Abstract
Background & Aims
The cancer stem cells (CSCs) have important therapeutic implications for multi-resistant cancers including hepatocellular carcinoma (HCC). Among the key pathways frequently activated in liver CSCs is NF-kB signaling.
Methods
We evaluated the CSCs-depleting potential of NF-kB inhibition in liver cancer achieved by the IKK inhibitor curcumin, RNAi and specific peptide SN50. The effects on CSCs were assessed by analysis of Side Population (SP), sphere formation and tumorigenicity. Molecular changes were determined by RT-qPCR, global gene expression microarray, EMSA, and Western blotting.
Results
HCC cell lines exposed to curcumin exhibited differential responses to curcumin and were classified as sensitive and resistant. In sensitive lines, curcumin-mediated induction of cell death was directly related to the extent of NF-kB inhibition. The treatment also led to a selective CSC-depletion as evidenced by a reduced SP size, decreased sphere formation, down-regulation of CSC markers and suppressed tumorigenicity. Similarly, NF-kB inhibition by SN50 and siRNA against p65 suppressed tumor cell growth. In contrast, curcumin-resistant cells displayed a paradoxical increase in proliferation and expression of CSC markers. Mechanistically, an important component of the CSC-depleting activity of curcumin could be attributed to a NF-kB-mediated HDAC inhibition. Co-administration of the class I/II HDAC inhibitor trichostatine sensitized resistant cells to curcumin. Further, integration of a predictive signature of curcumin sensitivity with human HCC database indicated that HCCs with poor prognosis and progenitor features are most likely to benefit from NF-kB inhibition.
Conclusions
These results demonstrate that blocking NF-kB can specifically target CSC populations and suggest a potential for combined inhibition of NF-kB and HDAC signaling for treatment of liver cancer patients with poor prognosis.
Keywords: liver cancer, cancer stem cells, hepatocarcinogenesis, NF-kB, histone deacetylases
Introduction
Sequential evolution of liver cancer is a complex process resulting in phenotypic and molecular tumor heterogeneity which hampers the progress in clinical management of HCC. Despite the considerable advance in the treatment and diagnosis of HCC, recent efforts in developing novel effective second-line treatments are overall discouraging [1, 2]. Further, HCC is considered highly chemoresistant tumor, with Sorafenib being the only approved current therapy for advanced HCC in patients with Child-Pugh A cirrhosis. The adverse characteristics of HCC, such as high recurrence rate and chemoresistance, are the primary domains of the tumor-initiating cells (TICs, also referred to as cancer stem cells, CSCs) which emphasizes the urgent need of TIC-directed therapies [3]. In line with this, a recent clinical trial using individualized therapy against MET receptor, a molecule of critical importance for liver stem cells as well as in hepatocarcinogenesis, showed promising results underlining the potential of targeting stemness molecules as a novel therapeutic approach against HCC [4, 5].
The relevance of TICs for tumor development and progression is an emerging theme in cancer research, and their existence has been convincingly demonstrated in a variety of solid and hematological malignancies [6]. By definition, TICs possess high proliferative and metastatic capacity as well as high activity of multi-drug resistance transporters which makes these cells an important therapeutic target [3, 7]. Therefore, the understanding of molecular mechanisms and signaling pathways specific for TICs is a subject of intense investigations [8]. In particular, stemness pathways including WNT/β-Catenin, TGFβ, Hedgehog and NOTCH signaling are emerging as attractive targets for specific eradication of TICs [6]. However, many of these pathways are potentially indispensable for normal stem cell activity and difficult to target [9].
Recently, we have demonstrated that epigenetic modulation of liver cancer cells using the DNA methyltransferase-1 inhibitor zebularine enriched the putative TIC population. This facilitated TIC isolation and allowed generation of the hepatic TIC gene signature [10]. Notably, the TIC signature possessed prognostic significance for HCC and other tumors, and exhibited a tight interaction with a variety of key oncogenic and stemness pathways. Among these was a common activation of NF-kB signaling, one of the key-signaling pathway for inflammatory cancers including HCC [11, 12]. Despite some controversies, the frequent up-regulation of NF-kB signaling during chronic liver diseases and hepatocarcinogenesis is well documented [13, 14] and shown to confer survival benefit to cancer cells by protecting them against stress-induced cell death [15-18]. Moreover, activation of NF-kB signaling has been frequently associated with progression of HCC [19-23]. Further, constitutive activation of this pathway in epithelial cells was sufficient to drive intestinal tumorigenesis and to increase expression of known stemness genes [24]. Importantly, disruption of NF-kB signaling has been shown to induce TIC development in liver cancer [25].
Given this information, we aimed to investigate whether NF-kB could be a potential target for a TIC-directed therapy in liver cancer. To achieve an efficient and selective inhibition of NF-kB we used curcumin, an effective IKK inhibitor, RNAi against p65 and selective peptide SN50. The specific objectives of the study were (i) to evaluate the effect of curcumin on the frequency and functional properties of putative liver TICs; (ii) to define the molecular mechanisms underlying the TIC-depletion (iii); to confirm the importance of NF-kB in this process by a direct targeting p65; and (iv) to predict a subclass of human HCCs likely to respond to NF-kB treatment. Our results demonstrate that curcumin targeted TIC fraction in a selected subgroup of the sensitive human HCC cell lines by the NF-kB mediated inhibition of histone deacetylases (HDACs). The HDAC inhibitor trichostatine A (TSA) sensitized unresponsive hepatoma cells to the curcumin treatment. Furthermore, integrative analysis identified a subclass of HCCs with progenitor cell features as likely responders to a targeted NF-kB treatment.
Materials and methods
Cell Lines and compounds
PLC/PRF5 (referred to as PLC), and WRL68 obtained from the American Type Culture Collection (ATCC), and Huh7 from Riken Cell Bank. KMCH is a gift from Dr. G.J. Gores (MAYO Clinic, USA), and Pitts1, is a freshly isolated, non-clonal, AFP-negative primary HCC cell line obtained from David Geller (UPMC, Pittsburgh, USA). Cells were grown in DMEM, supplemented with 2 mM L-glutamine, 1 unit/mL penicillin/streptomycin, and 10% FCS. Cells were treated for 3 days. Curcumin (Sigma Aldrich), SN50 (Merck Millipore) and trichostatine A (TSA, Sigma Aldrich) were used at indicated concentrations selected based on the published data, toxicity profiles and median IC50 across the different cell lines.
Viability and apoptosis
The cell viability was measured by WST-1 assay according to the manufacturer's protocol (Roche Applied Sciences). 5 × 103 cells were plated on 96-well plates and after overnight incubation treated with the indicated drugs for 72 hours. Cell viability defined as the absorbance in the treatment group compared to the control group was expressed as percent mean change ± SD (n = 4). Apoptosis was assessed using differential staining with acridine orange (green) (2 μg/ml; Sigma-Aldrich) incorporated by all cells and ethidium bromide (red) (2 μg/ml; Promega) incorporated only by apoptotic cells with compromised cell membrane. Quantification of apoptotic cells co-stained with acridine orange and ethidium bromide was performed on the images taken with confocal microscope Zeiss NLO710 as described [26]. Four replicate experiments were performed. Cells were counted in 5 independent random view fields.
Colony formation and matrigel-based sphere assays
5×102-103 cells were plated on 6-well plates for colony formation or on 48-well plates for sphere assays after resuspending in 100 μl of medium and Matrigel (vol/vol) (BD Biosciences, Bedford, MA). Colony and sphere formation was monitored for 7-14 days as indicated. The colony and sphere forming potential was calculated as number of colonies or spheres per seeded cells for each fraction. All experiments were performed in 3 independent replicates.
SP analysis
SP analysis was performed as described [10]. Cells were incubated at 37°C for 90 minutes with 15 μg/mL of Hoechst-33342 (Invitrogen). A parallel sample was stained with Hoechst-33342 in the presence of 50 μmol/L of the ABCG2 inhibitor Fumitremorgin C (Sigma) as a control to identify the SP cells. Cell viability was ensured by 7-AAD exclusion.
Tumorigenicity
All procedures were performed with approval of and in accordance with the guidelines of the National Institutes of Health animal care committee. After 72 h of curcumin treatment, viability was assessed by trypan blue. Viable hepatoma cells (102-103) were mixed with Matrigel (1:1) Matrigel (BD Bioscience) and transplanted subcutaneously into non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice as described [10]. Tumor growth was monitored weekly for a total of 12 weeks. Animals were euthanized when tumor size exceeded 15 mm.
siRNA treatment
p65 (RELA) siRNAs used for in vitro studies were chemically synthesized by Ambion (Austin, TX, USA) and evaluated as described before [27]. Negative control siRNA by Ambion (Austin, TX).
Western blotting and EMSA
Whole cell lysates (100 μg) were prepared from frozen cells using M-PER Tissue Extraction Buffer (Pierce) containing Complete Protease Inhibitor Cocktail (Roche), separated by SDS-PAGE and transferred onto Invitrolon PVDF (Invitrogen) [28]. Membranes were probed with the indicated antibodies. Immune complexes were detected using the enhanced chemiluminescence system (Pierce). Quantification of expression levels was performed by densitometric analysis using ImageJ
RNA extraction
Total RNAs was extracted using the Qiagen RNEasy mini Kit (Qiagen GMBH, Hilden, Germany) following the manufacturer's instructions. RNA quantity and purity were estimated using a Nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE), and integrity was assessed by Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Microrarray analysis
A total of 200 ng RNA from three independent experiments were linearly amplified as recommended by the manufacturer (Ambion, Austin, TX) and analyses were performed as described before [10]. The microarray datasets have been deposited to Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo, accession number GSE59713). Gene expression values were adjusted by subtracting background noises in each spot by GenomeStudio (illumina ®), and normalized by quantile normalization method across all samples. Signal intensity with a detection P > 0.05 was treated as a missing value, and only genes with sufficient representation across the samples were included in further data analysis (presence of 50% of samples required). Differentially expressed genes between treated and untreated cells from the individual cell lines were identified by the Bootstrap t-test with 10,000 repetitions (Neuhauser and Jockel, 2006). Genes with a Bootstrap P-value ≤0.05 were considered significantly different. All other two-group comparisons were performed using BRB ArrayTools V4.3.0 software package (Biometric Research Branch, National Cancer Institute) with a P-value ≤0.001 using a random variance model with 10,000 permutations. Hierarchical cluster analyses were based on Euclidean distance, and average linkage was performed with Cluster 3.0, including a filter of 80% presence for each gene. Results were visualized with TreeView 1.60 (Michael Eisen Laboratory, Lawrence Berkeley National Laboratory and University of California, Berkeley; http://rana.lbl.gov/eisen/).
RT-qPCR
A two-step RT-qPCR, cDNA synthesis using SuperscriptIII (Invitrogen), SYBR Green Master-Mix (Bio-Rad) and iQ5 or CFX Connect System was performed. Oligonucleotide primers were designed using Primer3 v.0.4.0 (http://frodo.wi.mit.edu/primer3/) as described before [10]. The amplification protocol was as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 seconds and 1 minute at 60°C, completed by a dissociation curve to identify false positive amplicons. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference. The relative expression level of each gene was normalized to untreated cells and calculated using the formula 2(−ΔΔCt).
Statistics, databases and patient integration
Statistical analysis was performed using Student's t-test, 1-way ANOVA test for multiple group comparisons or Mann Whitney U test for the apoptosis assay. P-values ≤ 0.05 were considered statistically significant. Results are presented as means ± SD or means ± SEM as indicated. Ingenuity Pathway Analysis (Ingenuity Systems Inc.) and GeneGo pathways analysis (Pathway Analysis MetaCore - GeneGo Inc., St. Joseph, MI) tools were used for functional classification and network analyses. The significance of each network, function and pathway was determined by the scoring system provided by Ingenuity Pathway Analysis tool. Gene Set Enrichment analysis (GSEA) was performed using GSEA software provided by Broad Institutes (http://www.broad.mit.edu/gsea/) [29]. All human gene sets from the MSigDB database were tested and gene sets with a NOM P-value <0.05 and FDR <0.25 were considered significantly enriched in a priori defined set of genes. Connectivity Map was used to screen for therapeutic drugs the common gene signature from sensitive lines as described before. For integration of patients, microarray expression data from 53 HCC tumor specimens generated by illumina beadchips was used [26].
Results
Growth suppressive effects of curcumin on hepatoma cells are dependent on the extent of NF-kB inhibition
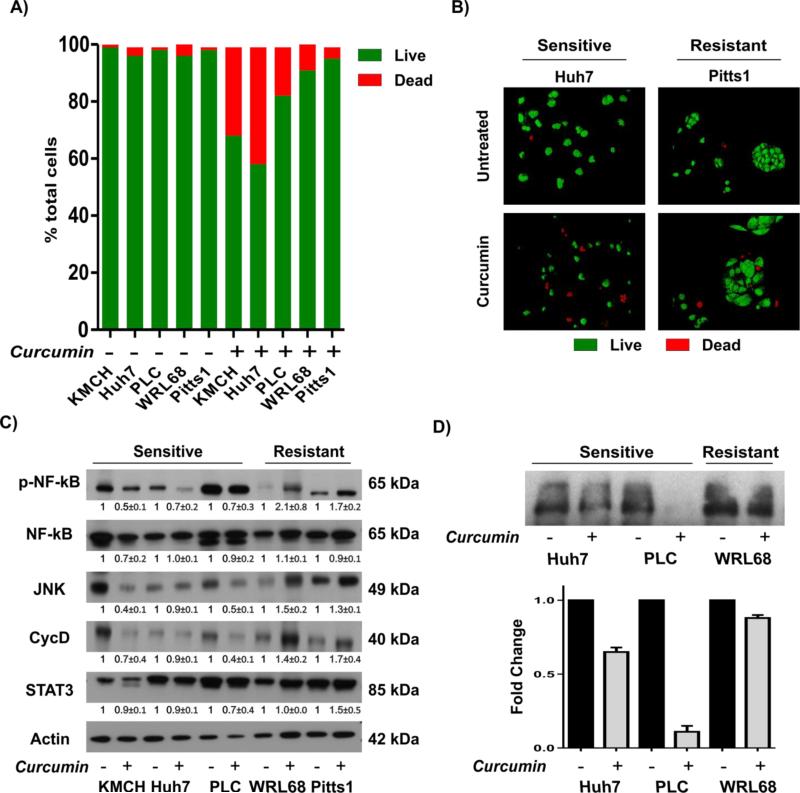
We have recently demonstrated that activation of NF-kB is a specific and common characteristic of putative TICs during malignant progression of liver cancer [10] by comparing the transcriptomic profile of the zebularine-treated Side Population (SP) and non-SP cells (Supplementary Fig. 1A). Furthermore, enrichment of the known NF-kB-dependent gene sets was detected in HCCs with progenitor cell origin and poor prognosis (Supplementary Fig. 1B). These results suggested that NF-kB inhibition may specifically target liver TICs. To test this hypothesis, we utilized an effective IKK inhibitor curcumin, known to block NF-kB signaling [30], and 5 liver cancer cell lines including one primary HCC cell line referred to as Pitts1. The results showed that a 3-day treatment with 25 μM curcumin reduced cell viability (not shown) and significantly increased cell death (Fig. 1A-B), albeit to a different degree. The induction of apoptosis was more significant in KMCH, Huh7, and PLC (18%-41%; P < 0.001) which were defined as sensitive, and modest in WRL68 and Pitts1 (4%-9%; P < 0.001 for WRL68, Pitts1, n.s.), defined as resistant, indicating a differential response to curcumin across the HCC cell lines.
Fig. 1. Growth suppressive effect of curcumin is dependent on NF-kB inhibition.
(A) Quantification of the apoptotic response. Indicated cell lines were grown in the absence or presence of 25 μM curcumin for 3 days and stained with ethidium bromide (EtBr, red) and acridine-orange (AO, green). Viable (green) and apoptotic (red) cells were counted in 5 independent images from 4 replicate experiments taken with confocal microscope at X200 and expressed as a percent of the total population. (B) Representative confocal microscopy images of the double immunofluorescence staining with ethidium bromide (EtBr, red) and acridine-orange (AO, green) show increased number of apoptotic cells in sensitive Huh7 (P < 0.001) cells while in Pitts1 cells, the induction of apoptosis was modest (n.s.). (C) Western blots for NF-kB and downstream targets. Actin was used as a loading control. Monolayer cultures of indicated cell lines were treated with 25 μM curcumin for 3 days. Representative images of two independent experiments are shown. The numbers show the results of quantitative analysis by densitometry normalized to the corresponding actin expression versus untreated cells and shown as means ± SD. (D) DNA binding of p65 determined by EMSA. Images are representative of one of two independent experiments, quantification by densitometry versus control cells (means ± SD) shown below.
In the sensitive cell lines, the growth-inhibitory effect of curcumin was associated with a repression of NF-kB activity as evidenced by down-regulation of phosphorylated p65 (p-p65), JNK, Cyclin D1 and STAT3 (Fig. 1C). Conversely, the resistant tumor cells retained high expression levels of NF-kB signaling (Fig. 1C,D). Importantly, the dynamic of NF-kB activation in response to TNF-alpha stimulation was comparable in the representative sensitive (Huh7) and resistant (WRL68) cell lines (Supplementary Fig. 2) indicating that both resistant and sensitive cells possessed a competent NF-kB signaling. Together these results show that curcumin has a differential effect on viability of hepatoma cells which is associated with NF-kB inhibition in the sensitive but not resistant tumor cell lines.
Curcumin exerts TIC depleting activity
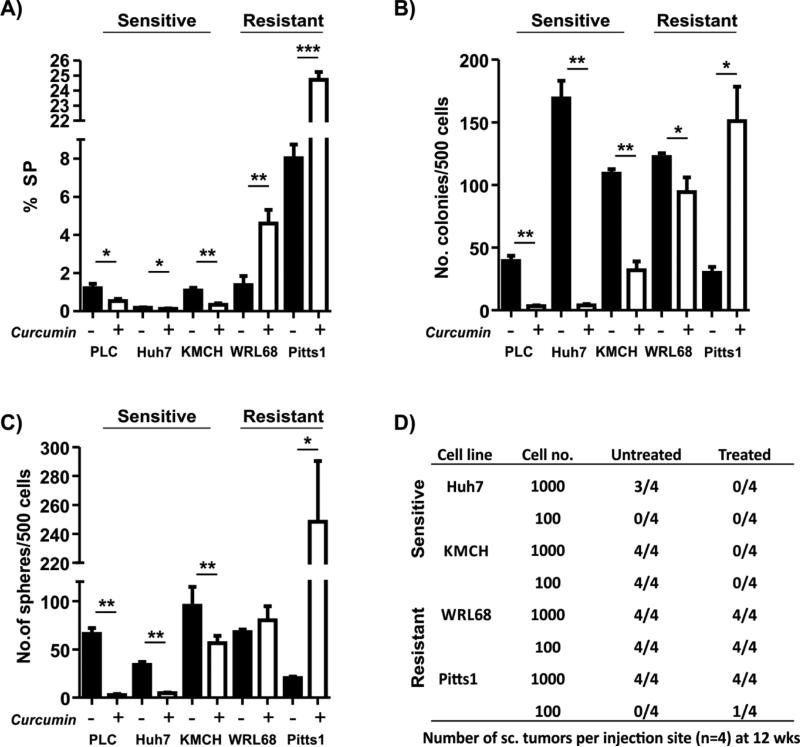
Next we assessed the effect of curcumin on the putative TICs using side-population (SP) approach and standard in vitro and in vivo assays. In agreement with the differential effects of curcumin on hepatoma cell growth (Fig. 1A,B), a 3-day curcumin exposure caused a significant (28-69% range) reduction in the SP fraction only in the sensitive cells (Fig. 2A). Conversely, the curcumin treatment of the resistant tumor cells increased the SP size (Fig. 2A). Consistently, the effect of curcumin on colony and sphere forming ability was also cell line-dependent. Both parameters were strongly elevated in Pitts1 and remained unaffected in WRL68, but were significantly reduced in the sensitive cells (Fig. 2B,C). Accordingly, the curcumin-treated resistant cells displayed a stronger and more consistent up-regulation of the selected stem/cancer stem cell markers, including, cKIT, EpCAM, CD133, BMI1, and NANOG (Supplementary Fig. 3).
Fig. 2. Effect of curcumin on hepatic TICs.
(A) Analysis of side population (SP). SP cells were identified using Hoechst 33342 staining and co-staining with Fumitremorgin C as a negative control. The data are means ± SD of three independent experiments. (B, C) Colony (B) and sphere (C) frequency. The data are means ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) The tumor frequency at 12 weeks after subcutaneous injection of tumor cells into NOD/SCID mice. Cells were exposed to 25 μM curcumin for 3 days before transplantation. Shown are the numbers of tumors per injection site.
To validate these findings in vivo, curcumin sensitive and resistant cell lines were exposed to curcumin for 3 days in vitro and injected subcutaneously into NOD/SCID mice. Limiting dilution analysis showed that curcumin pretreatment completely abolished the tumorigenicity of the sensitive Huh7 and KMCH cells (P <0.01 and <0.001, respectively) while the tumor forming ability of the curcumin-resistant cell lines was slightly increased (Pitt1) or remained unaffected (WRL68) (Fig. 2D). Pretreatment of Pitts1 cells also caused a significant reduction in tumor latency (Supplementary Fig. 4A) and increased tumor weight (Supplementary Fig. 4B). Together these results demonstrate that curcumin exerts a TIC-depleting activity in sensitive but not in the resistant liver cancer cells.
Specific inhibition of NF-kB effectively reduces TIC properties of hepatoma cells
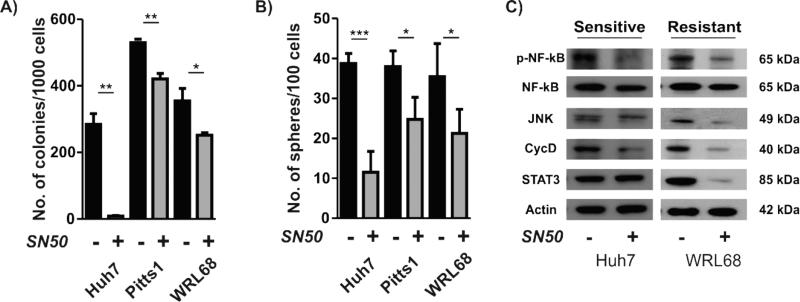
To confirm that the TIC-depleting capacity of curcumin was directly related to its ability to inhibit NF-kB, representative curcumin-sensitive and -resistant cell lines were treated with a known cell-permeable NF-kB inhibitor peptide SN50 for 3 days [31]. The results showed that SN50 caused a significantly stronger suppression of the colony (Fig. 3A) and sphere (Fig.3B) formation in curcumin-sensitive Huh7 cells as compared to the resistant Pitts1 and WRL68 cells. Anti-tumorigenic properties of SN50 were related to the extent of the inhibition of NF-kB signaling (Fig. 3C). To further confirm that the observed effects were NF-kB dependent, we used p65 siRNA to block NF-kB signaling in PLC and WRL68 (Supplementary Fig. 5A). Again, the effective knockdown of p65 led to a significant reduction in the size of the SP (Supplementary Fig. 5B). Furthermore, we found a drastic suppression of the NF-kB target gene expression resulting in significantly reduced colony and sphere forming potential indicative of TICs repression (Supplementary Fig. 5 C,D). These results corroborate that the observed TIC-depleting activity of curcumin is dependent on the NF-kB inhibition at least in part and underscore the potential of NF-kB targeting to effectively deplete TICs in liver cancer.
Fig. 3. Specific NF-kB inhibition by SN50 inhibitory peptide.
(A) Colony and (B) sphere frequency after a 3-day exposure to 75 μM SN50. The data are means ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Western blots for NF-kB and downstream targets in Huh7 and WRL68. Actin was used as a loading control. Monolayer cultures of indicated cell lines were treated with 75 μM SN50 for 3 days.
Molecular mechanisms underlying curcumin effects
To assess the molecular mechanisms of the differential impact of curcumin on hepatic TICs propagation in vitro and in vivo, we examined global genetic alterations caused by a 3-day curcumin exposure in 2D cultures. A total of 224 genes (209 down-regulated and 15 up-regulated) were significantly different at a fold change greater than 2 between curcumin-sensitive and -resistant hepatoma cell lines (Supplementary Fig. 6A and Supplementary Table 1). These genes were highly efficient in separating the resistant from sensitive cell lines. Subsequent Ingenuity Pathway Analysis (IPA) of the 224-gene curcumin-resistance signature showed that the dominant functional networks were involved in the regulation of cell cycle (CDKN1B, BRAF, MYC, E2F3, JUNB), DNA-damage (PKMYT1, LIG3, TP53BP1, MYC) as well as tissue remodeling and wound repair (TIMP2, CREBBP, GSK3B, AXIN1, RHOC, PLK1, CLO7A1) (Supplementary Fig. 6B). Notably, there was a strong induction of key oncogenic signaling pathways, such as c-MYC, PDGF, JUN-B as well as NF-kB, providing additional evidence that the resistance of a subgroup of liver cancer cell lines to tumor-suppressive effects of curcumin may be related to its inability to inhibit NF-kB (Supplementary Fig. 6C-E).
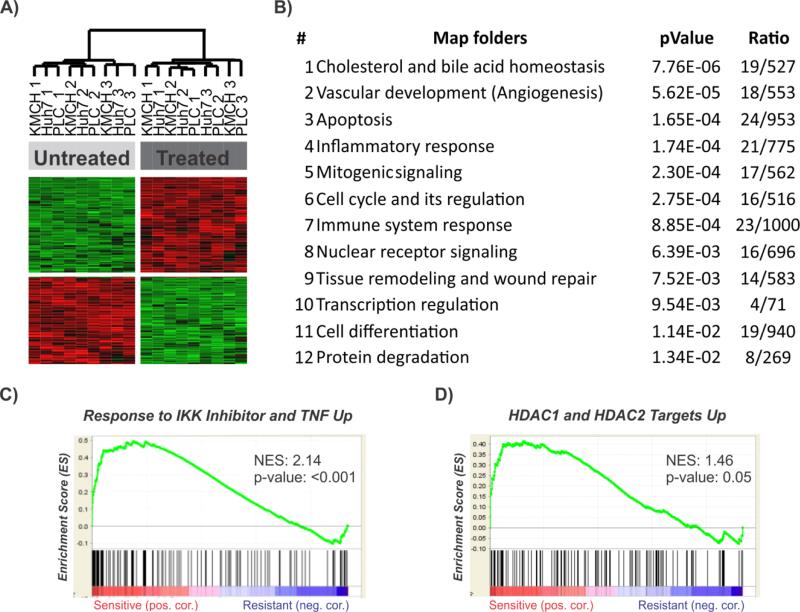
To determine the molecular mechanisms conferring to curcumin sensitivity, we then compared the transcriptome of three curcumin-sensitive cell lines before and after drug treatment. A high number of the differentially expressed genes could be detected in the sensitive cell lines, indicating that curcumin induces drastic transcriptomic changes (Huh7: 4397 genes; KMCH: 4025 genes; PLC: 4942 genes). Of note, the same analysis revealed significantly less genes in the resistant lines (WRL68: 1809 genes; Pitts1: 141 genes). While many of the induced changes were cell line specific, a total of 226 genes were identified to be commonly deregulated among the 3 sensitive cell lines. Hierarchical clustering of these common genes effectively separated treated from untreated cells (Fig. 4A). A significant proportion of the common genes were associated with cell cycle regulation and apoptosis (CDKN1A, CDKN1B, JUN, BIRC3, HSPA1A/B, MAPK3) as well as pathways involved in lipid metabolism (CAV1, CYP1A1, DHCR7, FDFT1, SCD, BMPR2, SREBF1, BMPR2), vascular development, immune response and tissue remodeling (CD68, MET, JUN, CAV1, FGFR4, ADM, TUBB2A/B, IRF1) (Fig. 4B). Gene set enrichment analysis (GSEA) identified an abundance of gene sets involved in IKK inhibition, validating that curcumin exerts its action partly via NF-kB inhibition in sensitive cell lines (Fig. 4C). Furthermore, gene sets surrounding targets of histone deacetylases (HDAC) I and II were significantly enriched in the sensitive cells (Fig. 4D).
Fig. 4. Common molecular characteristic of curcumin-sensitive cells.
(A) Unsupervised hierarchical cluster analysis of curcumin sensitive hepatoma cell lines based on the 226 commonly regulated genes. Cells were cultured in the absence or presence of curcumin for 3 days. (B) The top functional networks identified by GeneGo analysis of the commonly regulated genes in the sensitive cells. (C,D) Gene set enrichment analysis (GSEA) of the genes involved in IKK inhibition (C) and HDAC1/ HDAC2 targets (D) in sensitive cells as compared to untreated cells. Enrichment score (ES) reflects degree of overrepresentation for each group at the peak of the entire set. Statistical significance calculated by nominal P value of the ES by using an empirical phenotype-based permutation test.
We next tested whether the disruption of the common genes conferring to curcumin sensitivity could induce a similar response in the resistant liver tumor cell lines. Therefore, we applied Connectivity Map analysis, a resource of gene expression profiles for drug responses, to the 226 gene sensitivity-signature to identify compounds that would induce the same molecular changes associated with curcumin sensitivity. The highest connectivity scores were obtained for vorinostat and TSA, two known HDACs inhibitors (HDACi) (Supplementary Table 3) suggesting a potential synergistic effect of curcumin and HDACi combination for treating the resistant hepatoma cell lines.
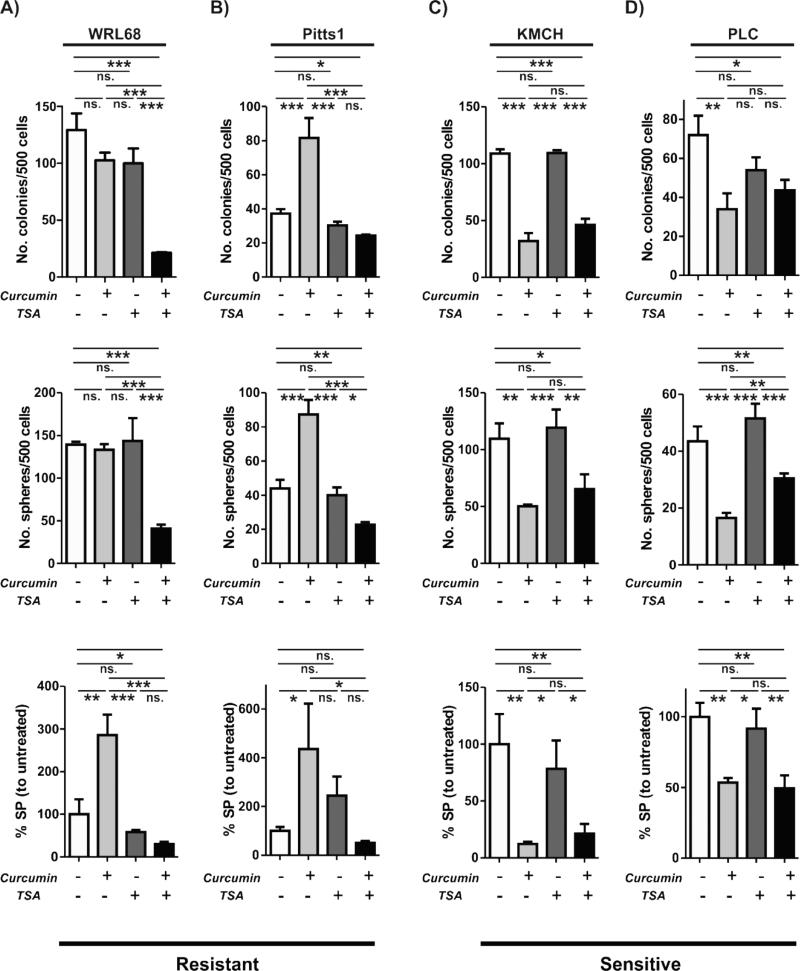
To address this possibility, we subjected the resistant (WRL68 and Pits1) and sensitive (KMCH and PLC) cell lines to treatment with TSA, curcumin and curcumin+TSA. Indeed, a combined treatment with curcumin and TSA was significantly more effective in reducing the SP size as well as colony and sphere forming ability of the resistant WRL68 cells and diminished the curcumin-induced increase in TIC properties of Pitts1 cells (Fig. 5A-B). In contrast, co-administration of TSA did not intensify the growth suppressive effects of curcumin on TIC properties in the sensitive KMCH and PLC cell lines (Fig. 5C-D).
Fig. 5. TSA sensitizes resistant cells to curcumin.
(A-D) Effect of curcumin, TSA or combined curcumin and TSA treatment on the putative TICs in the resistant (A, B) and sensitive (C, D) cell lines. Bar graphs show colony (top), sphere (middle) and side population (SP) (bottom) frequencies. The data are means ± SD of three independent experiments. *, P < 0.05; **, P < 0.01. ***, P < 0.001, ns. = not significant
Patients with HCC characterized by progenitor cell features might benefit from curcumin treatment
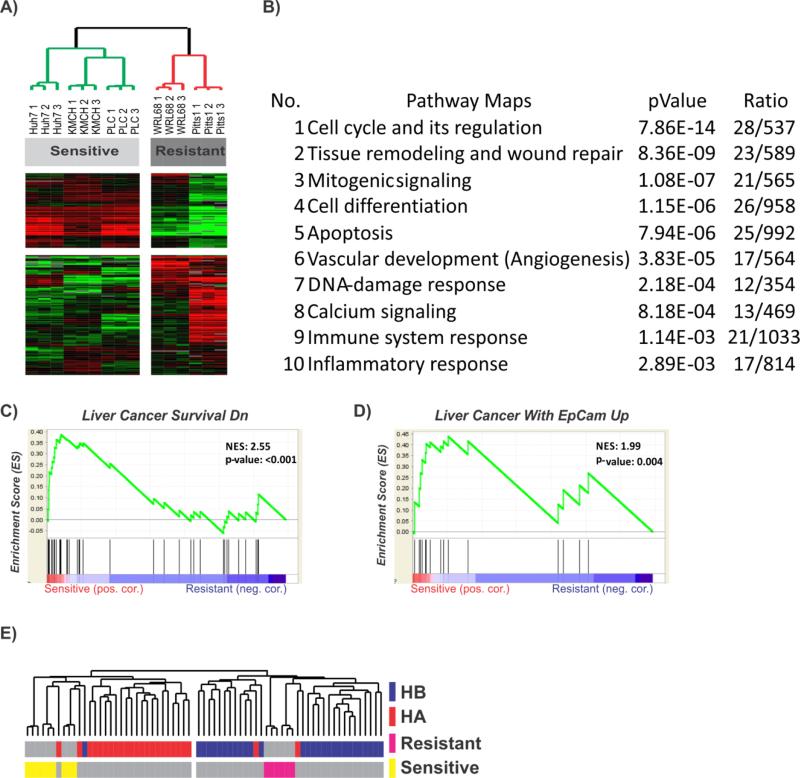
Finally, to identify patients who might respond to curcumin treatment, we performed an integrative analysis of untreated hepatoma cells with our database of 53 HCC [26]. First, we compared the transcriptomic profiles of untreated hepatoma cell lines and found a total of 217 differentially expressed genes between the curcumin sensitive and resistant lines (Fig. 6A and Supplementary Table 4). The major functional pathways were associated with the genes involved in known pro-oncogenic signaling such as cell cycle control (BRAF, CDKN2B, MAP2K1, IGFBP7, E2F3, KLF4, RAF1), apoptosis (CD24, ADAMTSL4, ADNP, ANTXR1, BAG6, BASP1, DLX1, HINT1, NFKBIA) as well as DNA-damage response (MST4, TP53BP1, NCL, Pi3K, SRP14, PKM, RNF32). Pathways involved in tissue remodeling, differentiation and neo-angiogenesis were also affected (Fig. 6B). GSEA demonstrated an activation of the gene sets associated with cancer cell survival and progenitor cell features (Fig. 6C-D). These results indicate that sensitive liver cancer cell lines possess molecular characteristics associated with malignant progression. In agreement with the enrichment of NF-kB in HCCs with progenitor cell features, integrative hierarchical clustering using 217-gene signature also revealed that the sensitive hepatoma cell lines were clustered with the previously identified poor prognostic HCC subtype A which also contains the high-risk hepatoblast-like HCC characterized by a progenitor cell origin and the worst prognosis (Fig. 6E) [32, 33]. These results suggest that poor prognostic HCC patients are likely to benefit from curcumin treatment, while a combined treatment with both curcumin and HDACi may be effective against the resistant tumors.
Fig. 6. Prediction of the response to curcumin treatment.
(A) Unsupervised hierarchical cluster analysis of untreated curcumin sensitive and resistant hepatoma cell lines based on the 217 differentially expressed genes. (B) GeneGo analysis of the top functional networks. (C-D) Gene set enrichment analysis (GSEA) of the genes associated with liver cancer survival (C) and upregulation of EpCAM-positive TICs in resistant versus sensitive cells. Enrichment score (ES) reflects degree of overrepresentation for each group at the peak of the entire set. Statistical significance calculated by nominal P value of the ES by using an empirical phenotype-based permutation test. (E) Integration of untreated curcumin sensitive and resistant cells with our previously published dataset of 53 HCC patients.[26] Unsupervised cluster analyses shows that resistant cell lines clustered with the better prognosis subtype B (HB), while sensitive cell lines were located in close proximity to the previously identified poor prognosis HCC subtype A (HA), potentially indicating that these patients might benefit from the curcumin treatment.
Discussion
The majority of HCCs develop on the basis of an inflammatory microenvironment. Among the most prominent molecular changes driving the inflammation-fibrosis-cancer axis during hepatocarcinogenesis is the activation of the NF-kB signaling [34]. Here, we provide evidence that targeting of NF-kB signaling by curcumin confers to anti-tumorigenic changes and affects TIC characteristics in several liver cancer cell lines. Interestingly, the TIC depleting capacity of curcumin was directly related to the extent of NF-kB inhibition. Mechanistically, gene expression analyses demonstrated that curcumin partly exerted its TIC depleting action via inhibition of histone deacetylases class I and II induced by the inhibition of NF-kB signaling. HDAC-inhibitor TSA sensitized resistant tumor cells to the curcumin treatment.
Constitutive NF-κB pathway activation has been reported in different cancers. Recent investigations have supported a role for NF-kB signaling in self-renewal and expansion of TICs in several hematological and solid tumors [35-37]. A work in acute myeloid leukemia demonstrated that NF-kB activation originates in the tumor-initiating cells that are linked to disease progression [38]. Furthermore, activation of this pathway is associated with decreased metastasis-free survival in breast-cancer [39]. Here, we report that curcumin can act as an effective NF-kB inhibitor in a subset of liver cancer cell lines by inducing anti-tumorigenic changes. A differential response to curcumin in sensitive and resistant hepatoma cell lines was directly related to the extent of inhibition of NF-kB and downstream signaling such as JNK, Cyclin D1 and STAT3. In sensitive cell lines, inhibition of NF-kB by curcumin caused a dramatic decrease in TIC properties as evidenced by diminished spherogenicity, tumorigenicity and a significant reduction of SP cells. In contrast, resistant hepatoma cell lines either retained their tumorigenicity or showed enhanced TICs characteristics. Mechanistically, the comparison of the transcriptome profiles of the curcumin sensitive and resistant cell lines demonstrated that curcumin induced global activation of pro-apoptotic and anti-tumorigenic signaling associated with repression of the functional networks involved in NF-kB and MYC signaling only in the sensitive cells. Conversely, both signaling pathways remained active in resistant cell lines. These results indicate that the anti-tumorigenic effect of curcumin in the sensitive cancer cells is dependent at least in part on NF-kB inhibition via direct inhibition of the downstream prooncogenic signaling such as MYC, Cyclin D1, JNK and STAT3 [25, 40]. Notably, resistance to curcumin in hepatoma cell lines was independent of ABCA1 expression (data not shown), an observation that was recently reported for breast cancer cells [41].
While the mechanisms of resistance to NF-kB inhibition by curcumin were cell line-specific, a common molecular feature of curcumin sensitivity was the NF-kB mediated decrease in histone deacetylase activity that significantly contributed to a reduction in TIC characteristics. The resistance against curcumin could be partly overcome by a co-administration with the pan-HDAC inhibitor TSA. The convergence of NF-kB and histone deacetylases signaling pathways was recently demonstrated by several studies [42-44]. NF-kB is well known to be involved in modifying histone function after cytokine exposure in different cellular contexts including cancer. The regulation of NF-kB by nucleosome as well as histone deacetylase remodeling is known to control inflammatory response homeostasis suggesting the potential for the combined targeting of both signaling pathways. A recent work showed that p65 acetylation significantly contributes to NF-kB activation in IKKβ-dependent manner that promotes p65 Ser-536 phosphorylation. Further, the p65 acetylation ultimately limited the HDACi efficiency in human multiple myeloma cells [45]. Another study showed that a MYC-dependent miR-29b repression and increased levels of the miR-29b could target Sp1 in KIT-driven leukemia. In this context, KIT overexpression responsible for tumor progression was dependent on the activity of a Sp1/NF-kB/HDAC/miR-29b complex that could be effectively targeted [46]. These results support our observation of an enhanced anti-oncogenic effect of curcumin in combination with TSA. Given the importance of efficient NF-kB depletion for the sensitivity to curcumin, further investigations are needed to determine if a sequential or combined administration would be the most effective. Several clinical trials strived to evaluate the potential of curcumin for the treatment of selected cancers [47]. Although curcumin is affordable, easy accessible and exhibits low to no toxicity at the moderate doses, the bioavailability remains generally low despite recent progress with structural analogues [48]. Additionally, it is well recognized that NF-kB inhibiting effect is highly context- and cell-type dependent [49]. Therefore, a more specific targeting with a precise delivery, e.g. by nanoparticles or RNAi, might be warranted.
Another key finding of the study is the potential of curcumin to diminish the cancer growth in HCC that display the worst prognosis and are most difficult to treat [32, 33]. Given that HCC with progenitor cell origin show the activation of NF-kB signaling, these patients are likely to respond to NF-kB modulation by curcumin. From the therapeutic perspective, the results suggest that specific NF-kB-depletion might be able to complement the current systemic chemotherapies for the HCC with low TIC activity, i.e. targeting both the tumor-initiating and bulk tumor cells [50]. Collectively these results demonstrate that specific disruption of the NF-kB pathway might be a potential therapeutic approach for prognostically adverse HCCs with progenitor features and activated NF-kB signaling.
Supplementary Material
Acknowledgments
This project was supported by the Intramural Research Program of the Center for Cancer Research, NCI and by internal funding of the University Hospital of Mainz (MAIFOR). J.U.M. is supported by a grant from the German Research Foundation (MA 4443/2-1). This project was supported by the Virtual Liver Network of the BMBF (to KB; FKZ 0315761). J.U.M. thanks Monika Herr for excellent technical support.
Aspects of this article are part of the doctoral thesis of LO. Arreguin Camacho.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors have no conflict of interest.
Author Contributions: Designed the experiments: JUM, SST, VMF; Performed the experiments: JUM, LGQ, LOAC, FP, YHL, MK, MPD, JBA; Analyzed the data: JUM, KB, EAC, PRG, JBA, VMF; Wrote the paper: JUM, SST, VMF. All authors discussed the results and critically commented on the manuscript.
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan CT, Guzman ML, Noble M. Cancer stem cells. NEnglJMed. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Alfa GK. Approaching the era of personalised therapy for liver cancer? The lancet oncology. 2013;14:7–8. doi: 10.1016/S1470-2045(12)70519-3. [DOI] [PubMed] [Google Scholar]
- 5.Marquardt JU, Thorgeirsson SS. Sall4 in “stemness”-driven hepatocarcinogenesis. N Engl J Med. 2013;368:2316–2318. doi: 10.1056/NEJMe1303026. [DOI] [PubMed] [Google Scholar]
- 6.Marquardt JU, Factor VM, Thorgeirsson SS. Epigenetic regulation of cancer stem cells in liver cancer: current concepts and clinical implications. J Hepatol. 2010;53:568–577. doi: 10.1016/j.jhep.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer research. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 8.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell stem cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Marquardt JU, Raggi C, Andersen JB, Seo D, Avital I, Geller D, et al. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology. 2011;54:1031–1042. doi: 10.1002/hep.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harbor perspectives in biology. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shostak K, Chariot A. NF-kappaB, stem cells and breast cancer: the links get stronger. Breast cancer research : BCR. 2011;13:214. doi: 10.1186/bcr2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsharkawy AM, Mann DA. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46:590–597. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- 14.Vainer GW, Pikarsky E, Ben-Neriah Y. Contradictory functions of NF-kappaB in liver physiology and cancer. Cancer letters. 2008;267:182–188. doi: 10.1016/j.canlet.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 15.Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links. Annals of the New York Academy of Sciences. 2009;1155:206–221. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 16.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell research. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavon I, Pikarsky E, Gutkovich E, Goldberg I, Bar J, Oren M, et al. Nuclear factor-kappaB protects the liver against genotoxic stress and functions independently of p53. Cancer research. 2003;63:25–30. [PubMed] [Google Scholar]
- 18.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capturing cancer stem cells. NatMed. 2008;14:814. doi: 10.1038/nm0808-814. [DOI] [PubMed] [Google Scholar]
- 20.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 21.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nature reviews Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 22.Nemeth J, Stein I, Haag D, Riehl A, Longerich T, Horwitz E, et al. S100A8 and S100A9 are novel nuclear factor kappa B target genes during malignant progression of murine and human liver carcinogenesis. Hepatology. 2009;50:1251–1262. doi: 10.1002/hep.23099. [DOI] [PubMed] [Google Scholar]
- 23.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 24.Vlantis K, Wullaert A, Sasaki Y, Schmidt-Supprian M, Rajewsky K, Roskams T, et al. Constitutive IKK2 activation in intestinal epithelial cells induces intestinal tumors in mice. The Journal of clinical investigation. 2011;121:2781–2793. doi: 10.1172/JCI45349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, et al. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen JB, Factor VM, Marquardt JU, Raggi C, Lee YH, Seo D, et al. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Sci Transl Med. 2010;2:54ra77. doi: 10.1126/scitranslmed.3001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YH, Andersen JB, Song HT, Judge AD, Seo D, Ishikawa T, et al. Definition of Ubiquitination Modulator COP1 as a Novel Therapeutic Target in Human Hepatocellular Carcinoma. Cancer research. 2010 doi: 10.1158/0008-5472.CAN-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marquardt JU, Seo D, Gomez-Quiroz LE, Uchida K, Gillen MC, Kitade M, et al. Loss of c-Met accelerates development of liver fibrosis in response to CCl(4) exposure through deregulation of multiple molecular pathways. Biochim Biophys Acta. 2012;1822:942–951. doi: 10.1016/j.bbadis.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralhan R, Pandey MK, Aggarwal BB. Nuclear factor-kappa B links carcinogenic and chemopreventive agents. Frontiers in bioscience. 2009;1:45–60. doi: 10.2741/S6. [DOI] [PubMed] [Google Scholar]
- 31.Gomez-Quiroz LE, Factor VM, Kaposi-Novak P, Coulouarn C, Conner EA, Thorgeirsson SS. Hepatocyte-specific c-Met deletion disrupts redox homeostasis and sensitizes to Fas-mediated apoptosis. The Journal of biological chemistry. 2008;283:14581–14589. doi: 10.1074/jbc.M707733200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 33.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 34.Luedde T, Schwabe RF. NF-kappaB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nature reviews Gastroenterology & hepatology. 2011;8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzman ML, Allan JN. Leukemia stem cells in personalized medicine. Stem cells. 2013 doi: 10.1002/stem.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. The Journal of clinical investigation. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kagoya Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S, et al. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. The Journal of clinical investigation. 2014;124:528–542. doi: 10.1172/JCI68101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Tan W, Wu X, Poustovoitov M, Strasner A, Li W, et al. A NIK-IKKalpha module expands ErbB2-induced tumor-initiating cells by stimulating nuclear export of p27/Kip1. Cancer cell. 2013;23:647–659. doi: 10.1016/j.ccr.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 41.Bachmeier BE, Iancu CM, Killian PH, Kronski E, Mirisola V, Angelini G, et al. Overexpression of the ATP binding cassette gene ABCA1 determines resistance to Curcumin in M14 melanoma cells. Molecular cancer. 2009;8:129. doi: 10.1186/1476-4598-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 43.Kuo HP, Wang Z, Lee DF, Iwasaki M, Duque-Afonso J, Wong SH, et al. Epigenetic roles of MLL oncoproteins are dependent on NF-kappaB. Cancer cell. 2013;24:423–437. doi: 10.1016/j.ccr.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pakala SB, Bui-Nguyen TM, Reddy SD, Li DQ, Peng S, Rayala SK, et al. Regulation of NF-kappaB circuitry by a component of the nucleosome remodeling and deacetylase complex controls inflammatory response homeostasis. The Journal of biological chemistry. 2010;285:23590–23597. doi: 10.1074/jbc.M110.139469. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Dai Y, Chen S, Wang L, Pei XY, Funk VL, Kramer LB, et al. Disruption of IkappaB kinase (IKK)-mediated RelA serine 536 phosphorylation sensitizes human multiple myeloma cells to histone deacetylase (HDAC) inhibitors. The Journal of biological chemistry. 2011;286:34036–34050. doi: 10.1074/jbc.M111.284216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Zhang T. Targeting cancer stem cells by curcumin and clinical applications. Cancer letters. 2014 doi: 10.1016/j.canlet.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Park W, Amin AR, Chen ZG, Shin DM. New perspectives of curcumin in cancer prevention. Cancer prevention research. 2013;6:387–400. doi: 10.1158/1940-6207.CAPR-12-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pikarsky E, Ben-Neriah Y. NF-kappaB inhibition: a double-edged sword in cancer? European journal of cancer. 2006;42:779–784. doi: 10.1016/j.ejca.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Kreso A, Dick John E. Evolution of the Cancer Stem Cell Model. Cell stem cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.