Abstract
The field of three-dimensional electron microscopy began more than 45 years ago with a reconstruction of a helical phage tail, and helical polymers continue to be important objects for three-dimensional reconstruction due to the centrality of helical protein and nucleoprotein polymers in all aspects of biology. We are now witnessing a fundamental revolution in this area, made possible by direct electron detectors, which has led to near-atomic resolution for a number of important helical structures. Most importantly, the possibility of achieving such resolution routinely for a vast number of helical samples is within our reach. One of the main problems in helical reconstruction, ambiguities in assigning the helical symmetry, is overcome when one reaches a resolution where secondary structure is clearly visible. However, obstacles still exist due to the intrinsic variability within many helical filaments.
Keywords: helical polymers, direct electron detectors, variable twist
Large amounts of protein in viruses, bacteria, archaea and eukaryotes exist in the form of helical polymers. For examples, these proteins may assemble into the capsids of filamentous viruses (Kendall et al., 2008; Namba et al., 1989), into flagellar filaments (Yonekura et al., 2003) or plasmid segregation filaments (Garner et al., 2004) in bacteria, and into cytoskeletal filaments in archaea (Duggin et al., 2014) or eukaryotes (Alushin et al., 2014; Galkin et al., 2015; von der Ecken et al., 2015). While the polymerization of protein in eukaryotic cells into helical filaments has been understood as an important element of motile processes, from muscle contraction to the segregation of chromosomes, it has become clear only recently that polymerization of proteins into helical assemblies is also an important aspect of signaling, innate immunity and inflammation (Berke et al., 2012; Ferrao et al., 2012; Lin et al., 2010; Lu et al., 2014a; Qiao et al., 2013; Wu et al., 2013; Wu et al., 2014). Further, proteins such as RecA in bacteria (Stasiak and DiCapua, 1982) or Rad51 in eukaryotes (Ogawa et al., 1993) assemble into helical nucleoprotein complexes that are crucial in many aspects of DNA repair using homologous recombination. While most structural biology of viruses has involved icosahedral viruses, many helical viruses exist (Bernal and Fankuchen, 1941; Kendall et al., 2008; Namba et al., 1989; Opella et al., 1980; Stubbs and Kendall, 2012; Wang et al., 2006), and nucleocapsids within other non-helical viruses are often helical (Bakker et al., 2013; Bhella et al., 2004; Communie et al., 2013; Desfosses et al., 2013; Schoehn et al., 2004). Methods to determine the structure of these biological polymers are thus of great importance.
Unfortunately, symmetry considerations dictate that unless a polymer has exactly two, three, four or six subunits per turn, it cannot be packed in a crystal so that every subunit is in an identical environment. This has limited the utility of x-ray crystallography in solving the structure of protein or nucleoprotein polymers. While it has been possible in at least one case (Chen et al., 2008) to form a polyprotein of subunits from a helical polymer that can be crystallized with the polyprotein as an asymmetric unit, this does not appear to be a general strategy that can provide atomic structures for a large number of filaments. X-ray fiber diffraction has led to atomic models for several rigid rod-like helical viruses (Namba and Stubbs, 1986; Wang et al., 1997; Wang and Stubbs, 1994), but the technique is quite laborious and difficult and fails for the more common flexible filamentous viruses (Kendall et al., 2008). Solid-state NMR has been used for structure determination of helical polymers such as filamentous bacteriophage (Colnago et al., 1987; Cross and Opella, 1985; Zeri et al., 2003) but since models are built from local interactions with no long-range constraints, they are not necessarily unique (Marvin et al., 2006; Straus et al., 2011; Straus et al., 2008).
Cryo-EM is ideal for helical polymers
Electron microscopy, on the other hand, is ideally suited to studying helical polymers, and can take advantage of the fact that in the projection of a helix many different views of identical subunits are provided. But until recently, only a few helical structures have been reconstructed at a near-atomic resolution (Ge and Zhou, 2011; Unwin, 2005; Yonekura et al., 2003). The symmetry inherent in helices led to the first three-dimensional reconstruction from electron micrographs (DeRosier and Klug, 1968), using a Fourier-Bessel formalism (Klug et al., 1958) which is based upon the fact that the Fourier transform of a helix can be readily decomposed into Bessel functions. The three-dimensional diffraction pattern of a helix is only non-zero on layer planes, and these become layer lines in the two-dimensional diffraction pattern. This formalism means that as long as only a single Bessel function is present on a layer line, a single image of a helix can provide all of the information needed to generate a three-dimensional reconstruction.
As has been extensively discussed (Egelman, 2000, 2007a, b, 2010), the limitations of the Fourier-Bessel approach are: 1) Filaments must be nearly crystalline, with long-range helical order; 2) This long-range order implies that filaments must be nearly perfectly straight; 3) Individual filaments must diffract strongly so that layer lines may be extracted from their Fourier transforms. Some biological polymers have been found which obey these three conditions, such as mutant flagellar filaments from Salmonella and reconstructions from these filaments at ∼ 4 Å allowed for building atomic models (Yonekura et al., 2003). While conditions (1) and (2) are quite rare but can be found, condition (3) is never met for low-dose cryo-EM from thin weakly scattering filaments. Some of the deviations from ideal helical symmetry (1 and 2) have been overcome with sophisticated approaches that have “relaxed” the traditional Fourier-Bessel methods (Beroukhim and Unwin, 1997; Unwin, 2005) and allowed for high resolution reconstructions of tubes formed by the acetylcholine receptor. Methods for straightening the images of slightly curved helical filaments have been developed (Egelman, 1986) and applied (Owen et al., 1996) using an assumption of a normal mode of bending (where one has uniform compression on the inside of a bend and uniform extension on the outside), but the resolution where these methods fail would depend upon individual specimens since one is basically assuming that a protein subunit compresses to the same extent as a helical groove. In any case, all of these modifications and enhancements of classical Fourier-Bessel approaches would still fail to surmount problem (3).
These limitations led to an alternate method (Egelman, 2000), Iterative Helical Real Space Reconstruction (IHRSR), which is based upon a single-particle approach. In any real polymer, long-range order may not exist due to the fact that all interactions are local. Thus, variability in the helical parameters, such as twist (Egelman and DeRosier, 1982; Egelman et al., 1982), accumulate and lead to a liquid-like state in which nearest-neighbor interactions are relatively ordered but order deteriorates the further one is away from any given reference subunit. In contrast, in a crystal one has a space group that maintains long-range order. The accumulation of disorder can be defined quantitatively in terms of a correlation length (Egelman and DeRosier, 1982), just as the flexural rigidity of a polymer can be defined in terms of its persistence length (Landau and Lifshitz, 1980), which is a measure of how far the correlation in direction persists for a filament undergoing thermally-driven flexing.
While the original implementation of IHRSR (Egelman, 2000) was in SPIDER (Frank et al., 1996), and many subsequent applications of IHRSR continue to use SPIDER, an implementation now exists (Behrmann et al., 2012) in SPARX (Hohn et al., 2007) and in RELION (Clemens et al., 2015). The IHRSR method surmounts many of the problems inherent in the Fourier-Bessel approach (Egelman, 2007a), but it does not solve the problem of intrinsic ambiguities present when attempting to reconstruct filaments at a limited resolution (Egelman, 2010; Egelman, 2014). A simple example is provided by Tobacco Mosaic Virus (TMV), which has been a model system in structural biology (Barrett et al., 1972; Bernal and Fankuchen, 1941; Desfosses et al., 2014; Franklin and Holmes, 1956; Ge and Zhou, 2011; Holmes and Franklin, 1958; Holmes et al., 1972; Holmes et al., 1975; Jeng et al., 1989; Mandelkow and Holmes, 1974; Namba and Stubbs, 1986; Sachse et al., 2007; Ward et al., 2003; Zhu et al., 2001) for the simple reason that it is anomalously crystalline!
If one has only 10 Å resolution, then the actual helical symmetry of TMV cannot be unambiguously determined from electron microscopic images (Egelman, 2010). One either needs to have prior knowledge, such as about the shape of the TMV subunit or accurate estimates of the mass per unit length from Scanning Transmission Electron Microscopy (Wall and Hainfeld, 1986), to make an informed choice about which of the possible symmetries is the correct one. In the published EM studies people started using the known symmetry of TMV determined by high-resolution x-ray fiber diffraction (Barrett et al., 1972; Franklin and Holmes, 1956; Holmes et al., 1975), a significant piece of prior knowledge.
Thus, determining the correct helical symmetry, usually based upon an analysis of power spectra from filaments (Egelman, 2010) is frequently the most difficult task and requires a thorough understanding of the Fourier-Bessel approach (Klug et al., 1958). While it might have been hoped that real-space methods would have surmounted the need for such knowledge, this has not proven to be the case. But no matter how great is one's understanding, intrinsic ambiguities frequently exist in symmetry determination from real specimens. If one has an array of atoms that are all at the same radius (Cochran, 1951) then indexing a helical diffraction pattern is very simple. But this is never the case for actual protein and nucleoprotein polymers, as subunits span a range of radial values. The consequence of this is that the relation between the Bessel order n on a given layer line and the distance of the first diffraction peak on that layer line from the meridian is not always simple (Fig. 1). One approach to determining helical symmetry that has been successfully applied (Bharat et al., 2014; Bharat et al., 2012; Briggs et al., 2009) involves tomograms of tubes. In this method, projection images that retain high resolution information from frozen-hydrated tubes are recorded first, followed by tomograms from the same tubes that can be analyzed at lower resolution to show the helical symmetry. Tomography of negatively-stained helical tubes (Heymann et al., 2013) was used to determine the helical hand, but as tubes and filaments become thinner such tomographic approaches will probably not work for determining helical symmetry.
Fig. 1.
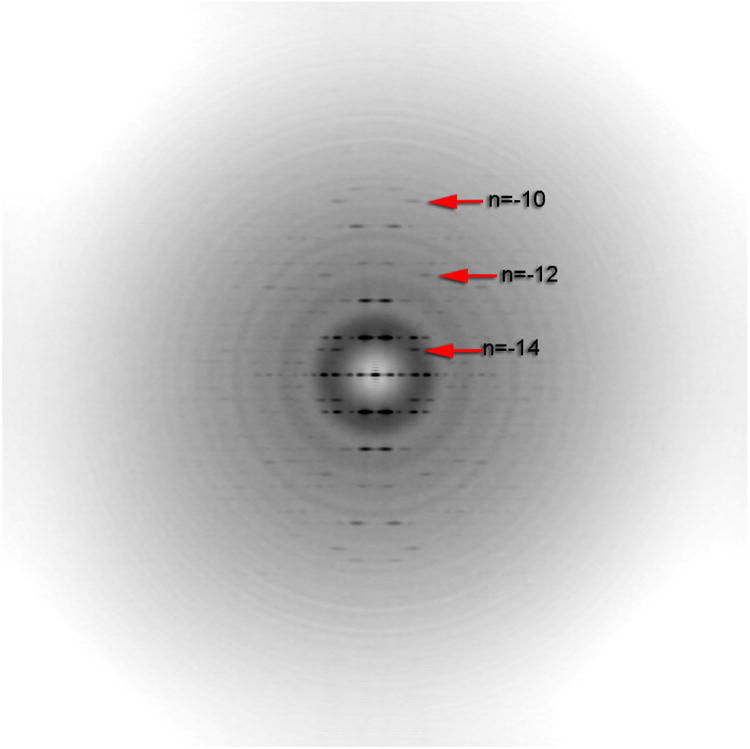
Averaged power spectrum showing that the distance of a peak from the meridian is not always simply related to the Bessel order n on any given layer line. For example, the first maxima on the layer line labeled n=-12 is at a larger distance R from the meridian than the peak on the layer line labeled n=-14. For a helical structure containing an array of atoms all at the same radius, the distance of these peaks from the meridian is a simple function of n, the Bessel order on that layer line. For a real structure, which extends over a range of radial values, this simple relationship breaks down.
Direct electron detectors solve many problems
The problem of symmetry ambiguities is completely eliminated when one has the resolution (perhaps better than 8 Å for α-helices and better than 5 Å for β-sheets) to see secondary structure. A map showing clear secondary structure must have the correct symmetry, while a map with the incorrect symmetry will be uninterpretable (Egelman, 2014). The difficulty, until very recently, has been how to get to such resolution, as most helical polymers do not have the order seen in such unusual filaments as TMV (Ge and Zhou, 2011) or mutant Salmonella flagellar filaments (Yonekura et al., 2003). Microtubules have now been reconstructed at ∼ 5 Å resolution in two states in a significant advance using film on a Titan microscope (Alushin et al., 2014). But few other polymers might reach such a resolution using film, and even fewer using conventional CCD cameras. The field has completely changed over the past two years with the introduction of direct electron detectors (Bai et al., 2013; Bammes et al., 2012; Bartesaghi et al., 2014; Li et al., 2013; Lu et al., 2014a; Lu et al., 2014b; Voorhees et al., 2014). I will focus in the remainder of this review on how these detectors have transformed cryo-EM helical reconstructions and the prospects for the near future. Prediction, goes an old Danish proverb, is hazardous, especially about the future. But given that a number of papers have recently been published on helical reconstructions with a near-atomic resolution using direct electron detectors, it is probably not too risky to suggest that such structures may soon prove to be routine, rather than exceptional.
While it might be imagined that the main benefit of the direct electron detectors is at the highest spatial frequencies, where the envelope function of either CCD-based detectors or film is lower, it appears that for helical filaments the greatest advantage of the direct electron detectors comes at an intermediate resolution, perhaps around 8-15 Å resolution. This can be seen clearly in Fig. 2, where a layer line at ∼ 1/(14 Å) is absent when using film on a Tecnai F20 (Fig. 2a) but very strong when using a Falcon II direct electron detector on a Titan Krios (Fig. 2b, blue arrow). If one only had the images used in Fig. 2a, one would be immediately confronted with the issue of ambiguities in indexing the pattern. Multiple symmetries might be possible, but none would yield a solution where secondary structure could be seen and hence the correct symmetry deduced. In fact, when one imposed the correct helical symmetry, a resolution of ∼ 18 Å was achieved generating a rather featureless blob for the small 91-residue subunit. In contrast, when one used the direct detector's higher resolution images (see power spectra, Fig 2b) and applied the correct symmetry, the all α-helical structure of the small subunit is seen providing evidence that the correct symmetry was, in fact, used (Lu et al., 2014a).
Fig. 2.
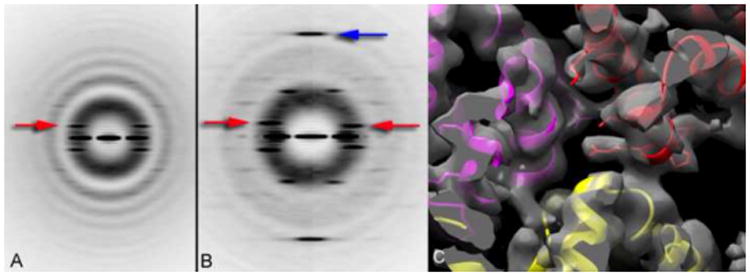
Power spectra from ASC PYD filaments (Lu et al., 2014a) recorded either on film using a Tecnai F20 microscope (A), or using a Falcon II direct electron detector on a Titan Krios (B). Both sets of images have been sorted based upon the pitch of the six-start helices (red arrows), with one twist state shown in (A), and two different twist states shown in (b). Although the twist is variable in the PYD filaments, the axial rise per subunit (which is actually three identical molecules, due to the C3 rotational symmetry in these filaments) is relatively fixed, and indicated by the blue arrow in (B) at ∼ 1/(14 Å). Surprisingly, this meridional layer line is not seen in (A) due to the much weaker modulation transfer function of the film on the F20 compared to the Falcon II on the Titan Krios. An atomic model can be built into the reconstruction (C), confirming that the correct symmetry has been used.
Similarly, imaging nanotubes formed by a 29-residue designed peptide (Egelman et al., 2015) one can see a strong layer line at 1/(9.0 Å) using a K2 direct electron detector on a Titan Krios (Fig. 3a) but this layer line was absent whether using a CCD camera on a Titan Krios or film on an F20. So without a direct electron detector the images would not even allow one to determine the helical symmetry of these nanotubes. But the images collected with the K2 allowed for building an atomic model of the structure (Fig. 3b) into the reconstruction that had a resolution of ∼ 3.6 Å (Egelman et al., 2015). This was in spite of the variable twist present in these tubes, which was overcome by simply sorting segments by twist and using a single subset having a similar twist in the reconstruction. Of course, this approach means that the more homogeneous the subset with respect to twist, the smaller it will be, and there will be a tradeoff between a worse signal-to-noise ratio due to the smaller sample and a potentially better resolution due to the greater homogeneity. So this is a classic optimization problem that can be best solved empirically by looking at the quality of the reconstruction as a function of the size of the subset.
Fig. 3.

A power spectrum from a thin helical nanotube formed by a 29-residue peptide (A), and the reconstruction of the nanotube with an atomic model (B) (Egelman et al., 2015). The layer line at 1/(9.0 Å) (A, arrows) is clearly visible when using a K2 direct electron detector on a Titan Krios, but absent when using either film on an F20 or a CCD camera on the Titan Krios. Most of the large side chains can be clearly seen in the density map (B).
The use of direct electron detectors over the past year has now led to a number of published applications to helical polymers (Clemens et al., 2015; Egelman et al., 2015; Galkin et al., 2015; Kudryashev et al., 2015; Lu et al., 2014a; von der Ecken et al., 2015; Wu et al., 2014; Xu et al., 2014). Two of these (Wu et al., 2014; Xu et al., 2014) involve the filament formed by the MAVS (Mitochondrial Anti-Viral Signaling protein) CARD (Caspase Activation and Recruitment Domain), and the comparison is quite interesting. One of these (Wu et al., 2014) is at a stated resolution of 3.6 Å while the other (Xu et al., 2014) is at a stated resolution of 9.6 Å, and a different helical symmetry has been applied in Xu et al. to that applied in Wu et al. In an extended Comment to the paper by Xu et al. Jiang argues that the helical symmetry is actually different in the filaments prepared by the two groups, and that the 3.6 Å reconstruction arises from harsh preparative procedures while their filaments represent a native state. The images used in Xu et al. are available, and this has allowed for an analysis (Egelman, 2014) showing that the filaments from both groups are consistent with the same symmetry, which is the one used in Wu et al., and that the different symmetry used in Xu et al. appears to have been mistakenly imposed due to a large degree of out-of-plane tilt. The analysis further shows that the reconstruction published by Xu et al. has little correlation with the model they built at anything better than 22 Å resolution, highlighting why existing resolution measures such as the Fourier Shell Correlation are not necessarily based upon any reality, only self-consistency (Yang et al., 2003).
Two of the papers (Galkin et al., 2015; von der Ecken et al., 2015) involve F-actin, where one was from naked actin filaments at 4.7 Å resolution (Galkin et al., 2015) while the other was from the actin-tropomyosin complex with the actin portion at 3.7 Å resolution (von der Ecken et al., 2015). The idea that the thin films used in cryo-EM exert large mechanical forces on filaments which cause structural changes in F-actin (Galkin et al., 2012) was further developed in Galkin et al. (2014) and it was shown that the variability in twist in F-actin (Egelman et al., 1982) could be modulated by the ice thickness, just as very thin ice reduced the subunit's structural heterogeneity (Galkin et al., 2010) present in thicker ice. A comparison between the two atomic models for this one state of F-actin (3J8I.PDB for the pure actin and 3J8A.PDB for the actin-tropomyosin complex) shows an rmsd of 1.0 Å for 332 Cα pairs and 1.3 Å over all Cα pairs, confirming how similar the two models are.
One of the highest resolutions achieved thus far in helical reconstruction (∼ 3.2 Å) comes from a cryo-EM study (Kudryashev et al., 2015) of the Type Six Secretion System sheath of Vibrio cholerae, the organism responsible for cholera (Fig. 4). The resolution enabled an ab initio chain trace of ∼ 600 amino acid residues in the asymmetric unit, a heterodimer of the VipA and VipB proteins. A related Type Six Secretion System contractile sheath from Francisella tularensis (the organism responsible for tularemia) has been solved at the same time at a resolution of ∼ 3.7 Å (Clemens et al., 2015), giving credibility to the suggestion that such resolutions will become the new normal. This resolution has allowed a full chain trace in both structures, revealing how heterodimers are intertwined at the level of secondary structure. At lower resolution such insights would simply not be possible.
Fig. 4.
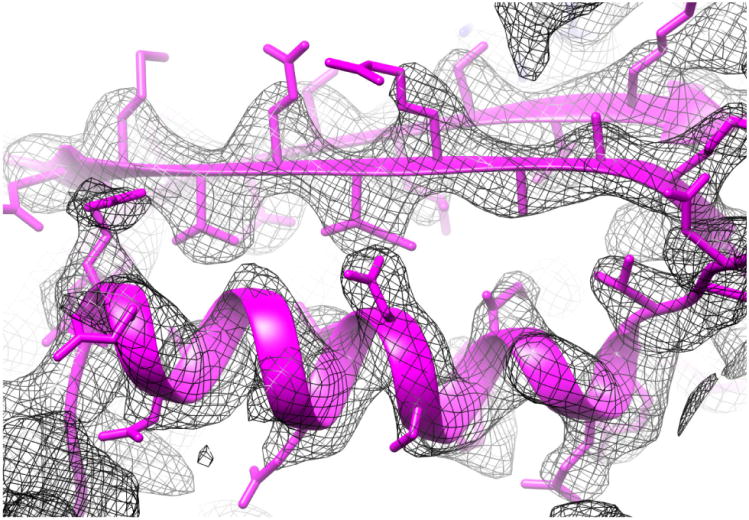
Map (mesh) and model (magenta) from the interior of the Type Six Secretion System sheath of Vibrio cholerae (Kudryashev et al., 2015). An α-helix is shown at the bottom, and two β-strands forming part of a β-sheet are seen at the top. The resolution of the map (∼ 3.2 Å in the most ordered parts) allowed for the ab initio tracing of > 600 residues in the two chains that form the heterodimer of the asymmetric unit.
Conclusion
It would have been difficult for anyone to predict several years ago where we are now with regard to the number of helical polymers that can be reconstructed at near-atomic resolution. While complexes such as the ribosome can be crystallized, they may now be solved at even higher resolution and more readily by cryo-EM (Fischer et al., 2015). But most helical polymers simply cannot be crystallized, so determining the atomic structure of these filaments has been almost impossible in the past. That has now changed. Not only does the new resolution available allow for atomic models, but it provides a validation of the correct symmetry that has frequently been absent previously. It is indeed an exciting time to be working on such helical filaments given their abundance in all aspects of biology.
Helical protein and nucleoprotein polymers are ubiquitous in biology.
Three-dimensional reconstruction in electron microscopy began with helices.
Intrinsic ambiguities in helical symmetry have been more common than realized.
Direct-electron detectors allow near-atomic resolution, resolving ambiguities.
Acknowledgments
This work has been supported by NIH EB001567
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-Resolution Microtubule Structures Reveal the Structural Transitions in alphabeta-Tubulin upon GTP Hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker SE, Duquerroy S, Galloux M, Loney C, Conner E, Eleouet JF, Rey FA, Bhella D. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. The Journal of general virology. 2013;94:1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. Journal of Structural Biology. 2012;177:589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AN, Leigh JB, Holmes KC, Leberman R, Mandelkow E, von Sengbusch P. An electron-density map of tobacco mosaic virus at 10 Angstrom resolution. Cold Spring Harb Symp Quant Biol. 1972;36:433–448. doi: 10.1101/sqb.1972.036.01.056. [DOI] [PubMed] [Google Scholar]
- Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci U S A. 2014;111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann E, Tao G, Stokes DL, Egelman EH, Raunser S, Penczek PA. Real-space processing of helical filaments in SPARX. J Struct Biol. 2012;177:302–313. doi: 10.1016/j.jsb.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal JD, Fankuchen I. X-Ray and Crystallographic Studies of Plant Virus Preparations. Iii. The Journal of general physiology. 1941;25:147–165. doi: 10.1085/jgp.25.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Unwin N. Distortion correction of tubular crystals: improvements in the acetylcholine receptor structure. Ultramicroscopy. 1997;70:57–81. doi: 10.1016/s0304-3991(97)00070-3. [DOI] [PubMed] [Google Scholar]
- Bharat TA, Castillo Menendez LR, Hagen WJ, Lux V, Igonet S, Schorb M, Schur FK, Krausslich HG, Briggs JA. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc Natl Acad Sci U S A. 2014;111:8233–8238. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA. Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature. 2012;487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- Bhella D, Ralph A, Yeo RP. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. Journal of Molecular Biology. 2004;340:319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. ProcNatlAcadSciUSA. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic Structure of T6SS Reveals Interlaced Array Essential to Function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WC, C FH, Vand V. The structure of synthetic polypeptides. I. The transform of atoms on a helix. Acta Crystallographica. 1951;5:581–586. [Google Scholar]
- Colnago LA, Valentine KG, Opella SJ. Dynamics of fd coat protein in the bacteriophage. Biochemistry. 1987;26:847–854. doi: 10.1021/bi00377a028. [DOI] [PubMed] [Google Scholar]
- Communie G, Habchi J, Yabukarski F, Blocquel D, Schneider R, Tarbouriech N, Papageorgiou N, Ruigrok RW, Jamin M, Jensen MR, et al. Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS pathogens. 2013;9:e1003631. doi: 10.1371/journal.ppat.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Opella SJ. Protein structure by solid state nuclear magnetic resonance. Residues 40 to 45 of bacteriophage fd coat protein. Journal of Molecular Biology. 1985;182:367–381. doi: 10.1016/0022-2836(85)90197-4. [DOI] [PubMed] [Google Scholar]
- DeRosier DJ, Klug A. Reconstruction of three-dimensional structures from electron micrographs. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- Desfosses A, Ciuffa R, Gutsche I, Sachse C. SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J Struct Biol. 2014;185:15–26. doi: 10.1016/j.jsb.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Desfosses A, Ribeiro EA, Jr, Schoehn G, Blondel D, Guilligay D, Jamin M, Ruigrok RW, Gutsche I. Self-organization of the vesicular stomatitis virus nucleocapsid into a bullet shape. Nature communications. 2013;4:1429. doi: 10.1038/ncomms2435. [DOI] [PubMed] [Google Scholar]
- Duggin IG, Aylett CH, Walsh JC, Michie KA, Wang Q, Turnbull L, Dawson EM, Harry EJ, Whitchurch CB, Amos LA, et al. CetZ tubulin-like proteins control archaeal cell shape. Nature. 2014 doi: 10.1038/nature13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. An algorithm for straightening images of curved filamentous structures. Ultramicroscopy. 1986;19:367–373. doi: 10.1016/0304-3991(86)90096-3. [DOI] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. Journal of Structural Biology. 2007a;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Single-particle reconstruction from EM images of helical filaments. Current Opinion in Structural Biology. 2007b;17:556–561. doi: 10.1016/j.sbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods in Enzymology. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. Helical Ambiguities. eLife. 2014;3:e04969. doi: 10.7554/eLife.04969.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, DeRosier DJ. The Fourier transform of actin and other helical systems with cumulative random angular disorder. Acta Crystallographica. 1982;A38:796–799. [Google Scholar]
- Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP. Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure. 2015;23:280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. SciSignal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by C-corrected cryo-EM. Nature. 2015 doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Franklin RE, Holmes KC. The helical arrangement of the protein subunits in tobacco mosaic virus. Biochim Biophys Acta. 1956;21:405–406. doi: 10.1016/0006-3002(56)90043-9. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Egelman EH. Actin filaments as tension sensors. Current Biology. 2012;22:R96–R101. doi: 10.1016/j.cub.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Schrîder GF, Egelman EH. Structural polymorphism in F-actin. NatStructMolBiol. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Vos MR, Schroder GF, Egelman EH. Near-atomic resolution for one state of f-actin. Structure. 2015;23:173–182. doi: 10.1016/j.str.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. ProcNatlAcadSciUSA. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Bartho JD, Rybakova D, Venugopal HP, Winkler DC, Sen A, Hurst MR, Mitra AK. Three-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila. J Biol Chem. 2013;288:25276–25284. doi: 10.1074/jbc.M113.456145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. Journal of Structural Biology. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Franklin RE. The radial density distribution in some strains of tobacco mosaic virus. Virology. 1958;6:328–336. doi: 10.1016/0042-6822(58)90086-2. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Mandelkow E, Leigh JB. The determination of the heavy atom positions in tobacco mosaic virus from double heavy atom derivatives. Die Naturwissenschaften. 1972;59:247–254. doi: 10.1007/BF00610199. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Stubbs GJ, Mandelkow E, Gallwitz U. Structure of tobacco mosaic virus at 6.7 a resolution. Nature. 1975;254:192–196. doi: 10.1038/254192a0. [DOI] [PubMed] [Google Scholar]
- Jeng TW, Crowther RA, Stubbs G, Chiu W. Visualization of alpha-helices in tobacco mosaic virus by cryo-electron microscopy. Journal of Molecular Biology. 1989;205:251–257. doi: 10.1016/0022-2836(89)90379-3. [DOI] [PubMed] [Google Scholar]
- Kendall A, McDonald M, Bian W, Bowles T, Baumgarten SC, Shi J, Stewart PL, Bullitt E, Gore D, Irving TC, et al. Structure of flexible filamentous plant viruses. Journal of Virology. 2008;82:9546–9554. doi: 10.1128/JVI.00895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A, Crick FH, Wyckoff HW. Diffraction by helical structures. Acta Crystallographica. 1958;11:199–213. [Google Scholar]
- Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. Structure of the Type VI Secretion System Contractile Sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau LD, Lifshitz EM. Statistical Physics. 3rd. Oxford: Pergamon Press; 1980. [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. NatMethods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014a;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, et al. Three-dimensional structure of human gamma-secretase. Nature. 2014b;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E, Holmes KC. The positions of the N-terminus and residue 68 in tobacco mosaic virus. J Mol Biol. 1974;87:265–273. doi: 10.1016/0022-2836(74)90148-x. [DOI] [PubMed] [Google Scholar]
- Marvin DA, Welsh LC, Symmons MF, Scott WR, Straus SK. Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. Journal of Molecular Biology. 2006;355:294–309. doi: 10.1016/j.jmb.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Namba K, Pattanayek R, Stubbs G. Visualization of protein-nucleic acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 A resolution by X-ray fiber diffraction. Journal of Molecular Biology. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- Namba K, Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986;231:1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Opella SJ, Cross TA, DiVerdi JA, Sturm CF. Nuclear magnetic resonance of the filamentous bacteriophage fd. Biophysical Journal. 1980;32:531–548. doi: 10.1016/S0006-3495(80)84988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen CH, Morgan DG, DeRosier DJ. Image analysis of helical objects: the Brandeis Helical Package. J Struct Biol. 1996;116:167–175. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, Bracken C, Rosen M, Melnick A, Egelman EH, et al. Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse C, Chen JZ, Coureux PD, Stroupe ME, Fandrich M, Grigorieff N. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. Journal of Molecular Biology. 2007;371:812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoehn G, Mavrakis M, Albertini A, Wade R, Hoenger A, Ruigrok RW. The 12 A structure of trypsin-treated measles virus N-RNA. Journal of Molecular Biology. 2004;339:301–312. doi: 10.1016/j.jmb.2004.03.073. [DOI] [PubMed] [Google Scholar]
- Stasiak A, DiCapua E. The helicity of DNA in complexes with RecA protein. Nature. 1982;229:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- Straus SK, Scott WR, Schwieters CD, Marvin DA. Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR. European biophysics journal : EBJ. 2011;40:221–234. doi: 10.1007/s00249-010-0640-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus SK, Scott WR, Symmons MF, Marvin DA. On the structures of filamentous bacteriophage Ff (fd, f1, M13) European biophysics journal : EBJ. 2008;37:521–527. doi: 10.1007/s00249-007-0222-7. [DOI] [PubMed] [Google Scholar]
- Stubbs G, Kendall A. Helical viruses. Adv Exp Med Biol. 2012;726:631–658. doi: 10.1007/978-1-4614-0980-9_28. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin--tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JS, Hainfeld JF. Mass Mapping with the Scanning Transmission Electron Microscope. AnnuRevBiophysBiophysChem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- Wang H, Culver JN, Stubbs G. Structure of ribgrass mosaic virus at 2.9 A resolution: evolution and taxonomy of tobamoviruses. J Mol Biol. 1997;269:769–779. doi: 10.1006/jmbi.1997.1048. [DOI] [PubMed] [Google Scholar]
- Wang H, Stubbs G. Structure determination of cucumber green mottle mosaic virus by X-ray fiber diffraction. Significance for the evolution of tobamoviruses. J Mol Biol. 1994;239:371–384. doi: 10.1006/jmbi.1994.1379. [DOI] [PubMed] [Google Scholar]
- Wang YA, Yu X, Overman S, Tsuboi M, Thomas GJ, Jr, Egelman EH. The structure of a filamentous bacteriophage. Journal of Molecular Biology. 2006;361:209–215. doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Ward A, Moody MF, Sheehan B, Milligan RA, Carragher B. Windex: a toolset for indexing helices. Journal of Structural Biology. 2003;144:172–183. doi: 10.1016/j.jsb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. eLife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yu X, Galkin VE, Egelman EH. Issues of resolution and polymorphism in single-particle reconstruction. Journal of Structural Biology. 2003;144:162–171. doi: 10.1016/j.jsb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Zeri AC, Mesleh MF, Nevzorov AA, Opella SJ. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. ProcNatlAcadSciUSA. 2003;100:6458–6463. doi: 10.1073/pnas.1132059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Carragher B, Kriegman DJ, Milligan RA, Potter CS. Automated identification of filaments in cryoelectron microscopy images. Journal of Structural Biology. 2001;135:302–312. doi: 10.1006/jsbi.2001.4415. [DOI] [PubMed] [Google Scholar]


