Abstract
Tight junctions (TJ) are multi-protein complexes located at the apicalmost tip of the lateral membrane in polarised epithelial and endothelial cells. Their principal function is in mediating intercellular adhesion and polarity. Accordingly, it has long been a paradigm that loss of TJ proteins and consequent deficits in cell-cell adhesion are required for tumour cell dissemination in the early stages of the invasive/metastatic cascade. However it is becoming increasingly apparent that TJ proteins play important roles in not just adhesion but also intracellular signalling events, activation of which can contribute to, or even drive, tumour progression and metastasis. In this review, we shall therefore highlight cases wherein the gain of TJ proteins has been associated with signals promoting tumour progression. We will also discuss the potential of overexpressed TJ proteins to act as therapeutic targets in cancer treatment. The overall purpose of this review is not to disprove the fact that loss of TJ-based adhesion contributes to the progression of several cancers, but rather to introduce the growing body of evidence that gain of TJ proteins may have adhesion-independent consequences for promoting progression in other cancers.
Keywords: Cancer, tight junction (TJ), junctional adhesion molecule A (JAM-A), claudin, zonula occludens (ZO) proteins, coxsackievirus and adenovirus receptor (CAR), tumorigenesis, adhesion, metastasis, over-expression, HER2
Introduction
Epithelial cells such as those lining the gastrointestinal and respiratory tracts are repeatedly exposed to environmental carcinogens and are therefore frequently damaged. This drives higher proliferative rates for cell renewal, and a higher probability of acquiring genomic mutations. The resulting vulnerability explains why approximately 90% of human cancers are carcinomas, originating from epithelial tissues. This review will first outline the importance of intercellular adhesion complexes for cellular polarity and epithelial homeostasis. We will then focus on proteins of the tight junction (TJ) complex and describe the implications of aberrant TJ protein expression in tumour progression and metastasis. As numerous excellent reviews have addressed the contribution of TJ protein loss to tumorigenesis (1-7), this review will dissect how TJ protein gain might drive tumorigenesis. Lastly, we will outline the potential of over-expressed TJ proteins to act as future therapeutic targets in human cancer.
Intercellular adhesion
Epithelial intercellular adhesion is essential for maintaining a functional barrier and the integrity of epithelial tissues. Adjacent cells are held together by multi-protein junctional complexes which maintain architectural structure and polarity. Cell-cell contacts are constantly remodelled in order to shed old or damaged cells and to incorporate newly-differentiated cells without loss of barrier integrity (8). However, dysregulation of adhesion proteins can impair barrier function, adhesion and polarity, as well as affecting intracellular signalling (9). Since disrupted polarity is an early histopathological indicator of carcinomas, many studies have outlined key roles for cellular adhesion proteins in cancer initiation or progression (10-19).
There are four major types of intercellular junctions, namely desmosomes, gap junctions, adherens junctions (AJs), and TJs. AJs and TJs are asymmetrically distributed at the apical end of the lateral membrane, forming the apical junctional complex. This review shall focus on TJs, and how upregulation of TJ proteins may contribute to certain types of cancer.
TJ structure and function
TJs are occluding junctions, acting as barriers that control paracellular permeability and regulate trans-epithelial water and solute movement (20). They also function in maintaining cellular polarity. These dynamically-adhesive structures are composed of integral transmembrane proteins that link adjacent cells through homophilic and heterophilic interactions. TJ transmembrane proteins also recruit cytoplasmic TJ adaptor proteins via binding sites in their intracellular C-terminal tails. Such adaptor proteins anchor TJs to the actin cytoskeleton (21) and can recruit cytosolic partners into signalling complexes that regulate, amongst other things, the expression of genes involved in proliferation and differentiation (9). In this manner, TJs form a connection between the extracellular milieu and the nucleus.
Tight junction (TJ) proteins
In 1963, Farquhar and Palade first described the ultrastructure of epithelial TJs (22). Since then >40 TJ proteins have been identified; including transmembrane proteins like occludin, claudins, the coxsackievirus and adenovirus receptor (CAR), and members of the junctional adhesion molecule (JAM) family. TJ adaptor proteins include members of the zonula occludens family (ZO-1, ZO-2 and ZO-3), AF6, Par3, CASK, MUPP1, MAGI-1 afadin, and PDZ-GEF2 (9,23). These proteins are dynamically regulated during processes such as wound repair, tissue remodelling and inflammation. Therefore TJ protein expression and localisation is strenuously regulated to maintain barrier function, and consequently TJ proteins are particularly vulnerable to aberrant modulation that may lead to cellular transformation.
Tight junction (TJ) proteins and cancer
TJs are well-recognized as major players in epithelial cell barrier function, by virtue of their adhesive functions. However, TJs also exert proliferative and differentiative functions which have only emerged in recent years. The current paradigm holds that loss of TJ protein expression and consequent deficits in cell-cell adhesion drive tumour cell detachment from a primary tumour, setting the stage for local invasion and later distant metastasis. Furthermore, loss of TJ adhesion compromises cellular polarity and stimulates dedifferentiation, another hallmark of cancer cells (13). Thus many studies have established that loss of TJ proteins (including occludin and Claudin-7) can enhance tumour progression (1,2). However this paradigm has recently been challenged by reports that the overexpression of many TJ proteins is associated with tumour growth and metastasis. This apparent contradiction highlights an under-appreciated complexity and plasticity in the functions of TJ proteins. Specifically it is likely that TJ proteins function not only in intercellular adhesion and polarity, but also in regulating intracellular signalling that controls proliferation and migration. Evidently, optimal expression of TJ proteins is important in maintaining normal physiological function and any imbalance could have pathological consequences. Therefore, we propose that aberrant TJ protein expression, rather than exclusively TJ protein loss, may promote tumorigenesis. This review will focus on reported overexpressions of TJ proteins in cancer, with a particular focus on members of the immunoglobulin-like superfamily (IgSF) and the claudin family. Table 1 summarises several studies which have reported roles for overexpression of members of these families in tumorigenesis.
Table 1. Table of TJ proteins whose overexpression has been reported to promote tumorigenesis in specific cancers.
| Proteins | Cancer | Reference |
|---|---|---|
| JAM-A | Brain | (24) |
| Breast | (25-30) | |
| Cancer stem cells | (31) | |
| Gastric | (32) | |
| Lung | (33) | |
| Nasopharyngeal | (34) | |
| JAM-C | Fibrosarcoma | (35) |
| Lung | (36) | |
| Melanoma | (37-39) | |
| Ovarian | (40) | |
| CAR | Breast | (41,42) |
| Endometrial | (43) | |
| Lung | (44) | |
| Oral | (45) | |
| Ovarian | (46) | |
| Thyroid | (47) | |
| Claudin-1 | Breast | (48-50) |
| Cervical | (51) | |
| Colorectal | (52-59) | |
| Gastric | (60) | |
| Liver | (61) | |
| Oral | (15) | |
| Ovarian | (62) | |
| Claudin-2 | Breast | (63-66) |
| Colorectal | (52,67) | |
| Lung | (68,69) | |
| Skin | (70) | |
| Claudin-3 | Breast | (71,72) |
| Colorectal | (73) | |
| Endometrial | (74,75) | |
| Gastric | (76) | |
| Kidney | (71) | |
| Lung | (71) | |
| Ovarian | (16,71,77-81) | |
| Prostate | (71) | |
| Uterine | (75) | |
| Claudin-4 | Breast | (71) |
| Endometrial | (74,82) | |
| Gastric | (71,76,83) | |
| Lung | (71) | |
| Kidney | (71) | |
| Nasopharyngeal | (84) | |
| Ovarian | (16,71,77,79) | |
| Pancreatic | (71,85) | |
| Uterine | (75) | |
| Claudin-7 | Cervical | (51) |
| Colon | (86) | |
| Gastric | (71) | |
| Liver | (71) | |
| Lung | (71) | |
| Nasopharyngeal | (84) | |
| Ovarian | (71,87) | |
| Pancreatic | (71,88) | |
| Thyroid | (71) | |
| Claudin-11 | Gastric | (89) |
| Claudin-16 | Breast | (90) |
| Ovarian | (91) | |
| Renal | (92) | |
| Claudin-20 | Breast | (93) |
Junctional adhesion molecules (JAMs)
The IgSF is a large group of proteins which share a structural feature with antibodies, in that they possess one or more immunoglobulin domains. Two members of the IgSF are expressed in TJs: the JAM family and CAR (94). JAMs are type I transmembrane glycoproteins expressed in a variety of cells and tissues, from endothelial and epithelial cells to leukocytes and platelets. The founder member of the family, JAM-A, was discovered in 1998 (94), and subsequently four additional members have joined (JAM-B, JAM-C, JAM-4, and JAM-L) (95-99). They share a similar structural organisation composed of two extracellular Ig-like domains in their N-terminal tail, a single transmembrane region, and a C-terminal cytosolic tail. The carboxyl terminus incorporates a PSD-95/Discs-large/ZO-1 (PDZ) binding domain responsible for intracellular interactions with scaffolding proteins like ZO-1, AF6, Par3, CASK, MUPP1, MAGI-1 afadin and PDZ-GEF2 (100). JAMs have important regulatory functions in numerous cellular processes including intercellular TJ assembly, cellular polarity, leukocyte transmigration, platelet activation, angiogenesis and cell morphology (101-106).
Given these dynamic functions, it is unsurprising that many studies have highlighted aberrant expression of JAM family members in carcinomas. While the role of JAM-A in breast tumorigenesis is best studied, evidence that JAM-A dysregulation contributes to glioblastoma (GBM), nasopharyngeal, gastric and lung cancer is also emerging (24,32-34). Initially JAM-A down-regulation was implicated in breast tumour progression, with a report that its expression decreased in breast tumour metastases (107). However, using larger clinical datasets, two independent groups subsequently demonstrated a strong correlation between JAM-A protein upregulation and poor prognosis in breast cancer patients (25,26,108). In addition, patients whose tumours expressed high levels of JAM-A were also more likely to develop recurrences within 5 years (25).
Regarding the mechanism by which JAM-A upregulation could influence cancer progression, an intriguing study in colonic epithelia had revealed that JAM-A dimerization initiates a signalling complex incorporating AF-6 and the Rap1 activator PDZ-GEF2 (guanine exchange factor), in turn promoting Rap1-GTPase activation, β1-integrin upregulation and enhanced cell migration (109). Confirmation of this mechanism in breast cancer cells, including primary breast tumour cultures (27), suggested that the link between JAM-A overexpression and increased risk of metastasis (25) may lie in the ability of JAM-A to promote cell migration, an early event in the metastatic cascade. This was further supported by a study demonstrating downregulation of microRNA miR-145 in breast tumour tissue relative to healthy tissue, in conjunction with the observation that miR-145 over-expression in breast cancer cells reduced JAM-A expression and decreased cell migration (28).
However the potential role of JAM-A in cancer progression may not be restricted to its regulatory influence on cell migration, but might also involve dynamic control of proliferation and apoptosis (29,108). One mechanism for this could involve its relationship with the breast cancer biomarker human epidermal growth factor receptor-2 (HER2). In a study examining the relationship between JAM-A expression and clinico-pathological variables, JAM-A was associated with increased HER2 gene and protein expression (25,26). Subsequent investigations revealed that JAM-A regulates HER2 expression in vitro, by putatively inhibiting the proteasomal degradation of HER2 protein (26). Taken together, this may explain reports that JAM-A expression promotes proliferation and inhibits apoptosis (29,108), two well-defined features executed by HER2 signalling via the PI3K and MAPK pathways. A role for JAM-A in apoptosis has been highlighted by in vivo studies in which JAM-A null mice developed smaller tumours than JAM-A positive mice, due to increased apoptosis in the former (108). Another in vivo murine study has also confirmed that JAM-A is involved in breast tumour proliferation, and, promisingly, that an anti-JAM-A monoclonal antibody can decrease murine breast tumour xenograft growth (29). However, contrary to the evidence that JAM-A levels positively regulate proliferation and negatively regulate apoptosis, one study has proposed the opposite. Specifically, loss of JAM-A expression reportedly enhanced intestinal epithelial cell proliferation through Akt-mediated activation of β-catenin transcription, while Akt inhibition reversed intestinal hyperproliferation in JAM-deficient mice (110). These results suggest JAM-A may exert tissue-specific control over proliferation. On the other hand, the dependence of these events on JAM-A dimerization (110) might suggest alternate regulation of JAM-A signalling between physiological versus pathophysiological settings. It is intriguing to speculate that, in the physiological setting of an intact epithelial barrier, JAM-A homodimerisation between adjacent epithelial cells would inhibit proliferation in order to maintain barrier homeostasis. By contrast, in pathophysiological settings [e.g., JAM-A upregulation in cancer (25) or wound repair (111,112)], over-expressed JAM-A distributed across the surface of less-polarised cells might engage in adhesion-independent signalling to promote cellular proliferation. Taken together, this suggests a complex spatial and temporal regulation of cellular phenotype by JAM-A, and offers hope that it could be selectively targeted in the pharmacological setting.
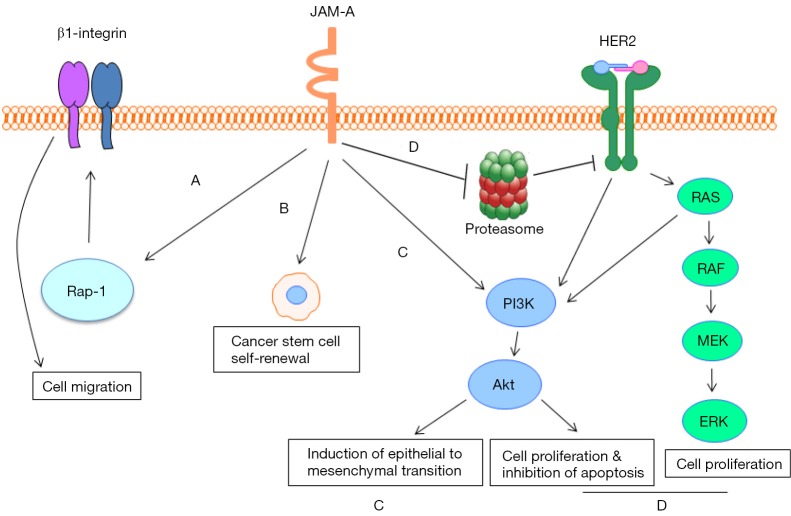
Accordingly, unbiased identification of JAM-A as a promising cancer drug target by a reverse-pharmacology approach (29) in conjunction with observations that JAM-A acts as a cell surface marker for triple-negative breast cancer (TNBC) cancer stem cells (CSCs) (30) support the idea that JAM-A inhibitors could be clinically useful. Similarly, the identification of JAM-A as an indispensable CSC adhesion protein and self-renewal promoter in patient-derived GBM cells (31) further suggests a pro-tumourigenic role for JAM-A in driving self-renewal and cellular dedifferentiation. Accordingly, overexpression of JAM-A in both GBM (31) and nasopharyngeal tumour tissue (34) has been correlated with poor patient prognosis, the latter potentially involving PI3K-mediated induction of epithelial to mesenchymal transition (EMT) (34). Recent studies have also implicated JAM-A in promoting proliferation and inhibiting apoptosis in both gastric and lung cancer (32,33); further suggesting that JAM-A upregulation may be a key molecular marker and novel therapeutic target in several cancers. A summary of mechanisms by which JAM-A may promote tumorigenesis is illustrated in Figure 1.
Figure 1.
Mechanisms by which JAM-A may promote tumorigenesis. (A) JAM-A dimerization leads to Rap1 activation and subsequent upregulation of β1-integrin, enhancing cell migration; (B) JAM-A overexpression promotes cancer stem cell (CSC) self-renewal in triple-negative breast cancer; (C) JAM-A induces epithelial to mesenchymal transition (EMT) in nasopharyngeal carcinoma cells via the PI3K and MAPK pathways; (D) JAM-A stabilises HER2 expression in breast cancer cells by putatively inhibiting HER2 proteasomal degradation, promoting signalling via the PI3K and MAPK pathways which results in cellular proliferation and the inhibition of apoptosis. Many of the arrows depict processes that require several steps.
Another member of the JAM family which has also been implicated in tumorigenesis is JAM-C. However, to date the primary role of JAM-C in aiding cancer progression has been reported to involve the promotion of cancer cell migration and angiogenesis rather than by directly influencing tumour cell proliferation or survival (36-38). JAM-C expression has been positively correlated with metastatic potential in melanoma cells (35), and JAM-C deficient mice with melanomas display decreased levels of lung metastasis (39). Homophilic JAM-C interactions in trans (between adjacent cells) appear to be responsible for JAM-C-dependent metastasis. However, interactions between JAM-C on melanoma cells and JAM-B on lung microvascular endothelial cells could also account for preferential metastasis to the lung (37). Recently, upregulated expression of JAM-C has also been shown to correlate with poor prognosis and reduced recurrence-free survival in non-small cell lung cancer patients, via enhanced lymph vessel formation and lymph node metastasis (36).
This role of JAM-C in promoting metastasis has been highlighted further in a study showing that JAM-C-overexpressing fibrosarcoma cells display increased migration and invasive potential, and that mice bearing these cells have shorter survival times than those implanted with JAM-C negative tumours (35). A complementary study also showed that conditional silencing of endothelial JAM-C in a murine ovarian cancer model hindered tumour growth via impairment of the tumour vasculature (40), indicating a vital role for JAM-C in driving metastasis via pro-angiogenic effects.
Coxsackievirus and adenovirus receptor (CAR)
CAR is another TJ protein in the Ig-superfamily. It is a 46-kDa transmembrane protein with two Ig-like domains in the extracellular region and, as the name indicates, it acts as a receptor for coxsackie- and adenoviruses (113). Overexpression of CAR has been reported in breast cancer compared to healthy tissue, with levels of expression positively correlating with tumour grade and metastasis (41). CAR expression has also been noted to increase after transition from precursor lesions to neoplastic breast tumours (42). Mechanistically, CAR overexpression has been shown to promote breast cancer cell survival following application of an apoptosis-inducing ligand, in a pathway involving Bcl-2 upregulation and reduced caspase-3 activation (42). Furthermore, levels of both transmembrane CAR and soluble CAR isoforms were shown to increase in breast cancer cells following tamoxifen treatment (114), suggesting that the protein may be upregulated in order to protect the cells from apoptosis. Augmented levels of soluble CAR isoforms (CAR 3/7 and 4/7) have also been reported to correlate with reduced overall survival in ovarian cancer patients (46). Furthermore CAR expression has been associated with tumour grade in endometrial adenocarcinoma and lung cancer patients (43,44), while CAR silencing decreased cellular adhesion, colony formation, and invasion in lung cancer cells (44). Studies in a thyroid tumour tissue microarray have also revealed over-expression of CAR in malignant compared with benign tumours, in addition to a correlation between CAR expression and larger tumour size (47). Previous studies have also shown that CAR expression is significantly upregulated in neuroblastomas and medullablastomas, the most common paediatric gliomas, compared to other central nervous system tumours (115). An anti-apoptotic role for CAR has also been postulated in oral squamous cell carcinoma, with gene silencing reported to suppress growth and induce apoptosis (45). Collectively these results suggest that CAR may represent a good drug delivery target for anti-cancer gene therapies, a concept which will be addressed more comprehensively in section “TJ proteins as therapeutic drug targets in cancer” of this review.
Claudins
The claudin family consists of 24 transmembrane proteins exhibiting distinct tissue- and development-specific distributions, with subtle differences in their extracellular loops which determine ionic selectivity (116). Claudins are expressed in both epithelial and endothelial cells and form a complex with occludin and/or JAM family members. They encode 20-27 kDa proteins with four transmembrane domains, two extracellular loops and a short carboxyl intracellular tail (117). Claudins expressed on adjacent cells interact via their extracellular loops, while those expressed in the same cell interact through their intracellular N-terminal domains (118). The last amino acids of this tail constitute highly conserved PDZ-binding motifs. Through these motifs, claudins are linked to the TJ PDZ-containing proteins ZO-1, ZO-2, ZO-3, PATJ and MUPP1 (119). Claudins have been shown to be essential and sufficient to induce TJ formation (119). Regardless of this critical role in normal physiology, overexpression of various claudin family members has also been associated with a number of cancers.
Claudin-1 and Claudin-2
Although Claudin-1 and -2 are frequently expressed in a similar pattern in cancers (52,120,121), some differences do exist. For instance, it has been reported that Claudin-1 levels decrease in skin squamous cell carcinoma while those of Claudin-2 increase (70). Elevated Claudin-1 expression has been noted in many cancers including ovarian, breast, colorectal, gastric, cervical and liver (15,48,52-58,60,61,122). Furthermore augmented Claudin-1 protein expression in colon cancer has been correlated with tumour growth, metastasis and poor patient prognosis (52,55,57). However, mechanisms underlying the upregulation of claudins in cancer are still unclear. One potential mechanism involves transcriptional activators such as Cdx-2, according to colon cancer model studies in conjunction with Claudin-1 and Cdx2 expressional data from colon cancer patients (54). However, it is likely that Claudin-1 upregulation in cancer could also reflect other mechanisms. Another study has revealed an association between Claudin-1 mRNA expression and histone deacetylase (HDAC)-2 mRNA in colon cancer patients; while HDAC inhibitors decreaseed Claudin-1 expression and inhibited colonic tumour cell invasion (59). Together, these results suggest that the upregulation of Claudin-1 in colon cancer is a dynamic process involving both transcriptional regulation and epigenetic modifications.
Post-transcriptional regulation of Claudin-1 has also been demonstrated in ovarian cancer-initiating cells (OCICs) (62). Specifically, Claudin-1 mRNA was shown to be upregulated in OCICs compared to ovarian cancer cells. Its expression correlated with downregulation of the microRNA miR-155, which directly targets Claudin-1 for degradation, thereby acting as a tumour-initiating cell suppressor (62). Other diverse mechanisms have also been reported to regulate Claudin-1 expression in a range of cancer types. For instance, elevation of Claudin-1 mRNA has been demonstrated following tamoxifen treatment in breast cancer cells, and linked with protection from apoptosis. This suggests that up-regulation of Claudin-1 may confer some degree of resistance to the drug (48). Other work has supported an anti-apoptotic role for Claudin-1 in breast cancer (49,50), with silencing of Claudin-1 shown to induce apoptosis in a number of cell lines (50). In addition, an increase in Claudin-1 expression and subsequent inhibition of apoptosis was observed following treatment with tumour necrosis factor α (TNFα), a cytokine which induces cancer cells to undergo apoptosis (49).
A role for Claudin-1 in inhibiting apoptosis has also been observed in colon cancer through suppression of anoikis via Src, Akt and Bcl-2-dependent pathways (58). Overexpression of Claudin-1 in colon cancer has also been shown to upregulate the transcription factor ZEB1 which represses E-Cadherin expression, resulting in augmented invasive capacity and reduced cell death (122). Furthermore, Claudin-1 has been linked with the induction of EMT in human hepatocellular carcinoma via ZEB1 and Slug acting upstream of MAPK signalling (61).
Although a similar role for Claudin-2 has not yet been demonstrated in liver cancer, Claudin-2 has nonetheless been associated with liver metastasis of breast cancer through the engagement of integrin complexes (63). Significantly, Claudin-2 is much upregulated in primary and secondary tumours that metastasise to the liver compared to other metastatic sites (63,64), and its expression shown to correlate with reduced metastasis-free survival (64). A recent study has suggested that Lyn, a member of the SRK family kinases, is responsible for increased Claudin-2 expression in primary breast tumours and their subsequent liver metastases (65). Preference for the liver as a metastasic site is thought to be exerted via interactions between Claudin-2 expressed on both breast cancer cells and primary hepatocytes (66).
Yet another alternative mechanism for Claudin-2 upregulation has been reported in lung cancer cells, with the demonstration that MMP-mediated release of EGF promotes Claudin-2 upregulation via the EGFR/MEK/ERK pathway (68). As EGFR is significantly upregulated in lung carcinoma tissue (123), this mechanism offers a potential explanation as to why Claudin-2 is expressed in human lung adenocarcinoma but absent in healthy lung tissue (68,69). When Claudin-2 is expressed in lung cancer cells, it has been shown to translocate to the nucleus, form a complex with ZO-1, ZONAB and cyclin D1 and consequently to promote proliferation (69). Similar interactions have also been reported in colorectal cancer, via a symplekin-dependent mechanism promoting Claudin-2 expression and subsequent enhancements in cyclin D1 expression and nuclear localisation of ZONAB to enhance proliferation (67).
Claudin-3 and Claudin-4
A myriad of studies have shown that Claudin-3 and -4 are frequently overexpressed in several carcinomas, including endometrial, breast, ovarian, lung, prostate, kidney, gastric and pancreatic (16,71-78,83,85). Gene expression profiling has demonstrated that both Claudins-3 and -4 are overexpressed in uterine serous papillary carcinoma (USPC), a highly aggressive and chemotherapy-resistant variant of endometrial cancer characterized by particularly poor prognosis (75). Complementary studies have also supported a role for Claudin-3 and -4 upregulation correlating with aggressive disease phenotypes and poor prognosis in endometrial cancer patients (74), with progressive expressional enhancements at both mRNA and protein levels along a scale of normal to hyperplastic and finally malignant endometrial tissue (82). The mechanism by which Claudins-3 and -4 are upregulated in cancer is not entirely understood. However, as discussed in the case of Claudin-1 upregulation in colon cancer (54), evidence from gastric cancer settings suggests that the transcriptional enhancer Cdx-2 may be involved (76).
Although much remains to be understood about the upstream regulators of claudin family members in carcinomas, some progress has been made on understanding the downstream events that drive claudin-dependent tumorigenesis. As already discussed for JAM-A (27), one way in which claudin upregulation could similarly influence cancer progression is via positive effects on cell motility at the start of the invasive/metastatic cascade. Claudin-4 overexpression in gastric cancer has been shown to increase cell invasion and migration, putatively via coordinate regulation of the expression or activation of matrix metalloproteinases (MMP)-2 and -9 (83). Similarly, studies have alluded to Claudin-3 and -4 overexpression in ovarian cancer cells promoting cell survival, invasion and motility via increased MMP-2 activity (79). Intriguingly, although the simultaneous overexpression of both Claudins-3 and -4 has been reported in ovarian cancer, findings suggest that Claudin-3 may be of more importance (77,78,80,81). Specifically, patient tissue analyses have revealed a significant correlation between tumours overexpressing Claudin-3 and a shorter survival time, suggesting that Claudin-3 alone may represent a novel therapeutic target for ovarian cancers (77). Thus, although these two proteins are usually analysed hand-in-hand and assumed to function together, this is not always the case. For instance, a study analysing Claudin-3 and -4 levels in a large cohort of breast cancer patients found that expression levels of the proteins act as differential prognostic factors in breast cancer (72). Specifically, overexpression of Claudin-3 in conjunction with downregulation of Claudin-4 was associated with poor prognosis in triple negative breast cancer, while the opposite was the case in luminal breast cancer subtypes (72). Such findings suggest crosstalk between these proteins and hormone receptor signalling in cells of the breast epithelium. Taken together, and in conjunction with the previously noted divergence between the utility of Claudins-1 and -2 as prognostic factors in skin cancer (70), these studies highlight a complex regulation of claudins in cancer which merits further unravelling.
Other claudins
Besides Claudins-1-4, several other members of the claudin family have also been linked with tumorigenesis in tissue-specific contexts. These include Claudin-7, Claudin-11, Claudin-16 and Claudin-20 (86-90,92,93). Although loss of Claudin-7 has been associated with tumorigenesis in a number of cancers including those of the lung, breast and pancreas (2,12,88), other studies have shown that Claudin-7 is upregulated in different cancers such as thyroid, liver, stomach, kidney and ovarian (51,71,86,87). For example, Claudin-7 expression has been shown to promote EMT and cancer cell motility and invasion in human colorectal cancer and has been associated with decreased survival in nasopharyngeal carcinoma patients (84,86). In ovarian cancer cells, Claudin-7 knockdown has been shown to drive differential expression of genes involved in proliferation, apoptosis and development (87). Claudin-16 protein has also been implicated in ovarian cancer, being identified as a human ovarian-cancer specific transcript during serial analysis of gene expression contributing to ovarian tumorigenesis (91). Interestingly, Claudin-16 is also highly expressed in renal cell carcinoma (92), while on the contrary loss of Claudin-16 appears to enhance tumour progression in breast cancer (124). Primary breast cancer tumour mRNA expression analysis has also revealed that high Claudin-20 expression correlates with poor patient prognosis (93). Overall it is clear that no simple paradigm exists in relation to the role of claudin family expression in cancer. However further elucidation of their spatial and temporal regulation is likely to reveal fascinating insights into their mechanistic contribution to cancer development and progression, in addition to exploring their feasibility as druggable targets.
TJ proteins as therapeutic drug targets in cancer
TJ proteins are barely accessible in well-structured normal epithelia but, due to abnormal function on tumour cells resulting in the disruption of cellular polarity, they become exposed (18). Therewith, as TJ proteins are overexpressed in several types of carcinomas, different approaches using monoclonal antibodies (mAbs), enterotoxins and therapeutic gene delivery have been created and tested as new promising candidates for anti-cancer drug therapy.
Currently, mAbs against cancer-specific antigens have shown enormous potential and efficacy as a new class of drugs. Specificity to the target, long stability in blood and several clinically relevant mechanisms of action are some of the features which make them interesting and useful (125). Briefly, mAbs bind to a specific receptor on the cell surface and induce one or more actions: firstly the recruitment and activation of immune cells resulting in cell lysis, and secondly the antagonism of ligands to interrupt signalling, normalize growth rates, induce apoptosis and sensitize tumours to chemotherapeutic agents (126).
Based on the advantages of using mAb therapy, a variety of pre-clinical studies have tested the efficacy of targeting TJ proteins via this approach in cancer. One study generated a dual-targeting mAb specific against Claudin-3 and -4, and found it to significantly inhibit tumour formation in cell lines of breast and ovarian cancer both in vitro and in vivo (127). Previously, another study succeeded in isolating a mAb which specifically bound to Claudin-4 and proved to be a promising therapeutic approach for pancreatic and ovarian cancers (19). Furthermore, an anti-JAM-C mAb was effective in reducing tumour growth by decreasing angiogenic vascularization of tumours (128) and a mAb against JAM-A resulted in tumour growth inhibition in several mouse models (29).
Although promising, there are some limitations to mAb therapy. In order to obtain maximum efficiency, high doses of therapeutic antibody for a prolonged time are needed, which could make the treatment prohibitively expensive. Moreover, pharmacokinetic and pharmacodynamic problems such as distribution and administration, individual variability (age, gender, other diseases) and safety, since the necessity for high concentrations leads to increased immunogenicity, are some of the problems with mAb therapy. Producing mAbs can also be particularly difficult. Besides the high cost, there is the challenge of large-scale antibody production from hybridomas, as it is limited by the quantity, quality and type of cell line used in the process (129).
Clostridium perfringens (C. perfringens) enterotoxin (CPE) mediated therapy represents a different approach which benefits from specific expression patterns of Claudins-3 and -4 in cancer cells. CPE is a toxin produced by C. perfringens bacteria which is associated with gastrointestinal symptoms of type A food poisoning. Another known characteristic of this 35 kDa polypeptide is its ability to rapidly lyse cells upon binding to its receptors, through its effects on membrane permeability (130).
Interestingly, Claudins-3 and -4 have been identified as natural CPE receptors, enabling the use of the enterotoxin to induce cell death in cancers which express those receptors. One study carried out with CPE-mediated therapy resulted in rapid and dose-dependent cytolysis exclusively in breast cancer cells that express Claudin-3 and -4. The same group also used the treatment to reduce significantly tumour volume in xenografts (131). Similarly, pancreatic (132), prostate (133), ovarian tumours (134) in addition to brain metastases (135) have also been shown to be sensitive to CPE treatment.
Although studies in mice have not demonstrated significant toxicity following intra-tumoural treatment with CPE, it must be noted that Claudins-3 and -4 are expressed in many normal tissues as well as being over-expressed in certain cancers. This may pose as a risk in the systemic utilization of this therapy, potentially restricting it to cancers accessible via local application. In turn this could lead to distribution problems of CPE in tumour tissue, principally in large solid tumour masses. The dependence of the expression of claudins and the need for multiple applications leading to immuno-sensitisation against the treatment are other limitations of CPE-mediated therapy (14,134).
With the establishment of therapeutic gene delivery, which consists of vector-driven insertion of DNA or RNA sequences into cells to correct a malfunctioning gene, switch it off or even cause cancer cell death (136), the search for mammalian receptors that enable virus attachment to target cells has increased. In this context, the involvement of the TJ protein CAR in attachment and infection by group B coxsackieviruses (CVB) and adenoviruses (Ad), has led CAR to be extensively studied in terms of cancer treatment (137). The fact that CAR over-expression has been reported in several cancers (as discussed previously) would suggest promise for some selectivity in this treatment approach. Paradoxically, however, some studies have shown that CAR expression is heterogeneous. Some types of cancer cell lines including glioma (138,139), bladder (140), prostate (141), rhabdomyosarcoma (142), colorectal (143), ovarian (144), lung (145) and breast (146), have low levels of CAR expression, limiting efficient adenovirus infection in those cancer cells and therefore the use of gene therapy. Thus, several studies have used pharmacological approaches to induce CAR expression in highly tumourigenic cancers which are normally CAR deficient. In some cases this has been shown to have growth-inhibitory effects, suggesting that the role of CAR in cancer progression is complex and likely tissue-specific (141,147-149). Accordingly this will influence its potential as a delivery target for gene therapy.
However, although most clinical trials have demonstrated adenoviral gene therapies to be safe, well-tolerated and minimally toxic, reasonable concerns still exist. The tragic death of a patient following a massive immune response to the viral vector in a Phase I trial sparked an FDA decision to investigate 69 US gene therapy trials in 2000 (150), and seriously set back research in the area. Besides negative press, the incidents exposed some problems related to the therapy such as purification of viral particles, measurement of vector concentration, selection of hosts (151) and indeed the choice of subjects in which to perform the trials.
Despite the limitations of each previously-mentioned form of therapy, the techniques described are already widely used in the treatment of various diseases including cancer; and many of the problems and side effects of the treatments have been well established. Thus, pending a greater mechanistic understanding of how TJ proteins contribute to the pathophysiology of diseases including cancer, the time is ripe to move forward with these therapeutic approaches using TJ proteins as targets.
To this end, it is necessary to expand our font of knowledge regarding the mechanistic contributions of TJ proteins to cancer. This in turn will drive the testing of new therapies including small molecule inhibitors of TJ protein signalling, and foster clinical trials to better understand treatment regimens and to maximise the safety and efficacy of TJ-targeted reagents as future anti-cancer therapies.
Conclusions
In conclusion, TJ proteins have a vast potential to both repress tumorigenesis (via the promotion of stable cell-cell adhesion) or to promote tumorigenesis (via adhesion-independent signal transduction events that control migration, proliferation and apoptosis). This review has highlighted a myriad of studies demonstrating TJ protein overexpression in specific cancers, and attempted to address mechanisms whereby such events would actively contribute to tumour progression or metastasis (rather than simply acting as passive biomarkers of the disease process). The highlighted findings regarding claudins, JAMs and CAR in tumorigenesis suggest great potential for these TJ proteins in particular to act as novel molecular targets in semi-personalised cancer strategies. The development of therapeutic reagents against these proteins will pose challenges, not least how to avoid interfering with their intrinsic roles in normal physiology while consciously interrupting their contributions to cancer pathophysiology. However several parallel avenues of promising pre-clinical data support the possibility that this is an exciting path towards a future of targeted cancer therapies, once it is firmly underpinned by solid mechanistic investigations into the biology of how TJ molecules contribute to disease pathogenesis.
Acknowledgements
AM Hopkins is grateful to Science Foundation Ireland (13/IA/1994; 08/RFP/NSC/1427), the Health Research Board of Ireland (HRA-POR-2014-545; HRA/2009/49; RP/2006/95), Breast Cancer Ireland, The Beaumont Hospital Cancer Research & Development Trust, Cancer Research Ireland (CRI05/HOP) for past or current support of relevant studies from her laboratory. AO Leech is funded by a Breast Cancer Ireland PhD studentship. RG Cruz is funded by a Brazil Science Without Borders PhD studentship (CAPES-Brasil/ Process 13306/ 13-8).
Footnotes
Conflicts of Interest: The authors have no conflict of interest to declare.
References
- 1.Martin TA, Mansel RE, Jiang WG. Loss of occludin leads to the progression of human breast cancer. Int J Mol Med 2010;26:723-34. [DOI] [PubMed] [Google Scholar]
- 2.Kominsky SL, Argani P, Korz D, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene 2003;22:2021-33. [DOI] [PubMed] [Google Scholar]
- 3.Sauer T, Pedersen MK, Ebeltoft K, et al. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology 2005;16:193-8. [DOI] [PubMed] [Google Scholar]
- 4.Seok SH, Kang SH, Lee SJ, et al. Reduced expression of claudin-7 correlates with invasiveness and nuclear grade of breast carcinomas. Korean J Pathol 2007;41:158-64. [Google Scholar]
- 5.Usami Y, Chiba H, Nakayama F, et al. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol 2006;37:569-77. [DOI] [PubMed] [Google Scholar]
- 6.Krämer F, White K, Kubbies M, et al. Genomic organization of claudin-1 and its assessment in hereditary and sporadic breast cancer. Hum Genet 2000;107:249-56. [DOI] [PubMed] [Google Scholar]
- 7.Osanai M, Murata M, Nishikiori N, et al. Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res 2006;66:9125-33. [DOI] [PubMed] [Google Scholar]
- 8.Tervonen TA, Partanen JI, Saarikoski ST, et al. Faulty epithelial polarity genes and cancer. Adv Cancer Res 2011;111:97-161. [DOI] [PubMed] [Google Scholar]
- 9.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol 2003;4:225-36. [DOI] [PubMed] [Google Scholar]
- 10.Balda MS, Matter K. Tight junctions at a glance. J Cell Sci 2008;121:3677-82. [DOI] [PubMed] [Google Scholar]
- 11.Bujko M, Kober P, Mikula M, et al. Expression changes of cell-cell adhesion-related genes in colorectal tumors. Oncol Lett 2015;9:2463-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z, Kim do H, Fan J, et al. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol Cancer 2015;14:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta 2009;1788:872-91. [DOI] [PubMed]
- 14.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res 2005;65:9603-6. [DOI] [PubMed] [Google Scholar]
- 15.Oku N, Sasabe E, Ueta E, et al. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res 2006;66:5251-7. [DOI] [PubMed] [Google Scholar]
- 16.Rangel LB, Agarwal R, D'Souza T, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res 2003;9:2567-75. [PubMed] [Google Scholar]
- 17.Runkle EA, Mu D. Tight junction proteins: from barrier to tumorigenesis. Cancer Lett 2013;337:41-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saeki R, Kondoh M, Kakutani H, et al. A novel tumor-targeted therapy using a claudin-4-targeting molecule. Mol Pharmacol 2009;76:918-26. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M, Kato-Nakano M, Kawamoto S, et al. Therapeutic antitumor efficacy of monoclonal antibody against Claudin-4 for pancreatic and ovarian cancers. Cancer Sci 2009;100:1623-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 2009;1:a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta 2008;1778:660-9. [DOI] [PMC free article] [PubMed]
- 22.Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol 1963;17:375-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin TA, Jiang WG. Tight junctions and their role in cancer metastasis. Histol Histopathol 2001;16:1183-95. [DOI] [PubMed] [Google Scholar]
- 24.Rosager AM, Dahlrot RH, Kristensen BW, et al. JAM-A – a novel prognostic cancer stem cell-related marker in glioblastoma. Brain Pathology 2014;24:93. [Google Scholar]
- 25.McSherry EA, McGee SF, Jirstrom K, et al. JAM-A expression positively correlates with poor prognosis in breast cancer patients. Int J Cancer 2009;125:1343-51. [DOI] [PubMed] [Google Scholar]
- 26.Brennan K, McSherry EA, Hudson L, et al. Junctional adhesion molecule-A is co-expressed with HER2 in breast tumors and acts as a novel regulator of HER2 protein degradation and signaling. Oncogene 2013;32:2799-804. [DOI] [PubMed] [Google Scholar]
- 27.McSherry EA, Brennan K, Hudson L, et al. Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res 2011;13:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Götte M, Mohr C, Koo CY, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 2010;29:6569-80. [DOI] [PubMed] [Google Scholar]
- 29.Goetsch L, Haeuw JF, Beau-Larvor C, et al. A novel role for junctional adhesion molecule-A in tumor proliferation: modulation by an anti-JAM-A monoclonal antibody. Int J Cancer 2013;132:1463-74. [DOI] [PubMed] [Google Scholar]
- 30.Thiagarajan PS, Hitomi M, Hale JS, et al. Development of a Fluorescent Reporter System to Delineate Cancer Stem Cells in Triple-Negative Breast Cancer. Stem Cells 2015;33:2114-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lathia JD, Li M, Sinyuk M, et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep 2014;6:117-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeo K, Oshima T, Shan J, et al. Junctional adhesion molecule-A promotes proliferation and inhibits apoptosis of gastric cancer. Hepatogastroenterology 2015;62:540-5. [PubMed] [Google Scholar]
- 33.Zhang M, Luo W, Huang B, et al. Overexpression of JAM-A in non-small cell lung cancer correlates with tumor progression. PLoS One 2013;8:e79173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Y, Tian Y, Zhang W, et al. Junctional adhesion molecule-A, an epithelial-mesenchymal transition inducer, correlates with metastasis and poor prognosis in human nasopharyngeal cancer. Carcinogenesis 2015;36:41-8. [DOI] [PubMed] [Google Scholar]
- 35.Fuse C, Ishida Y, Hikita T, et al. Junctional adhesion molecule-C promotes metastatic potential of HT1080 human fibrosarcoma. J Biol Chem 2007;282:8276-83. [DOI] [PubMed] [Google Scholar]
- 36.Hao S, Yang Y, Liu Y, et al. JAM-C promotes lymphangiogenesis and nodal metastasis in non-small cell lung cancer. Tumour Biol 2014;35:5675-87. [DOI] [PubMed] [Google Scholar]
- 37.Arcangeli ML, Frontera V, Bardin F, et al. The Junctional Adhesion Molecule-B regulates JAM-C-dependent melanoma cell metastasis. FEBS Lett 2012;586:4046-51. [DOI] [PubMed] [Google Scholar]
- 38.Ghislin S, Obino D, Middendorp S, et al. Junctional adhesion molecules are required for melanoma cell lines transendothelial migration in vitro. Pigment Cell Melanoma Res 2011;24:504-11. [DOI] [PubMed] [Google Scholar]
- 39.Langer HF, Orlova VV, Xie C, et al. A novel function of junctional adhesion molecule-C in mediating melanoma cell metastasis. Cancer Res 2011;71:4096-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leinster DA, Colom B, Whiteford JR, et al. Endothelial cell junctional adhesion molecule C plays a key role in the development of tumors in a murine model of ovarian cancer. FASEB J 2013;27:4244-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin TA, Watkins G, Jiang WG. The Coxsackie-adenovirus receptor has elevated expression in human breast cancer. Clin Exp Med 2005;5:122-8. [DOI] [PubMed] [Google Scholar]
- 42.Brüning A, Stickeler E, Diederich D, et al. Coxsackie and adenovirus receptor promotes adenocarcinoma cell survival and is expressionally activated after transition from preneoplastic precursor lesions to invasive adenocarcinomas. Clin Cancer Res 2005;11:4316-20. [DOI] [PubMed] [Google Scholar]
- 43.Giaginis CT, Zarros AC, Papaefthymiou MA, et al. Coxsackievirus and adenovirus receptor expression in human endometrial adenocarcinoma: possible clinical implications. World J Surg Oncol 2008;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Wang Q, Sun J, et al. Expression of the coxsackie and adenovirus receptor in human lung cancers. Tumour Biol 2013;34:17-24. [DOI] [PubMed] [Google Scholar]
- 45.Saito K, Sakaguchi M, Iioka H, et al. Coxsackie and adenovirus receptor is a critical regulator for the survival and growth of oral squamous carcinoma cells. Oncogene 2014;33:1274-86. [DOI] [PubMed] [Google Scholar]
- 46.Reimer D, Steppan I, Wiedemair A, et al. Soluble isoforms but not the transmembrane form of coxsackie-adenovirus receptor are of clinical relevance in epithelial ovarian cancer. Int J Cancer 2007;120:2568-75. [DOI] [PubMed] [Google Scholar]
- 47.Giaginis C, Zarros A, Alexandrou P, et al. Evaluation of coxsackievirus and adenovirus receptor expression in human benign and malignant thyroid lesions. APMIS 2010;118:210-21. [DOI] [PubMed] [Google Scholar]
- 48.Akasaka H, Sato F, Morohashi S, et al. Anti-apoptotic effect of claudin-1 in tamoxifen-treated human breast cancer MCF-7 cells. BMC Cancer 2010;10:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Wang L, Lin XY, et al. Anti-apoptotic effect of claudin-1 on TNF-α-induced apoptosis in human breast cancer MCF-7 cells. Tumour Biol 2012;33:2307-15. [DOI] [PubMed] [Google Scholar]
- 50.Achari C, Winslow S, Larsson C. Down Regulation of CLDND1 Induces Apoptosis in Breast Cancer Cells. PLoS One 2015;10:e0130300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JW, Lee SJ, Seo J, et al. Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol Oncol 2005;97:53-9. [DOI] [PubMed] [Google Scholar]
- 52.Kinugasa T, Huo Q, Higashi D, et al. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res 2007;27:3729-34. [PubMed] [Google Scholar]
- 53.Caruso M, Fung KY, Moore J, et al. Claudin-1 Expression Is Elevated in Colorectal Cancer Precursor Lesions Harboring the BRAF V600E Mutation. Transl Oncol 2014;7:456-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhat AA, Sharma A, Pope J, et al. Caudal homeobox protein Cdx-2 cooperates with Wnt pathway to regulate claudin-1 expression in colon cancer cells. PLoS One 2012;7:e37174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhawan P, Singh AB, Deane NG, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 2005;115:1765-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huo Q, Kinugasa T, Wang L, et al. Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal cancer. Anticancer Res 2009;29:851-7. [PubMed] [Google Scholar]
- 57.Resnick MB, Konkin T, Routhier J, et al. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol 2005;18:511-8. [DOI] [PubMed] [Google Scholar]
- 58.Singh AB, Sharma A, Dhawan P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis 2012;33:2538-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krishnan M, Singh AB, Smith JJ, et al. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene 2010;29:305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, Li J, Qu Y, et al. The expression of claudin 1 correlates with β-catenin and is a prognostic factor of poor outcome in gastric cancer. Int J Oncol 2014;44:1293-301. [DOI] [PubMed] [Google Scholar]
- 61.Suh Y, Yoon CH, Kim RK, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013;32:4873-82. [DOI] [PubMed] [Google Scholar]
- 62.Qin W, Ren Q, Liu T, et al. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett 2013;587:1434-9. [DOI] [PubMed] [Google Scholar]
- 63.Tabariès S, Dong Z, Annis MG, et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 2011;30:1318-28. [DOI] [PubMed] [Google Scholar]
- 64.Kimbung S, Kovács A, Bendahl PO, et al. Claudin-2 is an independent negative prognostic factor in breast cancer and specifically predicts early liver recurrences. Mol Oncol 2014;8:119-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabariès S, Annis MG, Hsu BE, et al. Lyn modulates Claudin-2 expression and is a therapeutic target for breast cancer liver metastasis. Oncotarget 2015;6:9476-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tabariès S, Dupuy F, Dong Z, et al. Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol Cell Biol 2012;32:2979-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchert M, Papin M, Bonnans C, et al. Symplekin promotes tumorigenicity by up-regulating claudin-2 expression. Proc Natl Acad Sci U S A 2010;107:2628-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ikari A, Sato T, Watanabe R, et al. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim Biophys Acta 2012;1823:1110-8. [DOI] [PubMed]
- 69.Ikari A, Watanabe R, Sato T, et al. Nuclear distribution of claudin-2 increases cell proliferation in human lung adenocarcinoma cells. Biochim Biophys Acta 2014;1843:2079-88. [DOI] [PubMed]
- 70.Hintsala HR, Siponen M, Haapasaari KM, et al. Claudins 1, 2, 3, 4, 5 and 7 in solar keratosis and squamocellular carcinoma of the skin. Int J Clin Exp Pathol 2013;6:2855-63. [PMC free article] [PubMed] [Google Scholar]
- 71.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 2006;6:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolokytha P, Yiannou P, Keramopoulos D, et al. Claudin-3 and claudin-4: distinct prognostic significance in triple-negative and luminal breast cancer. Appl Immunohistochem Mol Morphol 2014;22:125-31. [DOI] [PubMed] [Google Scholar]
- 73.de Souza WF, Fortunato-Miranda N, Robbs BK, et al. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PLoS One 2013;8:e74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konecny GE, Agarwal R, Keeney GA, et al. Claudin-3 and claudin-4 expression in serous papillary, clear-cell, and endometrioid endometrial cancer. Gynecol Oncol 2008;109:263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santin AD, Bellone S, Marizzoni M, et al. Overexpression of claudin-3 and claudin-4 receptors in uterine serous papillary carcinoma: novel targets for a type-specific therapy using Clostridium perfringens enterotoxin (CPE). Cancer 2007;109:1312-22. [DOI] [PubMed] [Google Scholar]
- 76.Satake S, Semba S, Matsuda Y, et al. Cdx2 transcription factor regulates claudin-3 and claudin-4 expression during intestinal differentiation of gastric carcinoma. Pathol Int 2008;58:156-63. [DOI] [PubMed] [Google Scholar]
- 77.Choi YL, Kim J, Kwon MJ, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol 2007;22:1185-95. [DOI] [PubMed] [Google Scholar]
- 78.Huang YH, Bao Y, Peng W, et al. Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc Natl Acad Sci U S A 2009;106:3426-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res 2005;65:7378-85. [DOI] [PubMed] [Google Scholar]
- 80.Sun C, Yi T, Song X, et al. Efficient inhibition of ovarian cancer by short hairpin RNA targeting claudin-3. Oncol Rep 2011;26:193-200. [DOI] [PubMed] [Google Scholar]
- 81.He ZY, Wei XW, Luo M, et al. Folate-linked lipoplexes for short hairpin RNA targeting claudin-3 delivery in ovarian cancer xenografts. J Control Release 2013;172:679-89. [DOI] [PubMed] [Google Scholar]
- 82.Pan XY, Wang B, Che YC, et al. Expression of claudin-3 and claudin-4 in normal, hyperplastic, and malignant endometrial tissue. Int J Gynecol Cancer 2007;17:233-41. [DOI] [PubMed] [Google Scholar]
- 83.Hwang TL, Changchien TT, Wang CC, et al. Claudin-4 expression in gastric cancer cells enhances the invasion and is associated with the increased level of matrix metalloproteinase-2 and -9 expression. Oncol Lett 2014;8:1367-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suren D, Yildirim M, Kaya V, et al. Expression patterns of claudins 1, 4, and 7 and their prognostic significance in nasopharyngeal carcinoma. J BUON 2015;20:212-7. [PubMed] [Google Scholar]
- 85.Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol 2004;121:226-30. [DOI] [PubMed] [Google Scholar]
- 86.Philip R, Heiler S, Mu W, et al. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget 2015;6:2046-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dahiya N, Becker KG, Wood WH, 3rd, et al. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS One 2011;6:e22119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alikanoglu AS, Gunduz S, Demirpence O, et al. Expression pattern and prognostic significance of claudin 1, 4 and 7 in pancreatic cancer. Asian Pac J Cancer Prev 2015;16:4387-92. [DOI] [PubMed] [Google Scholar]
- 89.Lin Z, Zhang X, Liu Z, et al. The distinct expression patterns of claudin-2, -6, and -11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagn Pathol 2013;8:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuo SJ, Chien SY, Lin C, et al. Significant elevation of CLDN16 and HAPLN3 gene expression in human breast cancer. Oncol Rep 2010;24:759-66. [DOI] [PubMed] [Google Scholar]
- 91.Rangel LB, Sherman-Baust CA, Wernyj RP, et al. Characterization of novel human ovarian cancer-specific transcripts (HOSTs) identified by serial analysis of gene expression. Oncogene 2003;22:7225-32. [DOI] [PubMed] [Google Scholar]
- 92.Men W, Martin TA, Ruge F, et al. Expression of claudins in human clear cell renal cell carcinoma. Cancer Genomics Proteomics 2015;12:1-8. [PubMed] [Google Scholar]
- 93.Martin TA, Lane J, Ozupek H, et al. Claudin-20 promotes an aggressive phenotype in human breast cancer cells. Tissue Barriers 2013;1:e26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martìn-Padura I, Lostaglio S, Schneemann M, et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J Cell Biol 1998;142:117-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aurrand-Lions M, Duncan L, Ballestrem C, et al. JAM-2, a novel immunoglobulin superfamily molecule, expressed by endothelial and lymphatic cells. J Biol Chem 2001;276:2733-41. [DOI] [PubMed] [Google Scholar]
- 96.Arrate MP, Rodriguez JM, Tran TM, et al. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J Biol Chem 2001;276:45826-32. [DOI] [PubMed] [Google Scholar]
- 97.Luissint AC, Lutz PG, Calderwood DA, et al. JAM-L-mediated leukocyte adhesion to endothelial cells is regulated in cis by alpha4beta1 integrin activation. J Cell Biol 2008;183:1159-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirabayashi S, Tajima M, Yao I, et al. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol Cell Biol 2003;23:4267-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mori H, Hirabayashi S, Shirasawa M, et al. JAM4 enhances hepatocyte growth factor-mediated branching and scattering of Madin-Darby canine kidney cells. Genes Cells 2004;9:811-9. [DOI] [PubMed] [Google Scholar]
- 100.Brennan K, Offiah G, McSherry EA, et al. Tight junctions: a barrier to the initiation and progression of breast cancer? J Biomed Biotechnol 2010;2010:460607. [DOI] [PMC free article] [PubMed]
- 101.Luo Y, Fukuhara M, Weitzman M, et al. Expression of JAM-A, AF-6, PAR-3 and PAR-6 during the assembly and remodeling of RPE tight junctions. Brain Res 2006;1110:55-63. [DOI] [PubMed] [Google Scholar]
- 102.Zhan L, Rosenberg A, Bergami KC, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 2008;135:865-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bradfield PF, Nourshargh S, Aurrand-Lions M, et al. JAM family and related proteins in leukocyte migration (Vestweber series). Arterioscler Thromb Vasc Biol 2007;27:2104-12. [DOI] [PubMed] [Google Scholar]
- 104.Sobocka MB, Sobocki T, Banerjee P, et al. Cloning of the human platelet F11 receptor: a cell adhesion molecule member of the immunoglobulin superfamily involved in platelet aggregation. Blood 2000;95:2600-9. [PubMed] [Google Scholar]
- 105.Naik TU, Naik MU, Naik UP. Junctional adhesion molecules in angiogenesis. Front Biosci 2008;13:258-62. [DOI] [PubMed] [Google Scholar]
- 106.Mandell KJ, Babbin BA, Nusrat A, et al. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on beta1 integrins and Rap1 activity. J Biol Chem 2005;280:11665-74. [DOI] [PubMed] [Google Scholar]
- 107.Naik MU, Naik TU, Suckow AT, et al. Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer Res 2008;68:2194-203. [DOI] [PubMed] [Google Scholar]
- 108.Murakami M, Giampietro C, Giannotta M, et al. Abrogation of junctional adhesion molecule-A expression induces cell apoptosis and reduces breast cancer progression. PLoS One 2011;6:e21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Severson EA, Lee WY, Capaldo CT, et al. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1A, regulate beta1 integrin levels, and enhance cell migration. Mol Biol Cell 2009;20:1916-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nava P, Capaldo CT, Koch S, et al. JAM-A regulates epithelial proliferation through Akt/β-catenin signalling. EMBO Rep 2011;12:314-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chatterjee S, Wang Y, Duncan MK, et al. Junctional adhesion molecule-A regulates vascular endothelial growth factor receptor-2 signaling-dependent mouse corneal wound healing. PLoS One 2013;8:e63674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu M, Ji S, Xiao S, et al. JAM-A promotes wound healing by enhancing both homing and secretion activities of mesenchymal stem cells. Clin Sci (Lond) 2015. [Epub ahead of print]. [DOI] [PubMed]
- 113.Reeh M, Bockhorn M, Görgens D, et al. Presence of the coxsackievirus and adenovirus receptor (CAR) in human neoplasms: a multitumour array analysis. Br J Cancer 2013;109:1848-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Auer D, Reimer D, Porto V, et al. Expression of coxsackie-adenovirus receptor is related to estrogen sensitivity in breast cancer. Breast Cancer Res Treat 2009;116:103-11. [DOI] [PubMed] [Google Scholar]
- 115.Persson A, Fan X, Salford LG, et al. Neuroblastomas and medulloblastomas exhibit more Coxsackie adenovirus receptor expression than gliomas and other brain tumors. Neuropathology 2007;27:233-6. [DOI] [PubMed] [Google Scholar]
- 116.Krause G, Winkler L, Piehl C, et al. Structure and function of extracellular claudin domains. Ann N Y Acad Sci 2009;1165:34-43. [DOI] [PubMed] [Google Scholar]
- 117.Morita K, Furuse M, Fujimoto K, et al. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A 1999;96:511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Piontek J, Winkler L, Wolburg H, et al. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J 2008;22:146-58. [DOI] [PubMed] [Google Scholar]
- 119.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol 2010;2010:541957. [DOI] [PMC free article] [PubMed]
- 120.Weber CR, Nalle SC, Tretiakova M, et al. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest 2008;88:1110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Halász J, Holczbauer A, Páska C, et al. Claudin-1 and claudin-2 differentiate fetal and embryonal components in human hepatoblastoma. Hum Pathol 2006;37:555-61. [DOI] [PubMed] [Google Scholar]
- 122.Singh AB, Sharma A, Smith JJ, et al. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology 2011;141:2140-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [DOI] [PubMed] [Google Scholar]
- 124.Martin TA, Harrison GM, Watkins G, et al. Claudin-16 reduces the aggressive behavior of human breast cancer cells. J Cell Biochem 2008;105:41-52. [DOI] [PubMed] [Google Scholar]
- 125.Mori K, Iida S, Yamane-Ohnuki N, et al. Non-fucosylated therapeutic antibodies: the next generation of therapeutic antibodies. Cytotechnology 2007;55:109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol 2010;10:317-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kato-Nakano M, Suzuki M, Kawamoto S, et al. Characterization and evaluation of the antitumour activity of a dual-targeting monoclonal antibody against claudin-3 and claudin-4. Anticancer Res 2010;30:4555-62. [PubMed] [Google Scholar]
- 128.Lamagna C, Hodivala-Dilke KM, Imhof BA, et al. Antibody against junctional adhesion molecule-C inhibits angiogenesis and tumor growth. Cancer Res 2005;65:5703-10. [DOI] [PubMed] [Google Scholar]
- 129.Samaranayake H, Wirth T, Schenkwein D, et al. Challenges in monoclonal antibody-based therapies. Ann Med 2009;41:322-31. [DOI] [PubMed] [Google Scholar]
- 130.Sarker MR, Carman RJ, McClane BA. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol 1999;33:946-58. [DOI] [PubMed] [Google Scholar]
- 131.Kominsky SL, Vali M, Korz D, et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol 2004;164:1627-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Michl P, Buchholz M, Rolke M, et al. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 2001;121:678-84. [DOI] [PubMed] [Google Scholar]
- 133.Long H, Crean CD, Lee WH, et al. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res 2001;61:7878-81. [PubMed] [Google Scholar]
- 134.Santin AD, Cané S, Bellone S, et al. Treatment of chemotherapy-resistant human ovarian cancer xenografts in C.B-17/SCID mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer Res 2005;65:4334-42. [DOI] [PubMed] [Google Scholar]
- 135.Kominsky SL, Tyler B, Sosnowski J, et al. Clostridium perfringens enterotoxin as a novel-targeted therapeutic for brain metastasis. Cancer Res 2007;67:7977-82. [DOI] [PubMed] [Google Scholar]
- 136.Patil SD, Rhodes DG, Burgess DJ. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J 2005;7:E61-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Coyne CB, Bergelson JM. CAR: a virus receptor within the tight junction. Adv Drug Deliv Rev 2005;57:869-82. [DOI] [PubMed] [Google Scholar]
- 138.Miller CR, Buchsbaum DJ, Reynolds PN, et al. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res 1998;58:5738-48. [PubMed] [Google Scholar]
- 139.Fuxe J, Liu L, Malin S, et al. Expression of the coxsackie and adenovirus receptor in human astrocytic tumors and xenografts. Int J Cancer 2003;103:723-9. [DOI] [PubMed] [Google Scholar]
- 140.Li Y, Pong RC, Bergelson JM, et al. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res 1999;59:325-30. [PubMed] [Google Scholar]
- 141.Okegawa T, Li Y, Pong RC, et al. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res 2000;60:5031-6. [PubMed] [Google Scholar]
- 142.Cripe TP, Dunphy EJ, Holub AD, et al. Fiber knob modifications overcome low, heterogeneous expression of the coxsackievirus-adenovirus receptor that limits adenovirus gene transfer and oncolysis for human rhabdomyosarcoma cells. Cancer Res 2001;61:2953-60. [PubMed] [Google Scholar]
- 143.Fechner H, Wang X, Wang H, et al. Trans-complementation of vector replication versus Coxsackie-adenovirus-receptor overexpression to improve transgene expression in poorly permissive cancer cells. Gene Ther 2000;7:1954-68. [DOI] [PubMed] [Google Scholar]
- 144.Kim JS, Lee SH, Cho YS, et al. Enhancement of the adenoviral sensitivity of human ovarian cancer cells by transient expression of coxsackievirus and adenovirus receptor (CAR). Gynecol Oncol 2002;85:260-5. [DOI] [PubMed] [Google Scholar]
- 145.Qin M, Chen S, Yu T, et al. Coxsackievirus adenovirus receptor expression predicts the efficiency of adenoviral gene transfer into non-small cell lung cancer xenografts. Clin Cancer Res 2003;9:4992-9. [PubMed] [Google Scholar]
- 146.Shayakhmetov DM, Li ZY, Ni S, et al. Targeting of adenovirus vectors to tumor cells does not enable efficient transduction of breast cancer metastases. Cancer Res 2002;62:1063-8. [PubMed] [Google Scholar]
- 147.Okegawa T, Pong RC, Li Y, et al. The mechanism of the growth-inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: a functional analysis of car protein structure. Cancer Res 2001;61:6592-600. [PubMed] [Google Scholar]
- 148.Kim M, Sumerel LA, Belousova N, et al. The coxsackievirus and adenovirus receptor acts as a tumour suppressor in malignant glioma cells. Br J Cancer 2003;88:1411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sachs MD, Rauen KA, Ramamurthy M, et al. Integrin alpha(v) and coxsackie adenovirus receptor expression in clinical bladder cancer. Urology 2002;60:531-6. [DOI] [PubMed] [Google Scholar]
- 150.Sibbald B. Death but one unintended consequence of gene-therapy trial. CMAJ 2001;164:1612. [PMC free article] [PubMed] [Google Scholar]
- 151.Vorburger SA, Hunt KK. Adenoviral gene therapy. Oncologist 2002;7:46-59. [DOI] [PubMed] [Google Scholar]