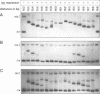
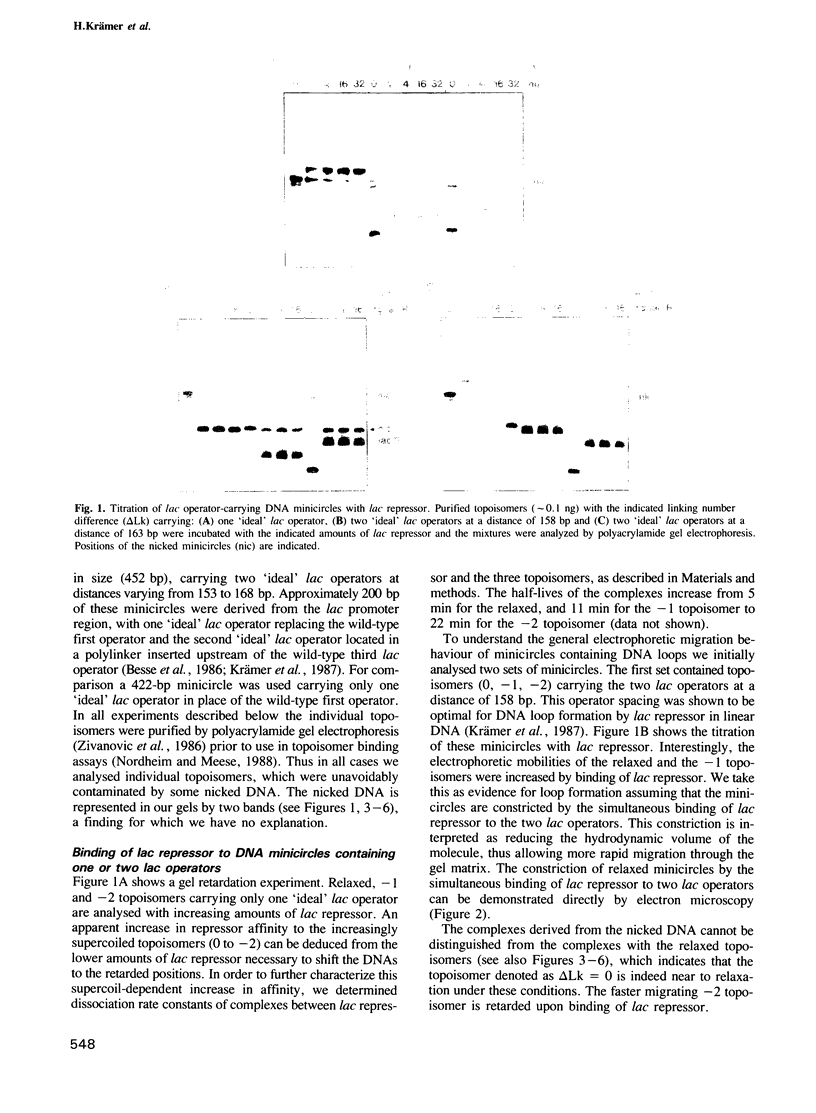
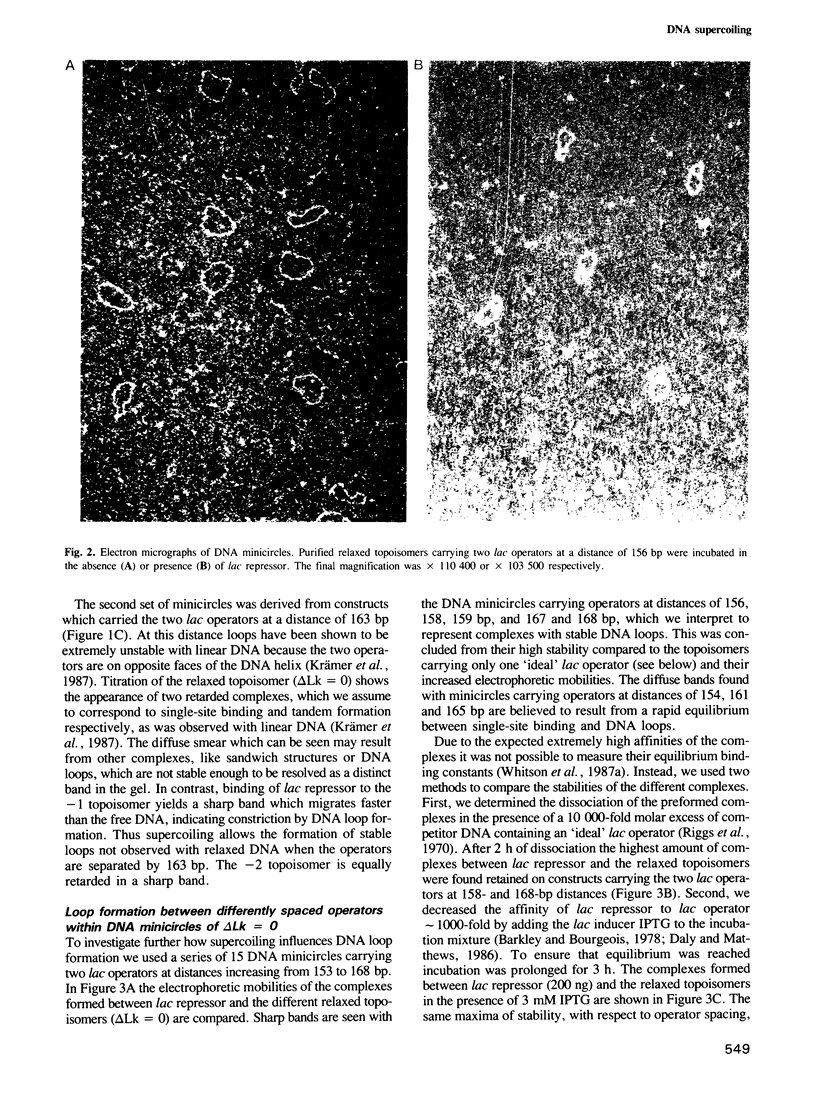
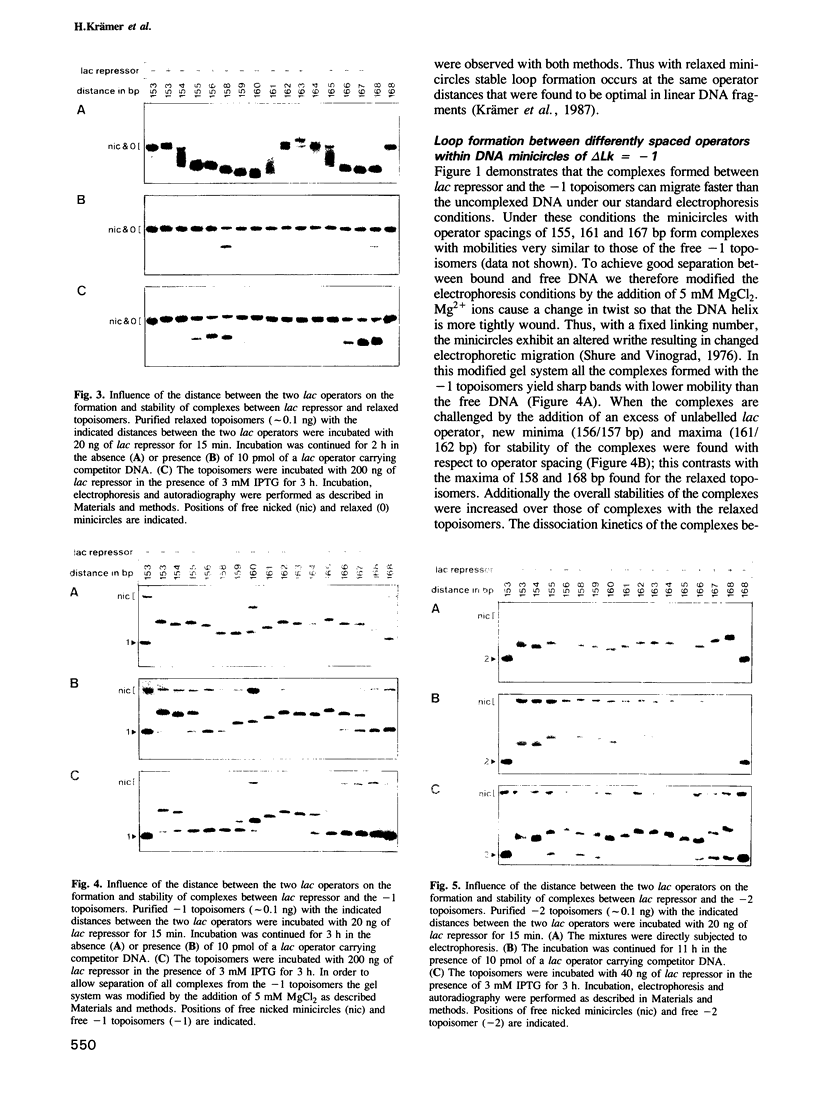
Abstract
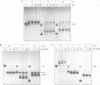
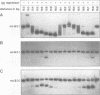
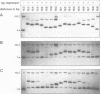
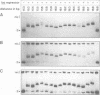
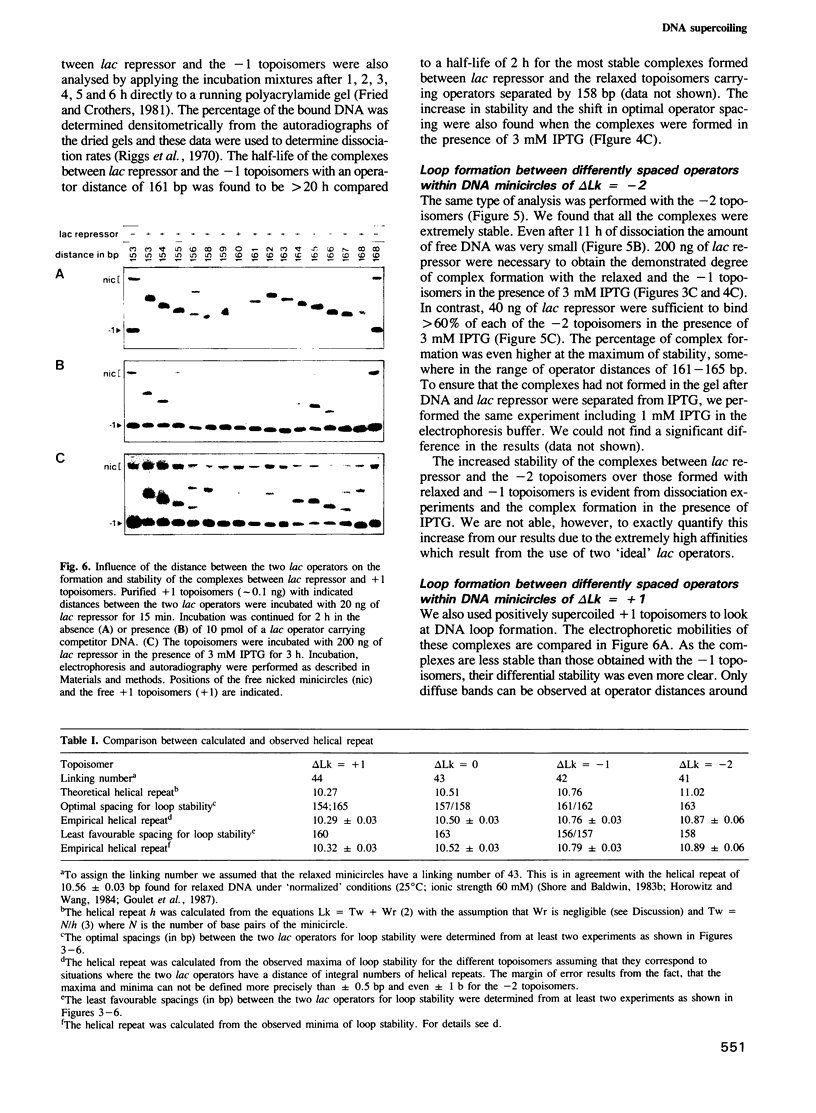
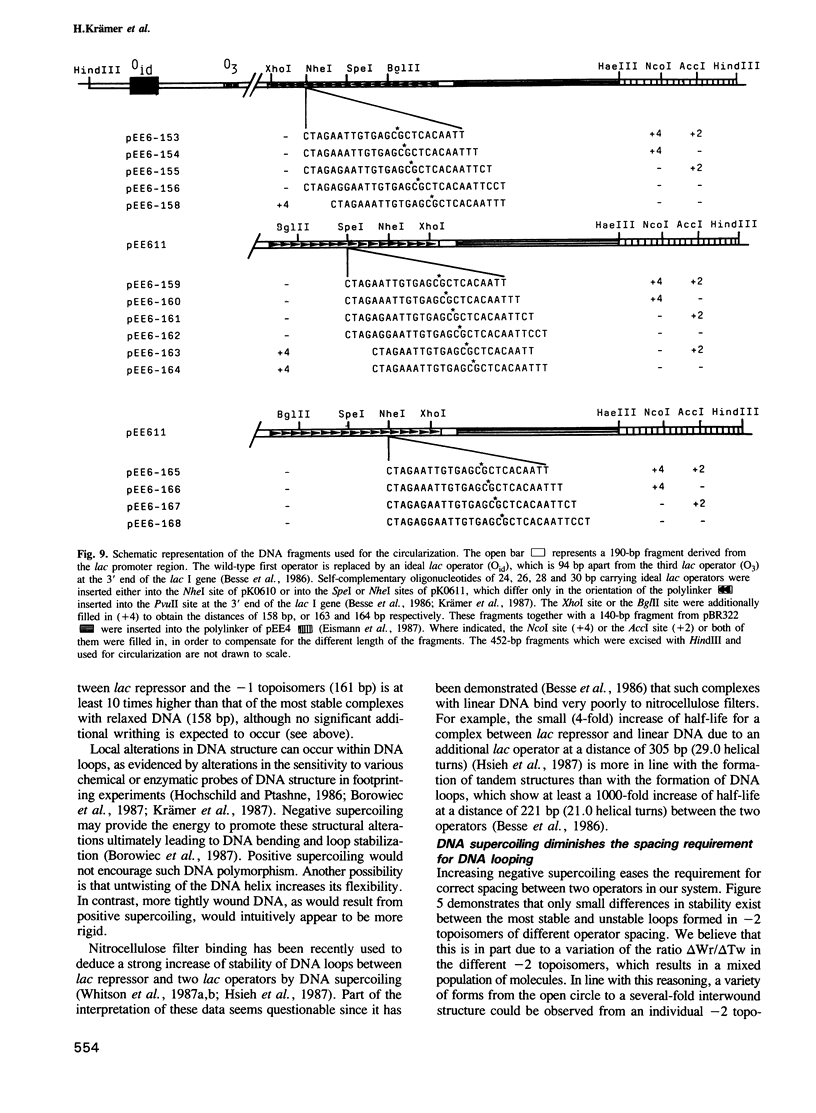
We have used a gel retardation assay to investigate the influence of DNA supercoiling on loop formation between lac repressor and two lac operators. A series of 15 DNA minicircles of identical size (452 bp) was constructed carrying two lac operators at distances ranging from 153 to 168 bp. Low positive or negative supercoiling (sigma = +/- 0.023) changed the spacing between the two lac operators required for the formation of the most stable loops. This reveals the presence of altered double helical repeats (ranging from 10.3 to 10.7 bp) in supercoiled DNA minicircles. At elevated negative supercoiling (sigma = -0.046) extremely stable loops were formed at all operator distances tested, with a slight spacing periodicity remaining. After relaxation of minicircle-repressor complexes with topoisomerase I one superhelical turn was found to be constrained in those minicircles which carry operators at distances corresponding to a non-integral number of helical turns. This indicates that DNA loop formation can define local DNA domains with altered topological properties of the DNA helix.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Besse M., von Wilcken-Bergmann B., Müller-Hill B. Synthetic lac operator mediates repression through lac repressor when introduced upstream and downstream from lac promoter. EMBO J. 1986 Jun;5(6):1377–1381. doi: 10.1002/j.1460-2075.1986.tb04370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. All three elements of the lac ps promoter mediate its transcriptional response to DNA supercoiling. J Mol Biol. 1987 May 5;195(1):89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Brady G. W., Satkowski M., Foos D., Benham C. J. Environmental influences on DNA superhelicity. The effect of ionic strength on superhelix conformation in solution. J Mol Biol. 1987 May 5;195(1):185–191. doi: 10.1016/0022-2836(87)90335-4. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Raymondjean M., Carranca A. G., Herbomel P., Yaniv M. Factors involved in control of tissue-specific expression of albumin gene. Cell. 1987 Aug 14;50(4):627–638. doi: 10.1016/0092-8674(87)90036-5. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culard F., Maurizot J. C. Lac repressor - lac operator interaction. Circular dichroism study. Nucleic Acids Res. 1981 Oct 10;9(19):5175–5184. doi: 10.1093/nar/9.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly T. J., Matthews K. S. Allosteric regulation of inducer and operator binding to the lactose repressor. Biochemistry. 1986 Sep 23;25(19):5479–5484. doi: 10.1021/bi00367a020. [DOI] [PubMed] [Google Scholar]
- Dandanell G., Valentin-Hansen P., Larsen J. E., Hammer K. Long-range cooperativity between gene regulatory sequences in a prokaryote. 1987 Feb 26-Mar 4Nature. 325(6107):823–826. doi: 10.1038/325823a0. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Eismann E., von Wilcken-Bergmann B., Müller-Hill B. Specific destruction of the second lac operator decreases repression of the lac operon in Escherichia coli fivefold. J Mol Biol. 1987 Jun 20;195(4):949–952. doi: 10.1016/0022-2836(87)90499-2. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Nash H. Communication between segments of DNA during site-specific recombination. 1987 Jan 29-Feb 4Nature. 325(6103):401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- Goulet I., Zivanovic Y., Prunell A. Helical repeat of DNA in solution. The V curve method. Nucleic Acids Res. 1987 Apr 10;15(7):2803–2821. doi: 10.1093/nar/15.7.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Griffith J., Hochschild A., Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986 Aug 21;322(6081):750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hendrickson W., Schleif R. Transcription of Escherichia coli ara in vitro. The cyclic AMP receptor protein requirement for PBAD induction that depends on the presence and orientation of the araO2 site. J Mol Biol. 1986 Apr 5;188(3):355–367. doi: 10.1016/0022-2836(86)90160-9. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Hsieh W. T., Whitson P. A., Matthews K. S., Wells R. D. Influence of sequence and distance between two operators on interaction with the lac repressor. J Biol Chem. 1987 Oct 25;262(30):14583–14591. [PubMed] [Google Scholar]
- Kania J., Müller-Hill B. Construction, isolation and implications of repressor-galactosidase - beta-galactosidase hybrid molecules. Eur J Biochem. 1977 Oct 3;79(2):381–386. doi: 10.1111/j.1432-1033.1977.tb11819.x. [DOI] [PubMed] [Google Scholar]
- Kim R., Kim S. H. Direct measurement of DNA unwinding angle in specific interaction between lac operator and repressor. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):451–454. doi: 10.1101/sqb.1983.047.01.053. [DOI] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bret M. Catastrophic variation of twist and writhing of circular DNAs with constraint? Biopolymers. 1979 Jul;18(7):1709–1725. doi: 10.1002/bip.1979.360180710. [DOI] [PubMed] [Google Scholar]
- Le Bret M. Twist and writhing in short circular DNAs according to first-order elasticity. Biopolymers. 1984 Oct;23(10):1835–1867. doi: 10.1002/bip.360231004. [DOI] [PubMed] [Google Scholar]
- Lehming N., Sartorius J., Niemöller M., Genenger G., v Wilcken-Bergmann B., Müller-Hill B. The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 1987 Oct;6(10):3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm A. D., Snounou G. Netropsin increases the linking number of DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):323–326. doi: 10.1101/sqb.1983.047.01.037. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossing M. C., Record M. T., Jr Upstream operators enhance repression of the lac promoter. Science. 1986 Aug 22;233(4766):889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B. Lac repressor and lac operator. Prog Biophys Mol Biol. 1975;30(2-3):227–252. doi: 10.1016/0079-6107(76)90011-0. [DOI] [PubMed] [Google Scholar]
- Nick H., Gilbert W. Detection in vivo of protein-DNA interactions within the lac operon of Escherichia coli. 1985 Feb 28-Mar 6Nature. 313(6005):795–798. doi: 10.1038/313795a0. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Meese K. Topoisomer gel retardation: detection of anti-Z-DNA antibodies bound to Z-DNA within supercoiled DNA minicircles. Nucleic Acids Res. 1988 Jan 11;16(1):21–37. doi: 10.1093/nar/16.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman R. B., Dunaway M., Matthews K. S. DNA binding characteristics of lactose repressor and the trypsin-resistant core repressor. J Biol Chem. 1980 Nov 10;255(21):10100–10106. [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E., Pfenninger O. Supercoils in prokaryotic DNA restrained in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1331–1335. doi: 10.1073/pnas.77.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Robertson M. Gene regulation: specificity and flexibility. Nature. 1987 Jun 11;327(6122):464–466. doi: 10.1038/327464a0. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Sasmor H., Betz J. L. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha R., Flanagan P., Fennewald M. Recombination by resolvase is inhibited by lac repressor simultaneously binding operators between res sites. J Mol Biol. 1987 Aug 5;196(3):505–516. doi: 10.1016/0022-2836(87)90028-3. [DOI] [PubMed] [Google Scholar]
- Schleif R. Gene regulation: why should DNA loop? Nature. 1987 Jun 4;327(6121):369–370. doi: 10.1038/327369a0. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983 Nov 15;170(4):983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Simons A., Tils D., von Wilcken-Bergmann B., Müller-Hill B. Possible ideal lac operator: Escherichia coli lac operator-like sequences from eukaryotic genomes lack the central G X C pair. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1624–1628. doi: 10.1073/pnas.81.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986 Jan 9;319(6049):121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- Théveny B., Bailly A., Rauch C., Rauch M., Delain E., Milgrom E. Association of DNA-bound progesterone receptors. Nature. 1987 Sep 3;329(6134):79–81. doi: 10.1038/329079a0. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Anshelevich V. V., Lukashin A. V., Frank-Kamenetskii M. D. Statistical mechanics of supercoils and the torsional stiffness of the DNA double helix. Nature. 1979 Jul 26;280(5720):294–298. doi: 10.1038/280294a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Barkley M. D., Bourgeois S. Measurements of unwinding of lac operator by repressor. Nature. 1974 Sep 20;251(5472):247–249. doi: 10.1038/251247a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- Whitson P. A., Hsieh W. T., Wells R. D., Matthews K. S. Influence of supercoiling and sequence context on operator DNA binding with lac repressor. J Biol Chem. 1987 Oct 25;262(30):14592–14599. [PubMed] [Google Scholar]
- Whitson P. A., Hsieh W. T., Wells R. D., Matthews K. S. Supercoiling facilitates lac operator-repressor-pseudooperator interactions. J Biol Chem. 1987 Apr 15;262(11):4943–4946. [PubMed] [Google Scholar]
- Zivanovic Y., Goulet I., Prunell A. Properties of supercoiled DNA in gel electrophoresis. The V-like dependence of mobility on topological constraint. DNA-matrix interactions. J Mol Biol. 1986 Dec 5;192(3):645–660. doi: 10.1016/0022-2836(86)90282-2. [DOI] [PubMed] [Google Scholar]