Abstract
Dengue viruses cause the most important human viral disease transmitted by mosquitoes. In recent years, a great deal has been learned about molecular details of dengue virus genome replication; however, little is known about genome encapsidation and the functions of the viral capsid protein. During infection, dengue virus capsid progressively accumulates around lipid droplets by an unknown mechanism. Here, we examined the process by which the viral capsid is transported from the ER membrane, where the protein is synthesized, to lipid droplets. Using different methods of intervention, we found that the GBF1-Arf1/Arf4-COPI pathway is necessary for capsid transport to lipid droplets, while the process is independent of both COPII components and Golgi integrity. The transport was sensitive to brefeldin A, while a drug resistant form of GBF1 was sufficient to restore capsid subcellular distribution in infected cells. The mechanism by which lipid droplets gain or lose proteins is still an open question. Our results support a model in which the virus uses a non-canonical function of the COPI system for capsid accumulation on lipid droplets, providing new ideas for antiviral strategies.
Keywords: dengue virus, capsid protein, COPI, GBF1, flavivirus, lipid droplets
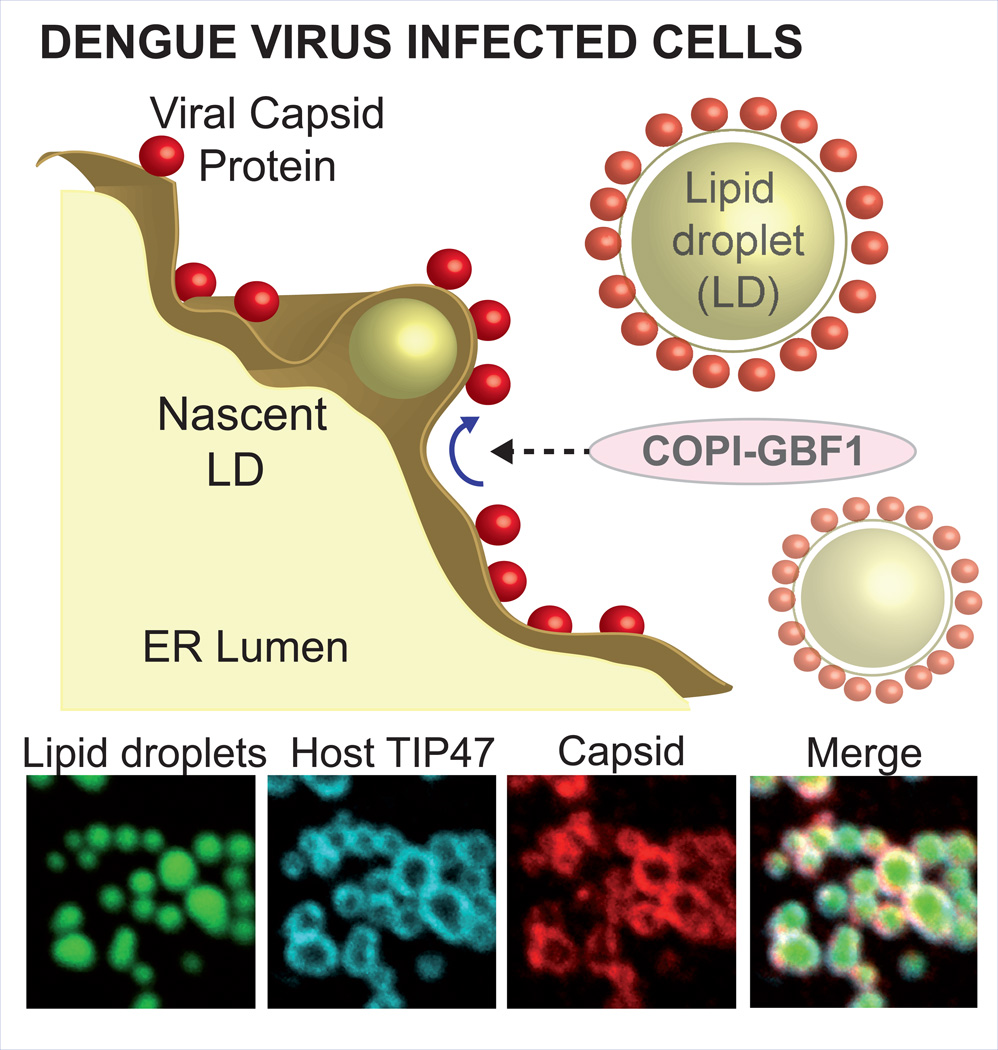
Brief synopsis of abstract figure
The dengue virus capsid protein accumulates progressively around cellular organelles named lipid droplets (LDs) during infection. Using different methods of intervention, we defined that capsid transport from virally-modified ER membranes to LDs requires a functional GBF1 protein. The process was found to be COPI dependent, while it was independent of both COPII components and Golgi integrity, supporting a non-canonical function of GBF1/ARF/COPI transport system.
INTRODUCTION
Dengue virus (DENV) is considered one of the most important arthropod-borne human pathogen worldwide. This virus is a member of the Flavivirus genus in the Flaviviridae family together with other important pathogens such as yellow fever virus (YFV), West Nile virus (WNV), Saint Luis encephalitis virus (SLEV), and Japanese encephalitis virus (JEV). DENV alone causes about 390 million infections per year (1). Despite this large public health problem and the urgent need to control DENV infections, vaccines and specific antivirals are still unavailable.
DENV, like other plus strand RNA viruses, induces a profound rearrangement of cellular membranes to provide platforms for viral replication (2–4). DENV RNA synthesis occurs in membranous structures called vesicle packets that function as viral genome factories, which are in close association to viral particle morphogenesis in the ER membrane (4). During this process, the viral capsid protein (C) is responsible for recruiting the viral genome during viral encapsidation, forming a nucleocapsid that buds into the ER lumen, acquiring membranes and the structural proteins E and prM (5, 6). The new viral particle travels through the secretory pathway to be released by exocytosis.
The viral C is a small, highly basic protein that forms homodimers in solution (7, 8). We have previously shown that C progressively accumulates around cellular lipid droplets (LDs) during DENV infection (9). Atomic force microscopy has been recently used to define biochemical properties of this C-LD interaction (10). LDs are dynamic organelles involved in lipid metabolism that regulate storage and turnover of neutral lipids. They contain mostly triacylglycerols and sterol esters surrounded by a phospholipid monolayer, decorated with proteins (11–13). Previous studies using leukocytes have demonstrated an increased amount of LDs per cell in samples from patients with dengue hemorrhagic fever when compared with that from healthy volunteers (14). Similar observations were reported in DENV infected human macrophages and in different infected cell lines, such as A549, Hep-G2 and BHK (9, 14–16). Interestingly, it has been shown that DENV infection triggers the redistribution of the rate-limiting enzyme in lipid biosynthesis, the fatty acid synthase (FASN) to the ER. It has been demonstrated that binding of FASN to the viral protein NS3 increases cellular fatty acid synthesis (17); and that pharmacological inhibition of FASN activity blocks induction of LDs during DENV infection (9). A relationship between autophagy-mediated lipid droplet degradation and DENV infection has been also observed (18). It has been suggested that lipid degradation was necessary during DENV RNA synthesis. However, inhibition of autophagy appears to affect different stages of viral infection. In this regard, destabilization of beclin-Vps34-Atg14, which inhibits autophagy, was shown to have a modest effect on viral RNA synthesis but a profound effect on particle infectivity, due to defects on peptide pr release from prM during particle maturation (19). Thus, it is likely that lipid biosynthesis and degradation are both necessary during different stages of the viral life cycle. More recently, the Ras-related protein in brain (Rab), GTPase Rab18, which participates in lipid exchange between ER and LD compartments, was shown to be necessary for efficient DENV replication and to have a positive role in LD induction (16). Even though different lines of evidence link DENV infection to LD metabolism, the functional significance and the mechanisms by which the virus manipulates LDs remain obscure. We have previously reported that C accumulation on LDs is necessary for efficient viral particle production; but the role of LDs in this process is still unknown. Budding viral particles in the ER have been observed next to the pores of vesicle packets, where the viral genome is replicated, suggesting that LDs are not the place of genome recruitment during encapsidation (4). It is possible that accumulation of C on LDs modulates the host lipid metabolism required for viral particle formation or serves as storage of the viral protein in early stages of viral replication (9).
To further examine the function of C on LDs and explore new tools for antiviral intervention, we investigated the process by which C is transported from the ER membrane to LDs during DENV infection. How LDs receive or deliver proteins is still an open question. In recent years, more than 200 cellular proteins have been reported that associate to LDs; including lipid metabolism enzymes, Rab GTPases, SNARE proteins and coatomer components (13). The most prominent lipid droplet associated proteins are the PAT proteins (Perilipin, Adipofilin/ADRP, and the Tail interacting protein/TIP47) (20). Interestingly, a function of components of the vesicle trafficking systems, Coat Protein II and I (COPII and COPI), has been reported for the transport of different cellular proteins to LDs (21–23). Normally COPII vesicles mediate transport of proteins and lipids from the ER to Golgi, while COPI mainly participate in the retrieval of proteins from the Golgi back to the ER. How COP components mediate delivery of proteins to LDs is unclear, but might involve novel functions of this class of proteins. Here, we investigated the involvement of these host factors in the accumulation of C on LDs. Because DENV requires an active vesicle transport system for viral maturation and egress, we restricted our studies to evaluate the C transport in a single viral replication cycle. Using different methods of intervention, we defined that C association to LDs requires a functional GBF1 protein. The process was found to be COPI dependent, COPII independent and did not involve the Golgi complex, supporting a non-canonical function of this cellular transport system. We believe that dissecting the host machinery used during DENV infection will provide novel ideas for employing host targets for antiviral approaches.
RESULTS
Delivery of capsid to LDs is BFA sensitive
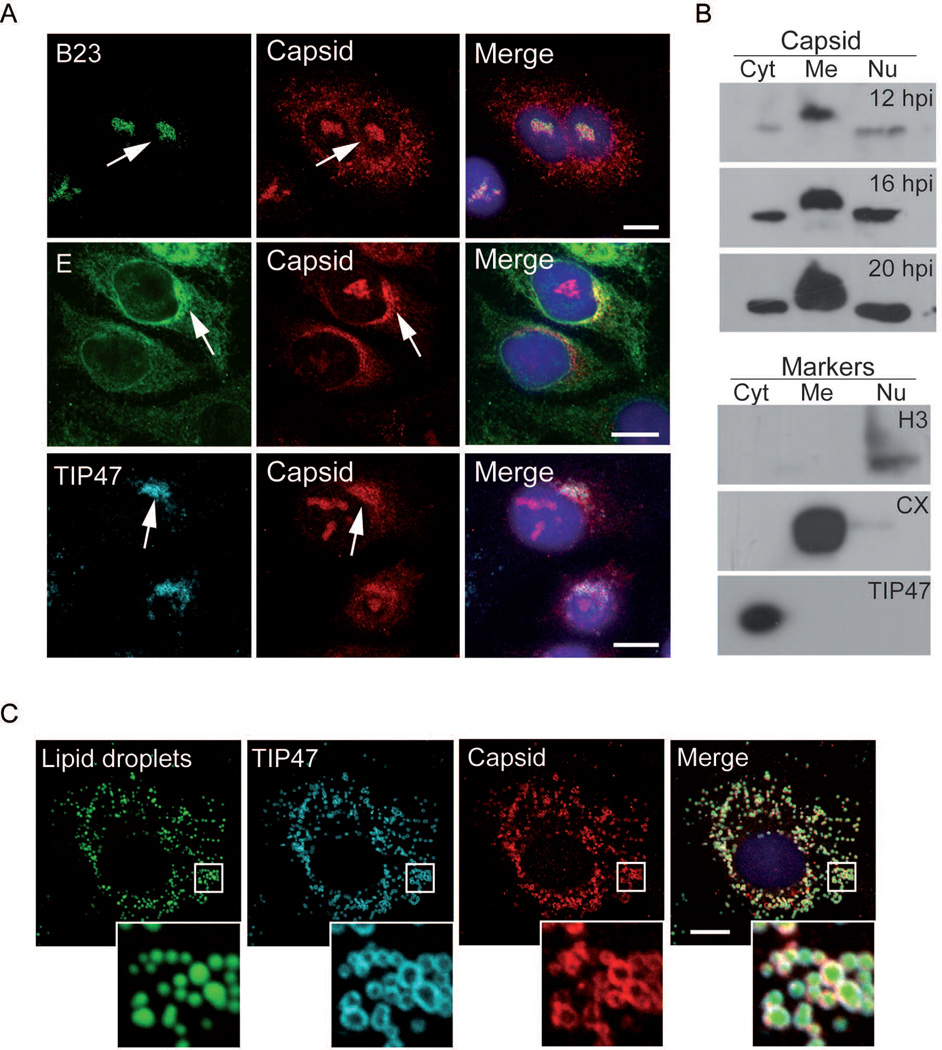
It has been previously reported that the DENV C protein associates to different cellular compartments during viral infection (9, 24, 25). Using immunofluorescence assays and subcellular fractionation of DENV infected cells, we observed C in the nucleus and cytoplasm (Fig.1A and 1B). During viral infection, C is translated as a polyprotein associated to ER membranes and it is proteolytically processed to release the mature protein. Mature C was detected by western blot at 12, 16 and 20 h post infection in membrane, cytoplasmic and nuclear fractions (Fig. 1B). The cytoplasmic fraction contained TIP47, indicating that the LDs were contained in this fraction. Immunofluorescence studies showed that the cytoplasmic C partially co-localized with the host protein TIP47 and the viral E protein, which associate to LDs and ER, respectively (Fig. 1A). In the nucleus, C co-localized with the nucleolar B23 protein (Fig. 1A). Visualization of proteins on LDs is limited when methanol is used as fixation method due to extraction of lipids (26). Because paraformaldehyde (PFA) preserves the integrity of LDs, this method was elected for detection of proteins on LDs (Fig. 1C). We conclude that in DENV infected cells, C accumulates in nucleolus, LDs and ER membranes.
Figure 1.
Subcellular localization of DENV C in A549 infected cells. A Immunofluorescence of C in infected cells using methanol as fixation method. A549 cells were infected with DENV and localization of C was analyzed in respect to different markers using antibodies anti-B23 (nucleolus), anti-DENV envelope E protein and anti-TIP47 (lipid droplets). Arrows indicate co-localization of C with markers for each case. Bar, 10 µm. B. Western blots with lysates of infected cells fractionated into cytoplasmic (Cyt), membranes (Me), and nuclear (Nu) fractions. The viral protein was detected with anti-C antibodies at 12, 16, and 20 hours post-infection, as indicated. The samples were obtained at 12 hours post-infection and immunobloted with anti-histone H3, anti-calnexin and anti-TIP47, as markers for each fraction. C. Immunofluorescence of C in infected cells using paraformaldehyde as fixation method allows lipid droplet visualization. Lipid droplets were stained with BODIPY and localization of viral C and the host TIP47 was determined with specific antibodies as indicated in each case. Bar, 10 µm.
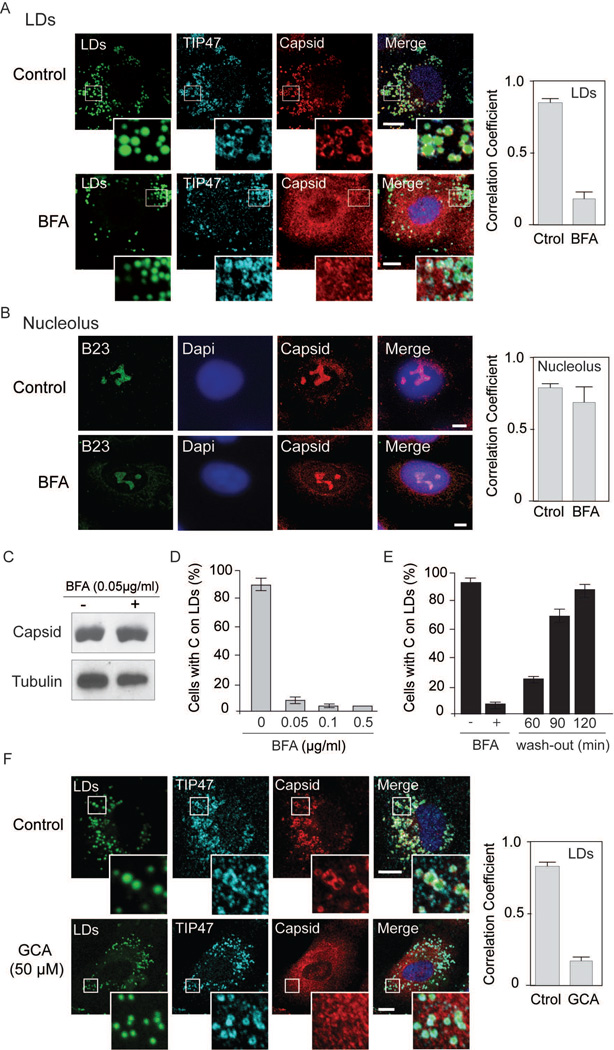
The mechanism by which host proteins are transported to LDs is still not fully understood. It is possible that C directly associates to nascent droplets after proteolytic processing or, alternatively, uses a specific transport system. In this regard, recent reports have implicated components of COPII and COPI complexes in the transport of some cellular proteins to LDs (21, 23). It has been shown that COPI and COPII are necessary for ADRP and ATGL accumulation on LDs, and this process was sensitive to brefeldin A (BFA), a GTP exchange factor inhibitor that blocks COPI activity. In contrast, other proteins such as TIP47, were found to be independent of COPI and COPII systems (21, 23). Thus, we examined the requirement of host proteins for C accumulation on LDs in DENV infected cells. Because BFA was reported to inhibit ADRP and ATGL accumulation on LDs, we first evaluated C distribution in cells treated or not with this drug. It is important to clarify that DENV particle morphogenesis and release requires a functional secretory pathway (27); and it has been demonstrated that BFA inhibits viral particle secretion (28). Therefore, to avoid new rounds of infections that would be sensitive to BFA, we examined the impact of the drug on C subcellular distribution in a single round of infection. To this end, cells were treated with different concentrations of BFA at 8h post-infection and pre-treated with oleate to promote formation of LDs. The correlation coefficient of C with a LD marker was drastically reduced in the presence of BFA, while no effect on C accumulation in nucleolus was observed (Fig.2A and 2B). The total amount of mature C detected in cells treated or not with BFA was similar as determined by western blot (Fig. 2C), indicating that BFA did not affect viral translation or RNA amplification in the conditions used. The effect of different concentrations of drug indicated that concentrations as low as 0.05 µg/ml greatly reduced C on LDs (Fig. 2D). Moreover, the effect was found to be reversible. Localization of C on LDs was recovered 2h after incubation of cells in media without BFA (Fig. 2E and S1). The results indicate that BFA did not significantly change the total amount of C or the nucleolar accumulation of the viral protein in infected cells; however, it greatly reduced protein association to LDs.
Figure 2.
Brefeldin A blocks C localization on LDs in DENV infected cells. A. Immunofluorescence showing the effect of BFA on C associated to LDs. Specific antibodies against C and TIP47 were used in infected cells as indicated on the top. LDs were stained with BODIPY. Representative confocal images are shown for each case in the absence or presence of 0.05 µg/ml of BFA. Bar, 5 µm. Quantification of the percentage of C that co-localizes with TIP47 (mean +/− standard deviation) was performed using ImajeJ (JACoP plug-in), Manders’ coefficient is shown. B. Immunofluorescence showing the effect of BFA on C accumulation in nucleolus using methanol as fixation method. Representative confocal images using antibodies against C and B23 (marker of nucleolus) are shown in the absence or presence of 0.05 µg/ml of BFA. Bar, 5 µm .Quantification of the percentage of C that co-localizes with B23 (mean +/− standard deviation) was performed using ImageJ (JACoP plug-in), Manders’ coefficient is shown. C. Total amount of C in cells treated or not with BFA. Cell extracts were used for immunoblots with specific anti-C antibodies. D. Effect of different concentrations of BFA on C association to LDs. Confluent A549 infected cells were treated at 8 h post-infection with 0, 0.05, 0.1 or 0.5 µg/ml of BFA. At 24 h post-infection cells were immunostained with anti-TIP47 and anti-C antibodies and the percentage of cells with C on LDs was determined. About 200 cells were scored; bars indicate the standard error of the mean. E. Confluent A549 infected cells were treated 8 h with BFA and then the BFA was removed. At 60, 90 and 120 min after washing, cells were fixed and stained. The percentage of cells with C on LDs is shown for each time. About 200 cells were scored in duplicates. Bars indicate the standard error of the mean. F. Golgicide A impairs C localization on LDs. Immunofluorescence showing the effect of 50 µM of Golgicide A on C association to LDs in A549 cells. Representative confocal images of cells treated or not with the drug (Control or GCA) are shown stained with BODIPY for LDs, or labeled with anti-TIP47 and anti-Capsid as indicated for each case. Bar, 10 µm. Right panel, quantification of the percentage of capsid that co-localizes with TIP47 (mean +/− standard deviation), Manders’ coefficient is shown.
BFA inhibits several GTP exchange factors (GBF1, BIG1 and BIG2). Because golgicide A (GCA) has been shown to be more specific for GBF1 inhibition, which is the GEF involved in the Arf1-COPI pathway (29), the effect of GCA on C association to LDs was also examined. GCA greatly reduced C accumulation on LDs (Fig. 2F), suggesting that transport of C to LDs is sensitive to GBF1 inhibition.
In these studies we could not evaluate the impact of the lack of C association to LDs by BFA on viral particle production because the drug inhibits viral particle secretion independently of C (15, 28). Accordingly, BFA greatly decreased viral titers in our experimental conditions (Fig. S2).
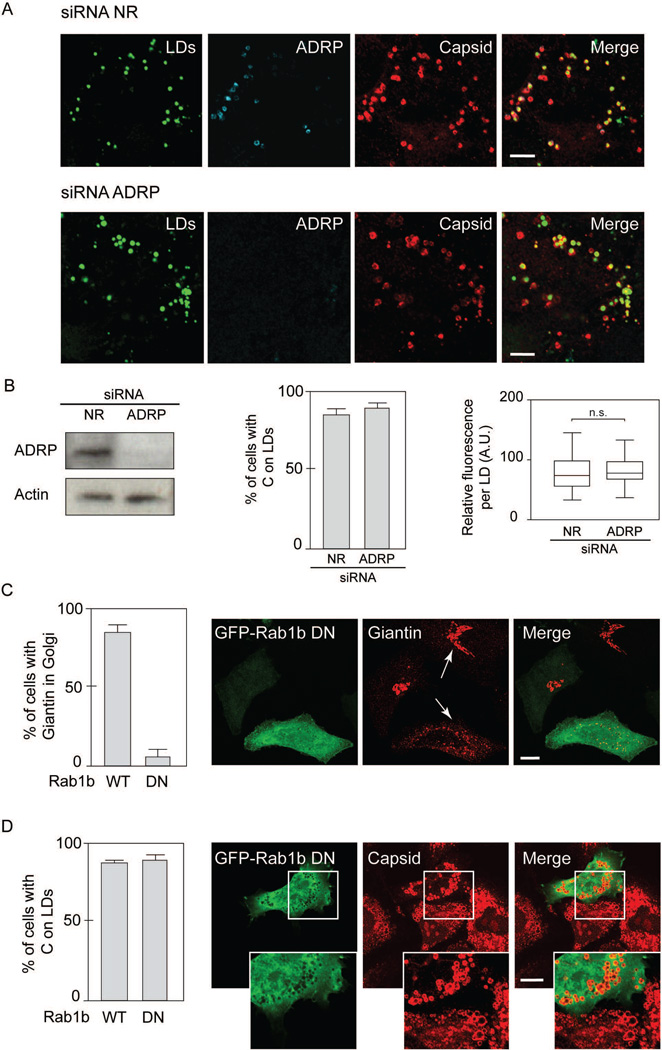
Because BFA impairs ADRP and ATGL accumulation on LDs, it is possible that alterations of LD composition could be responsible for C miss-localization in the presence of the drug. To evaluate this possibility, we analyzed C localization in infected cells in conditions in which ADRP was silenced or not. Cells were transfected with siRNA specific for ADRP or a non-related siRNA control, and then infected with DENV. The samples were then used to examine C localization by immunofluorescence (Fig.3A and 3B). Knockdown of ADRP was confirmed by western blots using specific antibodies (Fig. 3B). The results indicate that in conditions in which ADRP was undetectable on LDs, the viral C protein remain associated to this organelle (Fig.3A and 3B), suggesting that C accumulation on LDs is independent of ADRP.
Figure 3.
Neither ADRP nor Golgi integrity are required for C association to LDs. A. Immunofluorescence showing the effect of ADRP depletion on C distribution in DENV infected cells. A549 cells were transfected with a non-related siRNA directed to Renilla luciferase (siRNA NR) or siRNAs against ADRP, as indicated on the top of each panel. Cells were infected with DENV-2 and immune-stained with anti-ADRP and anti-C. Representative confocal images are shown. Bar, 5 µm. B. Immunoblot analysis showing ADRP depletion. Extracts of A549 cells transfected with siRNA ADRP were immunoblotted with specific antibodies. The same membranes were immunoblotted with anti-actin as a loading control. In the middle, quantification of the percentage of cells with C on LDs after interference with siRNA for ADRP, or a nonrelated siRNA control (NR). In each case, 100 cells were scored for the presence of C on LDs in three independent experiments. The bars indicate the standard error of the mean. On the right, the intensity staining obtained with anti-capsid antibody was measured in each lipid droplet using ImageJ and displayed as box-plots with 5 to 95% confidence intervals, n=250. Two-tailed unpaired student t test were performed to determine statistical relevance. Identical acquisition exposure-time conditions were used to capture images. C. Effect of Rab1b dominant negative on giantin distribution. On the left, quantification of cells with giantin in Golgi, expressing either the WT or the dominant negative (DN) form of Rab1b (N121I), as indicated. On the right, Representative confocal images showing the effect of expressing a DN form of GFP-Rab1b (N121I) on the cellular distribution of giantin. Arrows indicate cells expressing or not GFP-Rab1b (N121I), labeled with anti-giantin antibodies. Bar, 10 µm. D. On the left, quantification of cells with C on LDs, expressing either the WT or the DN form of GFP-Rab1b, as indicated. On the right, representative confocal images showing the effect of expressing the DN form of GFP-Rab1b (N121I) on the cellular distribution of the viral C protein. Bar, 10 µm.
Disruption of the Golgi complex does not alter capsid localization on LDs
Inhibition of GBF1 and COPI functions produces alterations in the secretory pathway, including Golgi disassembly, phenotype observed in the presence of BFA (23, 30). To examine whether the BFA effect on C distribution was the result of a general alteration on the host vesicular transport system, a dominant negative form of Rab1b was used. This protein is a GTPase that plays a role in ER to Golgi transport (31). It has been previously demonstrated that a dominant negative form of Rab1b causes Golgi reabsorption into the ER, analogous to that described for BFA treatment (30). Thus, we expressed wild type or dominant negative Rab1b (N121I) protein fused to GFP in DENV infected cells and evaluated C association to LDs. As a control for function of the dominant negative protein, localization of giantin, a Golgi associated protein, was examined. N121I that Rab1b was mainly cytosolic and induced Golgi disruption as observed by giantin redistribution (Fig. 3C), but did not alter C association to LDs (Fig. 3D and Fig. S3). Cells expressing Rab1b-GFP displayed the typical ring-like structures of C on LDs (Fig. 3D). The results suggest components of the ER-Golgi machinery are involved in C localization on LDs, independently of ADRP, Rab1b activity and Golgi integrity but sensitive to BFA.
Capsid accumulation on LDs requires COPI components but is independent of COPII function
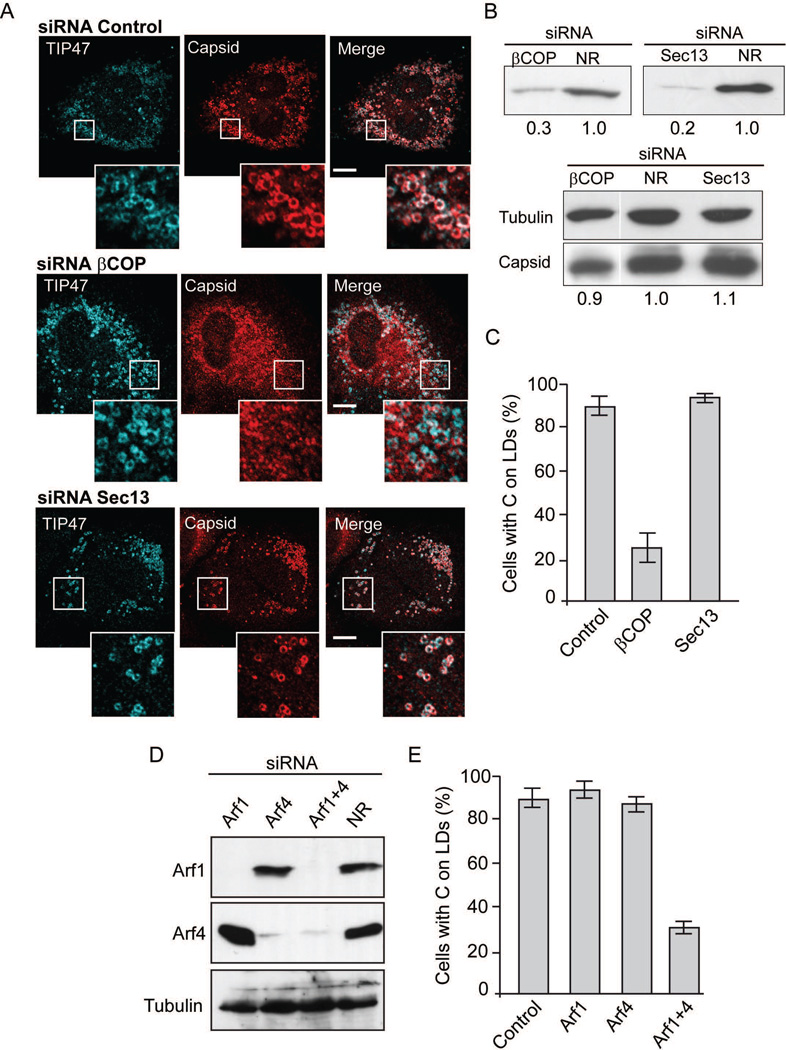
COPI and COPII systems have been implicated in the transport of host proteins to LDs (21–23, 32); therefore, we analysed the requirement of components of these systems on C transport to LDs during DENV infection. We used siRNAs directed to different subunits of COPI and COPII in A549 cells. Cells were transfected with siRNAs specific for βCOP subunit (COPI component), Sec13 (COPII component) or nonrelated control siRNAs, and then infected with DENV (Fig. 4A). Depletion of βCOP was about 70% and showed a drastic effect on C association to LDs without altering TIP47 localization (used as control). In contrast, depletion of Sec13 did not alter the amount of C nor its association to LDs, which remained co-localized with TIP47 (Fig. 4A, 4B and 4C). The results suggest that C transport to LDs in infected cells depends on COPI but is independent of COPII components.
Figure 4.
COPI but not COPII components are required for C localization on LDs. A. Immunofluorescence showing the effect of COPI depletion on C distribution in DENV infected cells. A549 cells were transfected with non-related siRNA directed to Renilla luciferase (siRNA NR), siRNAs against the β subunit of COPI (siRNA βCOP), or siRNAs against the COPII component Sec13 (siRNA Sec13), as indicated on the top of each panel. The cells were infected with DENV-2 and immune-stained with anti-TIP47 as control and anti-C. Representative confocal images are shown. Bar, 10 µm. B. Immunoblot analysis showing βCOP and Sec13 depletion. Extracts of A549 cells transfected with siRNA βCOP (on the left) and siRNA Sec13 (on the right), were immunoblotted with specific antibodies. The same membranes were immunoblotted with anti-tubulin as a loading control, and antibodies against the viral C protein, as indicated on the left of each panel. C. Quantification of the percentage of cells with C on LDs after interference with siRNA against βCOP, Sec13 or a nonrelated siRNA control (NR). In each case, 200 cells were scored for the presence of C on LDs in three independent experiments. The bars indicate the standard error of the mean. D. Redundancy of Arf1 and Arf4 for C association to LDs in DENV infected cells. Immunoblot showing cells treated with siRNA directed to Arf1, Arf4, Arf1+Arf4, or control (NR). Antibodies anti-Arf1, anti-Arf4 or anti-tubulin were used as indicated on each case. E. Effect of Arfs depletion on C accumulation on LDs. Quantification of the percentage of cells with C on LDs in cells interfered with a NR siRNA or siRNAs against Arf1, Arf4 or both. In each case, 200 cells were scored for the presence of C on LDs in three independent experiments. The bars indicate standard error of the mean.
The possible involvement of COPI on C association to LDs lead us to analyze a function of Arf1 in this process. To study this possibility, we evaluated the depletion of Arf1 on C distribution in DENV infections. Depletion of Arf1 in infected A549 cells was almost complete, while C and TIP47 remained associated to LDs (Fig. 4D, 4E and S4). This result was unexpected because Arf1 activity is proposed to be necessary for COPI recruitment. It is possible that COPI is directly recruited by GBF1, without Arf1 requirement (33). Alternatively, other Arf isoforms could participate in this process. Kahn and colleagues have previously reported a redundancy of Arf functions in the cell (34). Therefore, we analyzed depletion of different Arfs and found that simultaneous knock down of Arf1 and Arf4 largely reduced C transport to LDs, while cells lacking only Arf4 did not affect C subcellular distribution (Fig. 4D, 4E and S4). It has been shown previously that the double depletion of Arf1 and Arf4 inhibits COPI association with membranes (34). The mechanism is not known, but it has been shown that both Arfs can mediate COPI formation in vitro (35). These studies support the idea that localization of C on LDs requires recruitment of COPI by either Arf1 or Arf4.
BFA resistant GBF1 restores capsid delivery to LDs in DENV infected cells
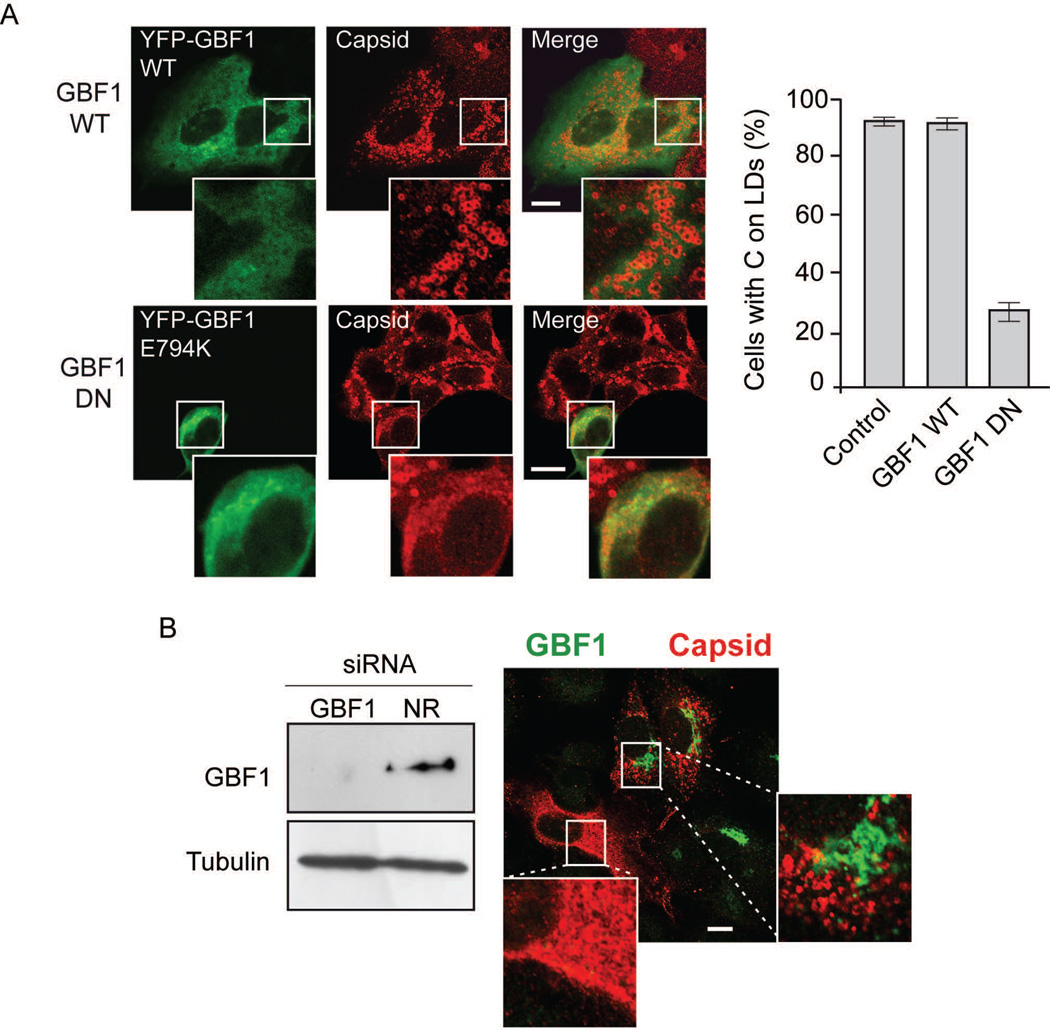
To further examine the role of GBF1 on C association to LDs, different methods were used. First, the effect of expressing a WT or a dominant negative form of GBF1 (E794K) was evaluated. A549 cells were infected with DENV and then transfected with expression constructs encoding GBF1 fused to YFP (Fig. 5A). While expression of the WT protein did not affect C association to LD, the E794K inactive protein reduced about 70% the amount of cells with C on LDs (Fig. 5A), suggesting the requirement of an active GBF1 protein. As control TIP47 was stained in cells expressing the different forms of GBF1 (Fig. S5).
Figure 5.
GBF1 function is necessary for C accumulation on LDs. A. On the left, representative confocal images showing the effect of expressing YFP-GBF1 WT or E794K (dominant negative, DN) on the cellular distribution of C. A549 cells were infected with DENV-2 and 4h later transfected with the GBF1 expressing plasmids. Immunofluorescence was performed 24 h post-infection using anti-capsid antibodies. Bar, 10 µm. On the right, the amount of YFP positive cells with C on LDs were scored and shown as percentage of cells with C on LDs. About 100 cells were scored in each case in duplicates. Errors bars indicate the standard error of the mean. B. Effect of GBF1 depletion on C accumulation on LDs. On the left, immunoblot showing GBF1 depletion. Extracts of A549 cells transfected with siRNAs for GBF1 or NR siRNAs were immunoblotted with anti-GBF1 antibodies. The same membrane was immunoblotted with anti-tubulin as a loading control, as indicated. On the left, representative confocal image showing A549 cells depleted or not of GBF1 using anti-GBF1 antibodies (green) and indicating the distribution of the viral C in infected cells by anti-C antibodies (red). Bar, 10 µm.
To confirm the requirement of GBF1, the effect of RNAi mediated depletion of this protein on C localization was also tested. The efficiency of GBF1 depletion was observed by western blots (Fig. 5B). Immunofluorescence analysis using antibodies to detect both GBF1 and C indicated that infected cells lacking GBF1 (without green signal in Fig. 5B) also lacked C localization on LDs, while cells expressing detectable levels of endogenous GBF1 showed a clear association of C to LDs.
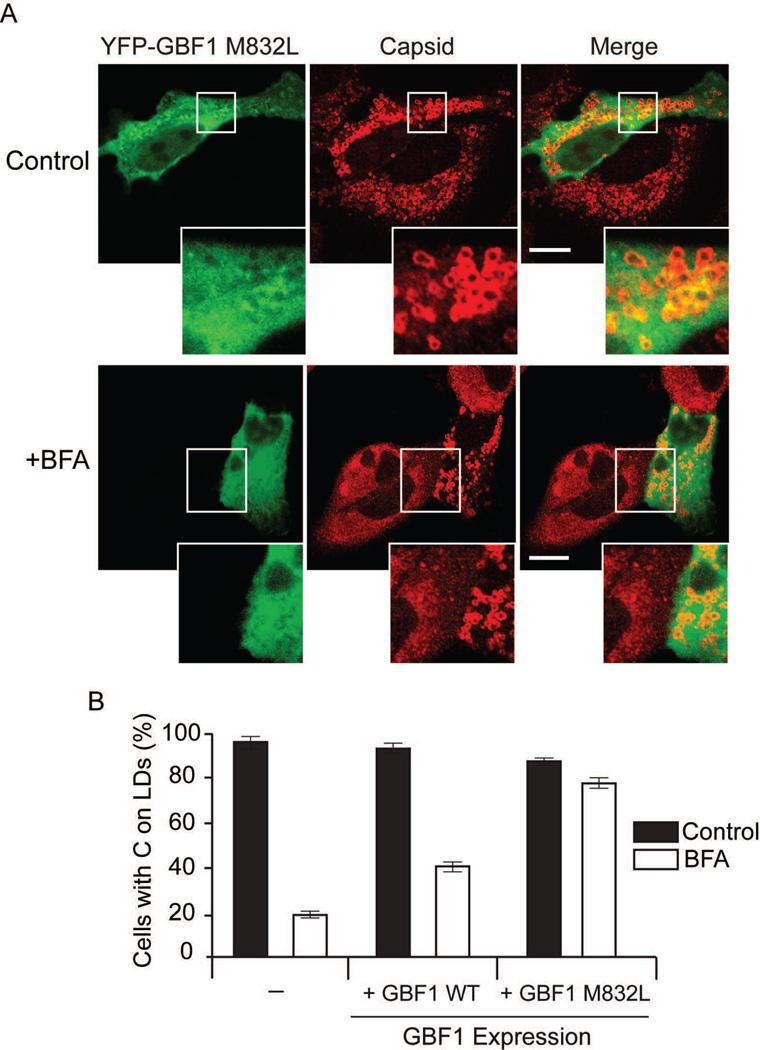
Taken together, it is likely that BFA inhibition of GBF1 is responsible for the phenotype observed. Thus, we examined whether a GBF1 mutant that is resistant to BFA was able to reverse the effect of the drug. Cells were infected with DENV, and then transfected with plasmids expressing YFP alone, GBF1-WT or BFA-resistant GBF1 (M832L) fused to YFP in the presence or absence of BFA (Fig. 6A). In the absence of BFA, between 90 to 95% of the cells that expressed YFP showed C on LDs, whether they were transfected with the control plasmid or either GBF1 constructs, suggesting that GBF1 overexpression had no effect on C localization (Fig. 6B). In the presence of BFA, the amount of cells expressing YFP control with C on LDs was reduced to 18%, whereas it was about 40% and 80% in WT or M832L GBF1 expressing cells (Fig. 6B). It was evident that cells expressing or not the BFA resistant form of GBF1 showed a remarkable difference on C distribution in the presence of drug (Fig. 6A). The results indicate that a BFA resistant form of GBF1 was sufficient to revert the effect of the drug, confirming the requirement of an active GBF1 for C subcellular distribution during DENV infection.
Figure 6.
BFA-resistant GBF1 reverts the effect of BFA on C localization. A. Representative confocal images showing the effect of expressing YFP-GBF1- M832L (BFA resistant) on the subcellular distribution of C in the presence and absence of BFA. Bar, 10 µm. A549 cells were infected with DENV-2 and 4h later transfected with the GBF1 expressing plasmids. Immunofluorescence was performed 24 h post-infection using anti-C antibodies. B. Percentage of C on LDs of cells expressing YFP control, YFP-GBF1 WT, or YFP-GBF1- M832L, in BFA-treated or non-treated cells, as indicated for each case. About 100 cells expressing YFP were scored for the presence of C on LDs in two independent experiments. Bars indicate standard error of the mean.
DISCUSSION
Different lines of evidence support a link between DENV replication and LD metabolism. Thus, it is important to further understand why and how the virus manipulates LDs. Here, we examined the requirement of host factors for C accumulation around LDs during infection with the final goal of identifying possible antiviral targets. We defined that an active GBF1-Arf-COPI pathway is necessary for C transport from the ER to LDs in human infected cells. The process was found to be dependent on COPI but independent of Golgi integrity, supporting a non-canonical function of components of this vesicle transport system.
During DENV infection C distributes between different cellular compartments: nucleolus, LDs and ER membranes. This distribution is observed early after infection, suggesting that it is not a consequence of cell damage during viral replication (Fig. 1). In the viral polyprotein, C is followed by an anchor peptide that directs prM-E to the ER lumen. It has been shown that the anchor peptide is not necessary for C accumulation on LDs (9). Release of C by the viral NS2B3 cleaves the anchor peptide rendering a mature protein that remains attached to the cytoplasmic side of the ER membrane (36), and then it is transported to LDs and nucleus. Why the protein accumulates in these cellular compartments is still unknown. We have previously shown that C could be toxic for viral RNA synthesis due to its high affinity for nucleic acids. In this regard, it is possible that C is sequestered in different organelles early during infection to avoid premature interaction with the viral RNA. However, more studies are necessary to define mechanistic aspects of C functions in the nucleus and LDs.
Previous studies using genome wide RNAi screens in drosophila, have linked COPI components with LD metabolism and composition (22, 23). It has been shown that COPI regulate lipid homeostasis, likely by promoting transport of enzymes, like ATGL, to the surface of LDs (23). Here, we used different methods to evaluate the involvement of components of this trafficking system on C transport. Because depletion or inhibition of these components could alter the composition and abundance of LDs, the experiments were performed in cells pretreated with oleate. Our results support the idea that an active GBF1 participates in the transport of C to LDs (Fig. 5 and 6). It is possible that the cellular protein accumulates on viral induced membranes and activates Arf1 and/or Arf4 to recruit COPI (Fig. 4). Depletion of COPI subunits reduced transport of C to LDs, while depletion of Sec13 of COPII did not alter C distribution in DENV infected cells (Fig. 4). Therefore, it is unlikely that C is first transported to a cis-Golgi compartment before it is directed to LDs. In this regard, a dominant negative form of Rab1b that induces Golgi disruption did not affect C subcellular distribution. We also observed that BFA greatly reduced C accumulation on LDs while it did not alter its transport to the nucleolus (Fig. 2). The effect of BFA was reversible by drug removal and its specificity on GBF1 was demonstrated by the recovery of C on LDs in the presence of a BFA resistant form of GBF1 (Fig 6). We propose that COPI/Arf/GBF1 system mediate transport of C from the place where it is synthesized on the ER membranes to LDs, independently of Golgi integrity.
The best described function of COPI vesicles is to mediate retrograde vesicle transport between Golgi cisternaes and from the Golgi to the ER, while COPII mediates anterograde transport mainly from the ER; however, it is evident that the canonical vesicular transport of cargo, as occurs in the secretory pathway, is not possible for protein transport to LDs. The monolayer of phospholipids that constitute the LD surface cannot fuse to a typical bilayer of a coated vesicle. Different possibilities that explain the transport of proteins have been suggested. For instance, lateral movement of proteins from ERES/ERGIC sites to LDs, or loading of nascent droplets with specific proteins (21, 37). A very recent report showed that cells lacking Arf1 and COPI have an increased amount of phospholipids on LDs, resulting in decreased LD surface tension and impairment to form bridges with the ER (38). Based on these recent findings, modulation of ER-LD membrane bridges, without including vesicle formation, was proposed as a new mechanism for controlling protein transport from the ER to LDs (38). Our results are in agreement with this model, supporting the idea that C is transported from the ER to LDs using new functions of COPI.
An emerging theme in pathogen biology is the interaction of bacteria, viruses, and protozoan with LDs (39–41). The mechanism by which these pathogens co-opt LDs remains to be elucidated. In the case of viruses, a link between hepatitis C virus (HCV) replication and LDs has been extensively documented (42–44). During HCV infection, the core protein, as well as other viral proteins, accumulates on the surface of LDs (45–47). This localization of core has been proposed to play an important role in viral encapsidation (42, 44, 46–51). Interestingly, a recent study analyzing the trafficking of core during HCV infection suggested that BFA treatment increase the amount of the protein on LDs (52). Thus, DENV C and HCV core appear to use distinct mechanisms for LD association, one sensitive and the other one resistant to BFA, respectively. Taking into account previous studies, there are at least two different pathways involved in delivering proteins to LDs, one dependent on COPI (and COPII in some cases), sensitive to BFA (such as ADRP, ATGL and DENV C); and the other one independent on these trafficking systems and resistant to BFA (such as TIP47 and possibly HCV core).
Different viruses subvert the GBF1/Arf/COPI system during infection. In several cases, components of this pathway are involved in non-canonical functions such as the case of GBF1 on picornavirus replication (53–56) and GBF1-Arf1 in Ebola virion formation (57). Here, we found that DENV uses this host trafficking system for C accumulation on LDs. Because the role of GBF1/Arf/COPI on LD protein transport is just emerging as a novel function of these proteins, further studies using different viruses that co-op LDs will help to uncover details of this cellular process and hopefully provide new ideas for antiviral approaches.
MATERIALS AND METHODS
Cells and viruses
A549 human epithelial lung cells were cultured in Dulbecco’s modified Eagle’s medium F-12 (Ham) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin. Stocks of DENV-2 16681 were prepared in mosquito cells (C6/36) and used to infect A549, as previously described (58).
Antibodies and immunofluorescence assays
For immunofluorescence and Western blot assays the following antibodies were used: mouse monoclonal anti-B23 (Abcam), Guinea pig polyclonal anti-TIP47 (Fitzgerald), Guinea pig polyclonal anti-ADRP (Fitzgerald), rabbit polyclonal anti-Sec 13 (Abcam), rabbit polyclonal anti-beta COP (Abcam), mouse monoclonal anti-Tubulin (Santa Cruz), mouse monoclonal anti-Arf1 (Abcam), rabbit polyclonal anti-Arf4 (Proteintech), rabbit polyclonal anti-Giantin (Abcam), and mouse monoclonal anti-GBF1 (BD Transduction Laboratories). Rabbit polyclonal and mouse polyclonal anti-DENV capsid were obtained in our laboratory. For DENV envelope detection, mouse monoclonal anti-E antibody E18 was used (59). For lipid droplets staining, cells were incubated with BODIPY493/503 (Molecular Probes). Immunofluorescence assays were performed as previously described (9). Briefly, A549 cells were seeded into 24-well plates containing glass coverslip. Twenty four hours later, the cells were infected with DENV-2 using a multiplicity of infection of 10. At 24 hours post-infection coverslip were collected and the cells were fixed with paraformaldehyde 4%, sucrose 4%, in PBS pH 7.4 at room temperature for 20 minutes. Alternatively, cells were fixed with methanol for 20 minutes at −20°C. PFA fixed cells were then permeated with 0.1% Triton X-100 for 4 minutes at room temperature. Images were obtained with a Carl Zeiss LSM 5 Pascal confocal microscope. For quantification of LD associated capsid, about 200 cells were scored for each experiment. Mean and standard deviation were indicated in each case. Three independent experiments were performed in each condition. Box graphs and statistical analysis was performed using GraphPad Prism 5.
Subcellular fractionation
For subcellular fractionation Qproteome Cell Compartment kit (QIAGEN) was used according to manufacturer’s protocol. The kit allows sequential isolation of proteins associated with the cytosol, membranes and nucleus from cell lysates. Different extraction buffers were added sequentially to an infected cell pellet, and the respective fractions were isolated by centrifugation. Each fraction was evaluated by Western blot with specific markers: anti-TIP47, anti-calnexin, and anti-Histone H3 antibodies. Additionally, each fraction was evaluated with anti-capsid antibodies at different times post-infection.
Brefeldin A and Golgicide A treatments
Confluent A549 cells were infected with DENV-2 16681 using a multiplicity of infection of 10. After 1 hour, cells were washed with PBS and DMEM-F12 medium supplemented with 10% serum was added. At 8 hours post-infection fresh medium with 0, 0.05, 0.1 or 0,5 µg/ml of Brefeldin A (Sigma), or 50 µM of Golgicide A (Sigma) was added. The medium was supplemented with 100 µM of oleic acid (Sigma) complexed with BSA (60). For immunofluorescence the cells were fixed at 24 hours post-infection.
Plasmids and transfection
YFP-tagged GBF1 constructs were kindly provided by J. Bonifacino and C. Jackson (61). GFP-Rab1b N121I mutant has been previously described (30). For expression of heterologous proteins, confluent A549 cells were infected with DENV-2 and at 4 hours post-infection transfected with different plasmids using Lipofectamine 2000 (Invitrogen), according to manufacturer’s protocol. At 24 hours post-infection the cells were fixed with paraformaldehyde and immunofluorescences were performed.
RNA interference
RNA interference experiments were carried out using siGENOME ON-TARGET plus SMART pool siRNA oligonucleotides (Dharmacon RNA Technologies, Lafayette, CO, USA). RNAi were directed to GBF1 (8729), Sec13 (6396), β COP (1314 and 1315), Arf1 (375) or Arf4 (378) and ADRP (123). The control no-related siRNA used was directed to Renilla luciferase. At 24 hours after seeded in 24-well plates, A549 cells were transfected with the corresponding siRNA using Oligofectamine (Invitrogen). Briefly, 25 pmol of siRNA in 50 µl of Opti-MEM (Invitrogen) were mixed with 2 µl of Oligofectamine in 50 µl of Opti-MEM and incubated for 20 minutes. The mix was added to a 50% confluent A549 cells monolayer and incubated overnight. Then, the medium was replaced with complete DMEM F12 medium. After 48 hours of transfection, except in cases that indicate otherwise, cells were infected with DENV-2 and incubated for 24 hours.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. Juan Bonifacino, Catherine Jackson and Richard Khan for useful discussions, technical advice, and reagents. We also thank members of Gamarnik’s laboratory for critical discussions. This work was supported by NIH (NIAID) R01.AI095175-01 and PICT 2010-1141 to AVG. NGI, CA, and AVG are members of the Argentinean Council of Investigation (CONICET).
Footnotes
Fig. S1. BFA Washout. The effect of BFA on C association to LDs is reversible. A549 infected cells were treated with 0.1 µg/ml of BFA during 8 hs; then, the BFA was removed and cells incubated with fresh media. Immunofluorescence of C at 0, 60 and 120 min are shown.
Fig. S2. Effect of BFA on DENV infectious particle production. Supernatants of A549 cells infected with DENV and treated with different concentration of BFA 8 h post infection. The concentration of drug was 0, 0.05, 0.1 or 0.5 µg/ml, as indicated. The amount of infectious particles was quantified by plaque assay in BHK cells. The errors bars indicate the standard error of the mean of two independent experiments.
Fig. S3. Representative confocal images showing the effect of expressing the WT form of GFP-Rab1b on the cellular distribution of the viral C protein. A549 cells were infected with DENV-2 and 4h later transfected with the Rab1b WT expressing plasmid. Immunofluorescence was performed 24h post-infection using anti-capsid antibodies. Scale bar, 5 µm.
Fig. S4. Redundancy of Arf1 and Arf4 activity for capsid association to LDs in DENV infected cells. Effect of Arf1, Arf4 and Arf1+Arf4 depletion on C accumulation on LDs. Representative confocal images showing TIP47 and viral capsid association to LDs in A549 cells depleted of Arf1 (siRNA Arf1), Arf4 (siRNA Arf4) or Arf1+Arf4 (siRNA Arf1+Arf4). Bar, 5 µm.
Fig. S5. GBF1 expression. Representative confocal images showing the effect of expressing YFP-GBF1-WT, E794K (dominant negative) or M832L on the cellular distribution of TIP47. A549 cells were infected with DENV-2 and 4h later transfected with the GBF1 expressing plasmids. Immunofluorescence was performed 24h post-infection using anti-TIP47 antibodies. Bars, 5 µm.
Supplementary methods
Virus titration
Infectious DENV particles were quantified by plaque assays. Approximately 3×104 BHK-21 cells per well were seeded in 24-well plates and grown overnight. Virus samples were serially diluted and 100 µl were added to the cells and incubated for 1h. Then, the cells were covered with 1 ml of medium supplemented with 0.8% methyl cellulose (MEM alpha medium, 2% FBS, 100U/ml of penicillin, 100 µg/ml streptomycin, and 0.8 % methyl cellulose). At 7 days post-infection, cells were fixed and stained with crystal violet.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, et al. The global distribution and burden of dengue. Nature. 2013 doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchil PD, Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. The Journal of biological chemistry. 2003;278(27):24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220(1):232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 4.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell host & microbe. 2009;5(4):365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorenz IC, Kartenbeck J, Mezzacasa A, Allison SL, Heinz FX, Helenius A. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. Journal of virology. 2003;77(7):4370–4382. doi: 10.1128/JVI.77.7.4370-4382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SH, Syu WJ, Hu ST. Identification of the homotypic interaction domain of the core protein of dengue virus type 2. The Journal of general virology. 2004;85(Pt 8):2307–2314. doi: 10.1099/vir.0.80067-0. [DOI] [PubMed] [Google Scholar]
- 8.Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein. Journal of virology. 2003;77(12):7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS pathogens. 2009;5(10):e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho FA, Carneiro FA, Martins IC, Assuncao-Miranda I, Faustino AF, Pereira RM, Bozza PT, Castanho MA, Mohana-Borges R, Da Poian AT, Santos NC. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. Journal of virology. 2012;86(4):2096–2108. doi: 10.1128/JVI.06796-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nature reviews Molecular cell biology. 2006;7(5):373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 12.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annual review of biochemistry. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. Journal of lipid research. 2010;51(2):262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assuncao-Miranda I, Amaral FA, Bozza FA, Fagundes CT, Sousa LP, Souza DG, Pacheco P, Barbosa-Lima G, Gomes RN, Bozza PT, Da Poian AT, Teixeira MM, Bozza MT. Contribution of macrophage migration inhibitory factor to the pathogenesis of dengue virus infection. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(1):218–228. doi: 10.1096/fj.09-139469. [DOI] [PubMed] [Google Scholar]
- 15.Scaturro P, Trist IM, Paul D, Kumar A, Acosta EG, Byrd CM, Jordan R, Brancale A, Bartenschlager R. Characterization of the mode of action of a potent dengue virus capsid inhibitor. Journal of virology. 2014;88(19):11540–11555. doi: 10.1128/JVI.01745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WC, Lin RJ, Liao CL, Lin YL. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. Journal of virology. 2014;88(12):6793–6804. doi: 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(40):17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell host & microbe. 2010;8(5):422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Jr, Yuan J, Kirkegaard K. Inhibition of cellular autophagy deranges dengue virion maturation. Journal of virology. 2013;87(3):1312–1321. doi: 10.1128/JVI.02177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. Journal of lipid research. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. Coatomer-dependent protein delivery to lipid droplets. Journal of cell science. 2009;122(Pt 11):1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS biology. 2008;6(11):e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SH, Syu WJ, Huang KJ, Lei HY, Yao CW, King CC, Hu ST. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. The Journal of general virology. 2002;83(Pt 12):3093–3102. doi: 10.1099/0022-1317-83-12-3093. [DOI] [PubMed] [Google Scholar]
- 25.Sangiambut S, Keelapang P, Aaskov J, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. Multiple regions in dengue virus capsid protein contribute to nuclear localization during virus infection. The Journal of general virology. 2008;89(Pt 5):1254–1264. doi: 10.1099/vir.0.83264-0. [DOI] [PubMed] [Google Scholar]
- 26.DiDonato D, Brasaemle DL. Fixation methods for the study of lipid droplets by immunofluorescence microscopy. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2003;51(6):773–780. doi: 10.1177/002215540305100608. [DOI] [PubMed] [Google Scholar]
- 27.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 28.Kudelko M, Brault JB, Kwok K, Li MY, Pardigon N, Peiris JS, Bruzzone R, Despres P, Nal B, Wang PG. Class II ADP-ribosylation factors are required for efficient secretion of dengue viruses. The Journal of biological chemistry. 2012;287(1):767–777. doi: 10.1074/jbc.M111.270579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, Haslam DB. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nature chemical biology. 2009;5(3):157–165. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Molecular biology of the cell. 2003;14(5):2116–2127. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia IA, Martinez HE, Alvarez C. Rab1b regulates COPI and COPII dynamics in mammalian cells. Cellular logistics. 2011;1(4):159–163. doi: 10.4161/cl.1.4.18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zehmer JK, Bartz R, Bisel B, Liu P, Seemann J, Anderson RG. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. Journal of cell science. 2009;122(Pt 20):3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Y, Golinelli-Cohen MP, Smirnova E, Jackson CL. A COPI coat subunit interacts directly with an early-Golgi localized Arf exchange factor. EMBO reports. 2009;10(1):58–64. doi: 10.1038/embor.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Molecular biology of the cell. 2005;16(10):4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popoff V, Langer JD, Reckmann I, Hellwig A, Kahn RA, Brugger B, Wieland FT. Several ADP-ribosylation factor (Arf) isoforms support COPI vesicle formation. The Journal of biological chemistry. 2011;286(41):35634–35642. doi: 10.1074/jbc.M111.261800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markoff L, Falgout B, Chang A. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology. 1997;233(1):105–117. doi: 10.1006/viro.1997.8608. [DOI] [PubMed] [Google Scholar]
- 37.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9(4):914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilfling F, Thiam AR, Olarte MJ, Wang J, Beck R, Gould TJ, Allgeyer ES, Pincet F, Bewersdorf J, Farese RV, Jr, Walther TC. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 2014;3:e01607. doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herker E, Ott M. Emerging role of lipid droplets in host/pathogen interactions. The Journal of biological chemistry. 2012;287(4):2280–2287. doi: 10.1074/jbc.R111.300202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saka HA, Valdivia R. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annual review of cell and developmental biology. 2012;28:411–437. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- 41.Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends in endocrinology and metabolism: TEM. 2010;21(1):33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nature cell biology. 2007;9(9):1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 43.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS pathogens. 2008;4(3):e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. The Journal of biological chemistry. 2007;282(51):37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 45.Moradpour D, Englert C, Wakita T, Wands JR. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222(1):51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]
- 46.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(4):1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hope RG, McLauchlan J. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. The Journal of general virology. 2000;81(Pt 8):1913–1925. doi: 10.1099/0022-1317-81-8-1913. [DOI] [PubMed] [Google Scholar]
- 48.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. The EMBO journal. 2002;21(15):3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boulant S, Targett-Adams P, McLauchlan J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. The Journal of general virology. 2007;88(Pt 8):2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- 50.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9(8):1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 51.Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. The Journal of biological chemistry. 2006;281(31):22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- 52.Counihan NA, Rawlinson SM, Lindenbach BD. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS pathogens. 2011;7(10):e1002302. doi: 10.1371/journal.ppat.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belov GA, Kovtunovych G, Jackson CL, Ehrenfeld E. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cellular microbiology. 2010;12(10):1463–1479. doi: 10.1111/j.1462-5822.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. Journal of virology. 2007;81(2):558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wessels E, Duijsings D, Lanke KH, van Dooren SH, Jackson CL, Melchers WJ, van Kuppeveld FJ. Effects of picornavirus 3A Proteins on Protein Transport and GBF1-dependent COP-I recruitment. Journal of virology. 2006;80(23):11852–11860. doi: 10.1128/JVI.01225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141(5):799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamayoshi S, Neumann G, Kawaoka Y. Role of the GTPase Rab1b in ebolavirus particle formation. Journal of virology. 2010;84(9):4816–4820. doi: 10.1128/JVI.00010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mondotte JA, Lozach PY, Amara A, Gamarnik AV. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. Journal of virology. 2007;81(13):7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nature medicine. 2005;11(5):522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brasaemle DL, Wolins NE. Isolation of lipid droplets from cells by density gradient centrifugation. Curr Protoc Cell Biol. 2006;3(3):15. doi: 10.1002/0471143030.cb0315s29. [DOI] [PubMed] [Google Scholar]
- 61.Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Molecular biology of the cell. 2005;16(3):1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.