Key Points
The VWF D′ domains are flexibly tethered entities projecting outside antiparallel dimers of the VWF D3 domain.
Extensive interactions between the VWF D′ domain and primarily the FVIII C1 domain mediate VWF-FVIII association.
Abstract
Binding to the von Willebrand factor (VWF) D′D3 domains protects factor VIII (FVIII) from rapid clearance. We performed single-particle electron microscopy (EM) analysis of negatively stained specimens to examine the architecture of D′D3 alone and in complex with FVIII. The D′D3 dimer ([D′D3]2) comprises 2 antiparallel D3 monomers with flexibly attached protrusions of D′. FVIII-VWF association is primarily established between the FVIII C1 domain and the VWF D′ domain, whereas weaker interactions appear to be mediated between both FVIII C domains and the VWF D3 core. Modeling the FVIII structure into the three-dimensional EM reconstructions of [D′D3]2-FVIII ternary and quaternary complexes indicates conformational rearrangements of the FVIII C domains compared with their disposition in the unbound state. These results illustrate the cooperative plasticity between VWF and FVIII that coordinate their high-affinity interaction.
Introduction
The strong association of plasma factor VIII (FVIII) with circulating von Willebrand factor (VWF) secures FVIII from rapid clearance in the blood. The VWF-FVIII complex forms through a high-affinity interaction between the FVIII light chain and the VWF D′D3 domains.1 Mutations within VWF that abrogate or abolish this high-affinity binding lead to type 2N von Willebrand disease, a condition characterized by reduced plasma levels of FVIII.2
The tertiary structure of mature VWF, particularly at the N-terminal D′D3 domains, regulates the affinity for FVIII. VWF circulates as a multi-subunit protein comprising repeated domains that distinctly facilitate VWF packaging and hemostasis.3 The VWF propeptide (domains D1and D2) catalyzes the multimerization of VWF via intermolecular disulfide bonds at the D3 domain (Figure 1A).4 In the absence of propeptide-dependent posttranslational modifications to the D′D3 domains, VWF binds FVIII with reduced affinity.5 Cleavage of the propeptide by furin facilitates FVIII stabilization in the circulation.6 We and others have previously reported that VWF fragments are sufficient to bind FVIII and that propeptide processing of these VWF fragments enhances the affinity for FVIII.7-9 Several of these VWF fragments were also sufficient to elevate FVIII levels in VWF-deficient mice.7
Figure 1.
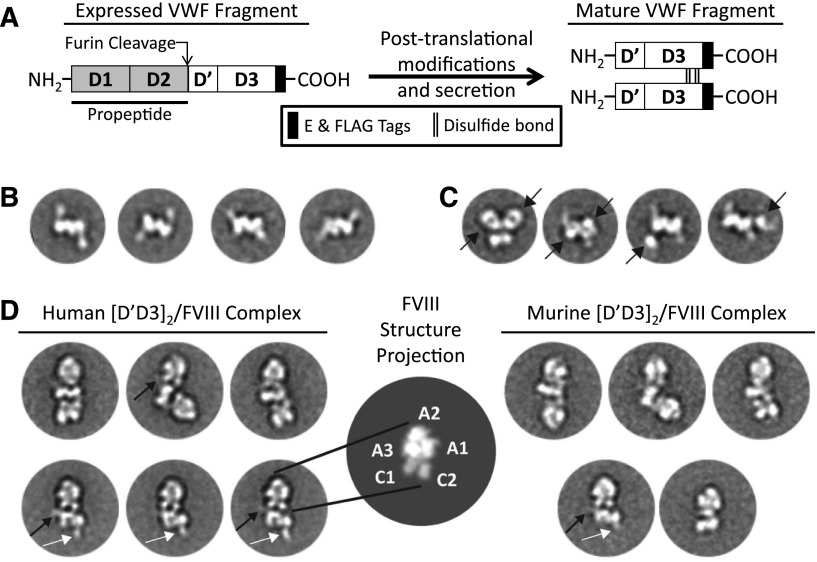
[D′D3]2 associates with FVIII primarily through the flexibly tethered D′. (A) The D1-D3 complementary DNA (cDNA) encodes the N-terminus (M1-P1247) of human or murine VWF and fuses C-terminal E and FLAG tags. Similar to VWF, D1-D3 undergoes posttranslational processing prior to its secretion as a dimer of the D′D3 domains. The propeptide catalyzes intermolecular disulfide bonds at the D3 C-terminus and is cleaved by furin to generate a new N-terminus at S764 of the D′ domain.3 (B) Class averages of murine [D′D3]2 reveal a bilobed central density with a flexibly tethered “handle” having dimensions in agreement with those of D′. (C) Fab fragments (black arrow) directed against the C-terminal FLAG tag decorate the body of [D′D3]2, distinguishing the central bilobed density as the D3 dimer and further confirming the “handle” density as D′. (D, left and right panels) Each molecule of human or murine [D′D3]2 may bind 1 to 2 human FVIII molecules and adopt various orientations. (D, center panel) Characteristic projection of a simulated map of FVIII crystal structure14 illustrates the FVIII domain architecture compared with the EM averages. In some class averages, the D′ handle can be seen in close, interacting proximity with one of the FVIII C domains (black arrows). Several averages of [D′D3]2 bound to a single FVIII clearly show a free D′ domain (white arrows) diametrically opposed to the FVIII-bound D′ domain (black arrows).
To further explore the association between VWF and FVIII, we used single-particle negative-stain electron microscopy (EM) to characterize the architecture of dimeric VWF D′D3 domains ([D′D3]2) alone and in complex with FVIII.
Study design
Protein expression, purification, and analyses are detailed in supplemental Data available on the Blood Web site.
Results and discussion
Structure of [D′D3]2
Mature D1-D3 (ie, [D′D3]2) recapitulates the FVIII-binding N-termini of circulating, multimeric VWF. Expression and maturation of D1-D3 results in a disulfide bonded dimer of the D′D3 domains concomitant with proteolysis of the propeptide (Figure 1A and supplemental Figure 1). [D′D3]2 was separable from pro-forms of D1-D3 (ie, uncleaved D1-D3) by size exclusion chromatography (supplemental Figure 1). Consistent with previous reports,10,11 class averages of negatively stained [D′D3]2 particles show an antiparallel dimer in a back-to-back configuration (Figure 1B and supplemental Figure 2). Within [D′D3]2, each monomer appears as an ovoid density along the dimer symmetry axis accompanied by a weaker elongated density, which we term the “handle,” in the periphery. The dimensions of the handle are ∼20 Å × ∼60 Å, in agreement with the dimensions of the D′ domain structure determined by nuclear magnetic resonance spectroscopy.9 This observation indicates that D3 forms the ovoid density (∼30 Å × ∼60 Å) and homodimerizes along the symmetry axis, consistent with a previous report identifying intermolecular disulfide bonds located at the C-terminus of the D3 domain (C1099-C1099 and C1142-C1142) that coordinate dimerization of VWF fragments comprising the D′D3 domains.4 To confirm this interpretation of the relative disposition of D′ and D3 domains, we labeled [D′D3]2 with an Ag-binding fragment (Fab) directed against the FLAG epitope tag located at the D3 C-terminus. Class averages of the Fab-labeled [D′D3]2 show distinct views of the complex with the central core of [D′D3]2, but not the handle, decorated with additional densities attributed to Fabs (Figure 1C and supplemental Figure 3). The densities corresponding to the D′ domain were often weak or partially averaged out, suggesting flexibility in its tethering and relative positioning to the more rigid D3 domain.
The [D′D3]2-FVIII complex
We previously reported that [D′D3]2 possesses an FVIII binding affinity comparable to that of multimeric VWF.7 Addition of [D′D3]2 to the culture medium of a stable cell line expressing 226N6 (an FVIII variant with improved secretion12) promoted the accumulation of a stable complex comprising partially proteolyzed 226N6 and disulfide-bonded dimers of the VWF D′D3 domains (supplemental Figure 4). FVIII preferentially co-immunoprecipitated with [D′D3]2 compared with pro-forms of D1-D3 (supplemental Figure 4), consistent with the requirement of proteolysis of the VWF propeptide by furin for FVIII binding.13 Negative stain EM class averages revealed an elongated architecture with a central [D′D3]2 flanked by 1 or 2 FVIII molecules at diametrically opposite sides, thereby resulting in ternary and quaternary complexes, respectively (Figure 1D and supplemental Figures 5 and 6). Particularly noticeable with quaternary complexes, this elongated configuration is often kinked with FVIII pivoting variably about the D3 core domains. This observation suggests that FVIII engages VWF primarily through the flexibly tethered density attributed to D′. Accordingly, some averages show a faint elongated density projecting from the D3 core and contacting FVIII. In addition, several averages indicate that FVIII may assume interactions with the D3 core. However, the overall observed variable juxtaposition of FVIII against D3 suggests that these interactions, if any, are weak and that the D′ interaction is the primary association component, consistent with FVIII binding solely to D′.9 Classification of similarly purified murine [D′D3]2 in complex with recombinant FVIII showed similar ternary and quaternary structures (Figure 1D and supplemental Figure 6), with the flexibly tethered D′ domain allowing the variable disposition of FVIII relative to the D3 dimer core.
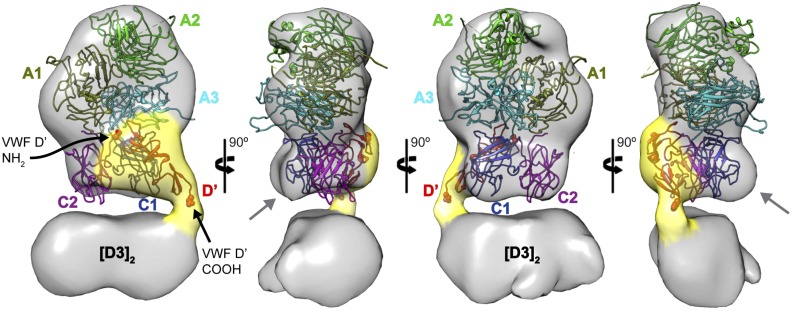
Compared with the distinct projection profile of the structure of FVIII,14 the averages show that FVIII interacts with D′D3 through its two globular “feet” attributed to the C1 and C2 domains (Figure 1D), consistent with previously reported biochemical15 and genetic16 data. To gain further insight into this association, we obtained negative-stain three-dimensional (3D) reconstructions of the ternary (Figure 2) and quaternary (supplemental Figure 7) complexes. The crystal structure of FVIII can be modeled into the 3D envelope with a unique orientation, whereby the FVIII A domains occupy the pointed, distal end(s) of the complex. In this configuration, the C1 and C2 domains are in close proximity to the density representing [D′D3]2. We also observed clear additional density that can accommodate the structure of D′ in association with FVIII. The low-resolution model shows that D′ interacts mainly with C1, but smaller interfaces may also be established with C2 and A3. Consistent with class averages of quaternary complexes (Figure 1D), the corresponding 3D envelope (supplemental Figure 7A) further indicates that both C domains may weakly interact with D3. Interestingly, the C domains of FVIII do not fit within the EM envelope as well as the FVIII A domains. Although we cannot exclude that this represents an artifact of the negative-stain preparation, it is quite possible that the C domains have undergone conformational rearrangements imposed by the binding of [D′D3]2 compared with their unbound form observed in the crystal structures.14,17 Highlighting this flexibility, the FVIII C2 domain makes only minor contacts with the A1 and C1 domains and may assume different positions relative to the remainder of FVIII.14,17 We also note that the contribution of the FVIII acidic a3 domain to FVIII’s high affinity for VWF15 remains unclear, and further detailed investigations will be needed to define this interaction as well as the mechanism(s) by which von Willebrand disease type 2N and hemophilia A mutations prohibit VWF-FVIII association.
Figure 2.
The D′ domain interfaces primarily with the FVIII C1 domain. 3D reconstruction of the murine [D′D3]2-FVIII ternary complex (determined from 962 projections) reveals the D′ density (suggested by the yellow shell) extending from the [D3]2 core and enmeshed with the region corresponding to the FVIII C domains. The structure of FVIII14 (Protein Data Bank: 2R7E) fits the EM density in a unique orientation (A1, olive; A2, green; A3, cyan; C1, blue; C2, purple). The suboptimal agreement between the EM envelope and the position of the C domains suggests that they likely undergo conformational changes upon interaction with VWF D′. Modeling of the VWF D′ domain (Protein Data Bank: 2MHP, red) within the corresponding EM density suggests that it interacts primarily with the FVIII C1 domain, although more limited interactions are possible with C2 and A3. The N- and C-termini of the VWF D′ domain are represented by spheres and indicated by black arrows.
Notwithstanding these limitations, the EM results provide a first architectural framework for the association of FVIII with VWF. Considering that the observed conformational variability of the [D′D3]2-FVIII complex likely poses a great challenge for its structure determination by x-ray crystallography, an orthogonal integration of results from different biophysical methods may be required to determine how VWF binding alters the conformation of the FVIII light chain while still allowing proteolytic activation of FVIII.
Acknowledgments
The authors thank Dr Steven W. Pipe for providing recombinant FVIII and the 226N6 stable cell line.
This work was supported by a Judith Graham Pool Postdoctoral Fellowship (A.Y.) from the National Hemophilia Foundation, by grant No. HL039693 from the National Institutes of Health, National Heart, Lung, and Blood Institute, and by the Pew Foundation (G.S.). D.G. is a Howard Hughes Medical Institute investigator.
Footnotes
Presented in abstract form at the 54th Annual Meeting of the American Society of Hematology, Atlanta, GA, December 8, 2012.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.Y., D.G., and G.S. designed the experiments; A.Y., C.A.K., and R.D.G. performed biochemical experiments; A.N.O., A.M.D., S.D., M.S., and G.S. imaged and analyzed the structures; A.Y. and G.S. interpreted the data; and A.Y., D.G., and G.S. wrote the manuscript.
Conflict-of-interest disclosure: D.G. receives royalty income from Boston Children’s Hospital related to the production of recombinant von Willibrand factor. The remaining authors declare no competing financial interests.
Correspondence: Georgios Skiniotis, University of Michigan, Life Sciences Institute, 210 Washtenaw Ave, Ann Arbor, MI 48109; e-mail: skinioti@umich.edu; David Ginsburg, University of Michigan, Life Sciences Institute, 210 Washtenaw Ave, Room 5028, Ann Arbor, MI 48109; e-mail: ginsburg@umich.edu.
References
- 1.Lollar P, Hill-Eubanks DC, Parker CG. Association of the factor VIII light chain with von Willebrand factor. J Biol Chem. 1988;263(21):10451–10455. [PubMed] [Google Scholar]
- 2.Lillicrap D. von Willebrand disease: advances in pathogenetic understanding, diagnosis, and therapy. Blood. 2013;122(23):3735–3740. doi: 10.1182/blood-2013-06-498303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yee A, Kretz CA. Von Willebrand factor: form for function. Semin Thromb Hemost. 2014;40(1):17–27. doi: 10.1055/s-0033-1363155. [DOI] [PubMed] [Google Scholar]
- 4.Purvis AR, Gross J, Dang LT, et al. Two Cys residues essential for von Willebrand factor multimer assembly in the Golgi. Proc Natl Acad Sci USA. 2007;104(40):15647–15652. doi: 10.1073/pnas.0705175104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendetowicz AV, Morris JA, Wise RJ, Gilbert GE, Kaufman RJ. Binding of factor VIII to von willebrand factor is enabled by cleavage of the von Willebrand factor propeptide and enhanced by formation of disulfide-linked multimers. Blood. 1998;92(2):529–538. [PubMed] [Google Scholar]
- 6.Hilbert L, Nurden P, Caron C, et al. INSERM Network on Molecular Abnormalities in von Willebrand Disease. Type 2N von Willebrand disease due to compound heterozygosity for R854Q and a novel R763G mutation at the cleavage site of von Willebrand factor propeptide. Thromb Haemost. 2006;96(3):290–294. doi: 10.1160/TH06-03-0157. [DOI] [PubMed] [Google Scholar]
- 7.Yee A, Gildersleeve RD, Gu S, et al. A von Willebrand factor fragment containing the D′D3 domains is sufficient to stabilize coagulation factor VIII in mice. Blood. 2014;124(3):445–452. doi: 10.1182/blood-2013-11-540534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster PA, Fulcher CA, Marti T, Titani K, Zimmerman TS. A major factor VIII binding domain resides within the amino-terminal 272 amino acid residues of von Willebrand factor. J Biol Chem. 1987;262(18):8443–8446. [PubMed] [Google Scholar]
- 9.Shiltagh N, Kirkpatrick J, Cabrita LD, et al. Solution structure of the major factor VIII binding region on von Willebrand factor. Blood. 2014;123(26):4143–4151. doi: 10.1182/blood-2013-07-517086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang RH, Wang Y, Roth R, et al. Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci USA. 2008;105(2):482–487. doi: 10.1073/pnas.0710079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou YF, Eng ET, Nishida N, Lu C, Walz T, Springer TA. A pH-regulated dimeric bouquet in the structure of von Willebrand factor. EMBO J. 2011;30(19):4098–4111. doi: 10.1038/emboj.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao HZ, Sirachainan N, Palmer L, et al. Bioengineering of coagulation factor VIII for improved secretion. Blood. 2004;103(9):3412–3419. doi: 10.1182/blood-2003-10-3591. [DOI] [PubMed] [Google Scholar]
- 13.Wise RJ, Dorner AJ, Krane M, Pittman DD, Kaufman RJ. The role of von Willebrand factor multimers and propeptide cleavage in binding and stabilization of factor VIII. J Biol Chem. 1991;266(32):21948–21955. [PubMed] [Google Scholar]
- 14.Shen BW, Spiegel PC, Chang CH, et al. The tertiary structure and domain organization of coagulation factor VIII. Blood. 2008;111(3):1240–1247. doi: 10.1182/blood-2007-08-109918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saenko EL, Scandella D. The acidic region of the factor VIII light chain and the C2 domain together form the high affinity binding site for von willebrand factor. J Biol Chem. 1997;272(29):18007–18014. doi: 10.1074/jbc.272.29.18007. [DOI] [PubMed] [Google Scholar]
- 16.Liu ML, Shen BW, Nakaya S, et al. Hemophilic factor VIII C1- and C2-domain missense mutations and their modeling to the 1.5-angstrom human C2-domain crystal structure. Blood. 2000;96(3):979–987. [PubMed] [Google Scholar]
- 17.Ngo JC, Huang M, Roth DA, Furie BC, Furie B. Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex. Structure. 2008;16(4):597–606. doi: 10.1016/j.str.2008.03.001. [DOI] [PubMed] [Google Scholar]