed] [Google Scholar]
- Li LP, Fang YC, Dong GF, Lin Y, Saito S. Depletion of invariant NKT cells reduces inflammation-induced preterm delivery in mice. J Immunol. 2012;188(9):4681–4689. doi: 10.4049/jimmunol.1102628. [DOI] [PubMed] [Google Scholar]
- Collins MK, Tay CS, Erlebacher A. Dendritic cell entrapment within the pregnant uterus inhibits immune surveillance of the maternal/fetal interface in mice. J Clin Invest. 2009;119(7):2062–2073. doi: 10.1172/JCI38714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Lesperance J, Kim M, Terskikh AV. Efficient propagation of single cells Accutase-dissociated human embryonic stem cells. Mol Reprod Dev. 2008;75(5):818–827. doi: 10.1002/mrd.20809. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wu X, Hu C, Wang P, Li X. Rho kinase inhibitor Y-27632 and Accutase dramatically increase mouse embryonic stem cell derivation. In Vitro Cell Dev Biol Anim. 2012;48(1):30–36. doi: 10.1007/s11626-011-9471-y. [DOI] [PubMed] [Google Scholar]
- Pang SC, Janzen-Pang J, Tse Y, Croy BA. Ch. 2. In: Yamada AT, Croy BA, DeMayo FJ, Adamson SL, editors. The Guide to Investigation of Mouse Pregnancy. Elsevier Academic Press; 2014. pp. 21–42. [Google Scholar]
- Zenclussen AC, et al. Murine abortion is associated with enhanced interleukin-6 levels at the feto-maternal interface. Cytokine. 2003;24(4):150–160. doi: 10.1016/j.cyto.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Mallidi TV, Craig LE, Schloemann SR, Riley JK. Murine endometrial and decidual NK1.1+ natural killer cells display a B220+CD11c+ cell surface phenotype. Biol Reprod. 2009;81(2):310–318. doi: 10.1095/biolreprod.109.076448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addio F, et al. The link between the PDL1 costimulatory pathway and Th17 in fetomaternal tolerance. J Immunol. 2011;187(9):4530–4541. doi: 10.4049/jimmunol.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shynlova O, et al. Infiltration of myeloid cells into decidua is a critical early event in the labour cascade and post-partum uterine remodelling. J Cell Mol Med. 2013;17(2):311–324. doi: 10.1111/jcmm.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchision DM, et al. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells. 2007;25(6):1560–1570. doi: 10.1634/stemcells.2006-0260. [DOI] [PubMed] [Google Scholar]
- Gartner S. The macrophage and HIV: basic concepts and methodologies. Methods Mol Biol. 2014. pp. 670–672. [DOI] [PubMed]
- Quan Y, et al. Impact of cell dissociation on identification of breast cancer stem cells. Cancer Biomark. 2012;12(3):125–133. doi: 10.3233/CBM-130300. [DOI] [PubMed] [Google Scholar]
- Gordon KM, Duckett L, Daul B, Petrie HT. A simple method for detecting up to five immunofluorescent parameters together with DNA staining for cell cycle or viability on a benchtop flow cytometer. J Immunol Methods. 2003;275(1-2):113–121. doi: 10.1016/s0022-1759(03)00009-7. [DOI] [PubMed] [Google Scholar]


