Abstract
Genetic Risk Scores are an increasingly popular tool for summarizing the cumulative risk of a set of SNPs with disease. Typically only the set of the SNPs that have reached genome-wide significance compose these scores. However recent work suggests that including additional SNPs may aid risk assessment. In this paper, we used the Atherosclerosis Risk in Communities Study (ARIC) cohort to illustrate how one can choose the optimal set of SNPs for a GRS. In addition to p-value threshold, we also examined linkage disequilibrium, imputation quality and imputation type. We provide a variety of evaluation metrics. Results suggest that p-value threshold had the greatest impact on GRS quality for the outcome of coronary heart disease, with an optimal threshold around 0.001. However, GRSs are relatively robust to both linkage disequilibrium and imputation quality. We also show that the optimal GRS partially depends on the evaluation metric and consequently the way one intends to use the GRS. Overall the implications highlight both the robustness of GRS and a means to empirically choose the best set of GRSs.
Keywords: Coronary Heart Disease, Risk Assessment, Risk Score
Introduction
Genome wide association (GWA) studies and follow-up meta-analyses have identified a plethora of susceptibility loci for many different common diseases (Welter et al., 2014). While the number of polymorphisms reaching genome wide significance remains modest, typically < 100 per disease, simulation and empirical studies suggest that many more common susceptibility variants remain to be discovered (Stahl et al., 2012; Visscher et al., 2012; Welter et al., 2014; Wray et al., 2011). These discoveries will undoubtedly result in a better understanding of the pathophysiology of chronic diseases and the development of novel therapeutic options over the long term. However, great interest exists in leveraging these findings to better predict the occurrence of chronic diseases over the short term particularly for diseases that have therapeutic options that can reduce risk irrespective of the source of excess genetic risk (Jostins and Barrett, 2011; Thanassoulis and Vasan, 2010). Such is the case for coronary heart disease (CHD) where a combination of optimal modifiable risk factors can virtually eradicate the probability of developing disease (Stamler et al., 1999; Yusuf et al., 2004).
Currently the most practical way to leverage recent GWAS findings for risk prediction is through the use of genetic risk scores (GRS) (Amin et al., 2009). To date, GRSs have been created for many complex diseases, including cardiovascular disease (Thanassoulis et al., 2012), schizophrenia (Purcell et al., 2009) and multiple sclerosis (De Jager et al., 2009). GRSs combine the modest effects of multiple SNPs into a single variable. They are typically calculated as a weighted sum of the number of high risk alleles, where the GRS for person i is:
RA is an indicator for the presence of risk alleles at allele k at SNP j (0 – 1) and wj is the weight of each risk allele, generally derived from the estimated log odds ratio for that allele from either a large GWAS or a meta-analysis involving multiple case-control studies. However, these weights can also be derived from the hazard ratio or relative risk of one or more cohort studies. Since the weights are based off of marginal associations, the two implicit assumptions in such a construction are that (1) the effect each SNP is independent and (2) the marginal effects capture the full effect for a given SNP, indicating the SNP does not interact with any other SNP in the GRS. While adjacent SNPs are expected to be in linkage disequilibrium (LD), one can remove such SNPs through LD pruning. While there may be interactions, main effects are likely to dominate overall (Hemani et al., 2014; Hill et al., 2008). Thus, while these assumptions may be violated, the impact is likely minimal. The range of GRS values is determined by both the number of SNPs involved and the sign (positive or negative) of the weight (based on the direction of effect of the coded allele). Since the raw value of the GRS itself is not particularly meaningful, standardizing it to a mean of 0 and variance of 1 facilitates its interpretation.
Recent work has focused on combining GRSs with clinical risk scores to demonstrate incremental value in risk prediction (Goldstein et al., 2014). While most investigators have tested scores that include only the few SNPs that have met GWA significance – typically p < 5 ×10−8 – others have constructed more expansive GRSs using many more SNPs (Purcell et al., 2009; Simonson et al., 2011), showing that doing so explains a greater percentage of the variance in the outcome. Others have considered the impact of study sample size (Chatterjee et al., 2013) and LD (Wu et al., 2013). Building off these, and other considerations, the aim of this analysis is to illustrate how one could empirically determine which SNPs to include in a GRS using the most reliable genetic association data available. We present our approach for the outcome of CHD using a large scale, publicly available GWAS but it can be easily applied to other chronic diseases and to association data that more reliably survey the lower frequency spectrum of mutations. However, we emphasize that our goal here is to illustrate which factors should be considered understanding that the specific findings in this study are not necessarily generalizable to other cohorts or to other outcomes. Instead, the optimal GRS depends on a number of factors including the true genetic architecture of the disease and the strength of the corresponding genetic association studies.
Methods
SNP Selection & Weighting
An important consideration in constructing a GRS is selecting the study from which the association results (i.e. weights) will be derived. We constructed our GRSs using SNPs from the most recent and largest GWAS for coronary artery disease conducted by the CARDIoGRAM. The GWAS included 22,233 cases and 64,762 controls of white/European ancestry (Schunkert et al., 2011).
Prospective Cohort for testing Genetic Risk Scores
We selected the Atherosclerosis Risk in Communities Study (ARIC) study to test the various GRSs we constructed. The ARIC Study is an ongoing prospective investigation of atherosclerosis and its clinical sequelae involving 15,792 white and black persons aged 45–64 years at recruitment (1987–1989). Detailed descriptions of the study designs, IRB consent process, sampling procedures, methods, definitions of cardiovascular outcomes, and approach to statistical analyses is published elsewhere (1989; White et al., 1996). ARIC is an ideal testing cohort for several reasons including the availability of individual level genome wide data for all participants through the National Institutes of Health (NIH) controlled access database of Genotypes and Phenotypes (dbGaP) (Mailman et al., 2007), a prolonged follow up with > 1000 incident outcomes of interest among participants of European descent, and no overlap of incident cases with prevalent cases that were included in the CARDIoGRAM consortium study.
All white/Europeans without a history of CHD, myocardial infarction, or heart failure at baseline were included. Incident CHD was defined by the recording for the first time of either non-fatal or fatal myocardial infarction (“mi04”, “fatchd04”), CHD related revascularization procedure (“in_by04p”), or silent MI detected by ECG (“in_04s”). The outcome of interest was incident CHD within 10 years. Those without a positive event who died or were lost to follow up prior to their 10th year anniversary of follow up were removed from analysis. All others were deemed event free at 10-years regardless of whether they developed incident CHD sometime after their 10 year anniversary of follow up.
Genotyping and 1000 genomes Imputation
The Affymetrix 6.0 array was used to genotype all participants of the ARIC study. We used MACH and Minimac to phase and impute the individual level genotype data from ARIC to the latest build of the 1000 genomes project to minimize the need to search for and use proxies used in the construction of the GRS (Howie et al., 2012; Li et al., 2010). We used GTOOL (Genetics Software Suite, (c) 2007, The University of Oxford) to convert Minimac dosage files to best guess genotype calls.
GRS Construction
We considered four different choices for choosing SNPs to be part of the GRS. These were: p-value for the marginal association (8 p-values: ≥ 5 × 10−8, ≥ 1 × 10−5, ≥ 0.001, ≥ 0.01, ≥ 0.05, ≥ 0.1, ≥ 0.2, and ≥ 0.5), LD between SNPs, (4 values ≤ 0.2, ≤ 0.5, ≤ 0.8 and ≤ 1.0 (all)), imputation quality (5 values: ≥ 0.9, ≥ 0.8,. ≥ 0.5, ≥ 0.3, and ≥ 0 (all)) and allele coding of imputed SNPs (genotype or probabilities). P-values were derived from the CARDIoGRAM GWAS. We used the PLINK software (Purcell et al., 2007) to calculate pairwise unphased correlation (LD) between SNPs and obtained the imputation quality score from MACH. To choose SNPs for the GRS we:
Removed all SNPs below the imputation quality score
Rank ordered the SNPs based on p-value
Chose the top SNP from the list
Removed any SNPs that were in LD with that SNP based on the LD threshold
Repeated steps 3 & 4 until the p-value threshold was reached.
Constructed the GRS using either genotype calls or dosages of the imputed SNPs
Standardized the GRS to have mean 0 and variance 1.
We performed a grid search assessing each permutation of GRS construction for a total of 256 GRSs per person.
Analysis
We first assessed the degree of similarity between the various GRSs by calculating their pairwise correlations. Next, we assessed the performance of each GRS by testing it as a predictor of CHD within 10 years in the ARIC cohort using log-linear regression to estimate the relative risk (RR) for a 1-unit change in the GRS. In the regression, we controlled for all clinical risk factors that are part of the Framingham Risk Score: age, gender, blood pressure, cholesterol (Total and HDL), smoking status and diabetes (Wilson et al., 1998). Using 10-fold cross-validation, we calculated the c-statistic and calibration slope for predicting CHD from each GRS. A calibration slope of 1.00 indicates perfect calibration while values less than 1.00 suggest over-fitting and above 1.00 less than perfect calibration (Crowson et al., 2014). Finally we calculated the change in the c-statistic using 2000 bootstrap replications (Obuchowski & Lieber, 1998).
All statistical analyses were performed in R version 3.01 (R Core Team, 2012).
Results
ARIC cohort
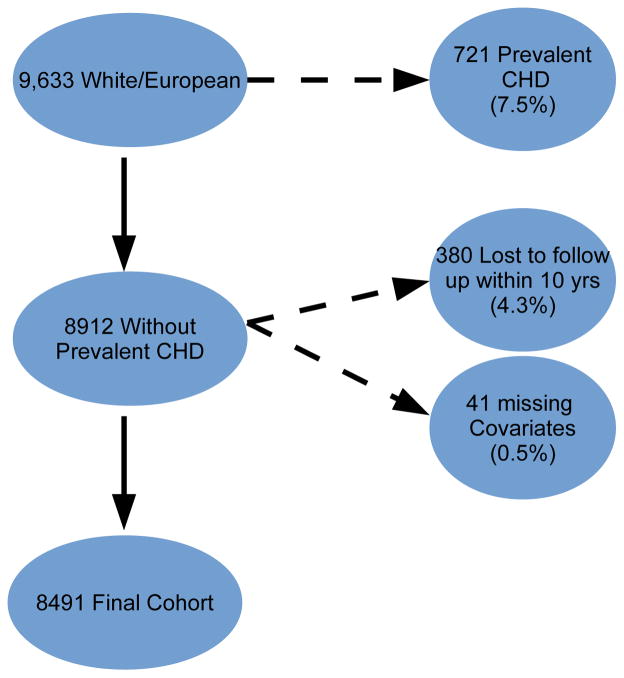
Of the 12,771 from the ARIC cohort with phenotypic and genotypic data, 9,633 (75%) were white/European (Figure 1). Among the remaining subjects, 721 (7.5%) had a history of CHD or CHF at baseline and were excluded from further analysis. Lastly, we excluded 380 people who were lost to follow-up or died of non-CHD related factors within 10 years and 41 people with missing covariate information, comprising a final cohort of 8,491 (Figure 1). Summary statistics of baseline characteristics for the ARIC subcohort used in our analyses are shown in Table 1.
Figure 1.
Flow diagram for inclusion of subjects in the study cohort
Table 1.
Characteristics of the ARIC subcohort used in analyses (n = 8491)
|
mean (IQR)
|
|
| Age (years) | 54 (49, 59) |
| SBP (mm/Hg) | 116 (106, 128) |
| DBP (mm/Hg) | 71 (65, 78) |
| HDL (mg/dL) | 48 (39, 61) |
| TC (mg/dL) | 211 (187, 238) |
|
count (%)
|
|
| white/European | 8491 (100) |
| Male | 3848 (45) |
| Diabetes | 626 (7.4) |
| Smoking status | |
| Current | 2010 (24) |
| Former | 2914 (34) |
| Never | 3567 (42) |
IQR = inter-quartile range, SBP = Systolic Blood Pressure, DBP = Diastolic Blood Pressure, HDL = High-Density Lipoprotein Cholesterol, TC = Total Cholesterol
SNP Data
Overall, 2,430,359 SNPs were in the CARDIoGRAM GWAS. Of these, 841,820 (35%) were directly genotyped within ARIC. Our 1,000 genomes imputation allowed us to imputed 30, 061,896 SNPs in ARIC, providing coverage of 2,393,551 (98%) of the CARDIoGRAM SNPs. Of these, 2,101,223 (88%) were common SNPs with MAF > 0.05 in ARIC.
GRS Scores and Improvement in Risk Prediction
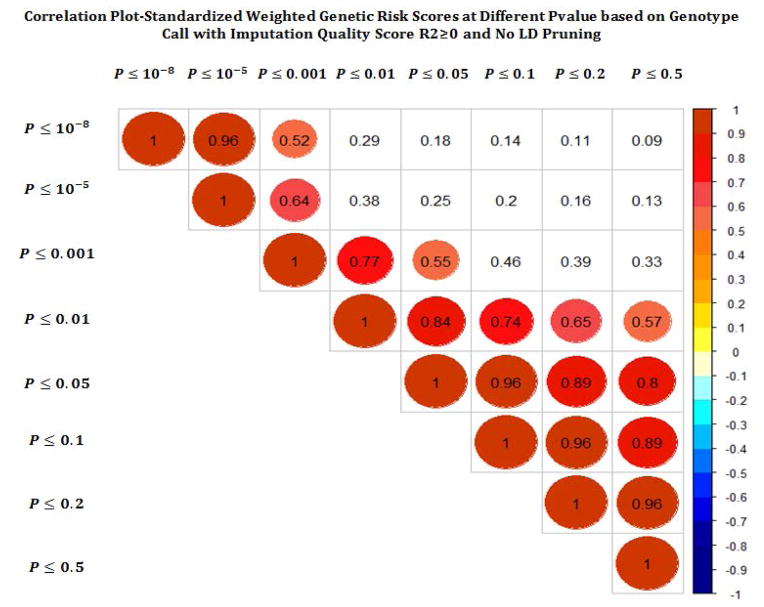
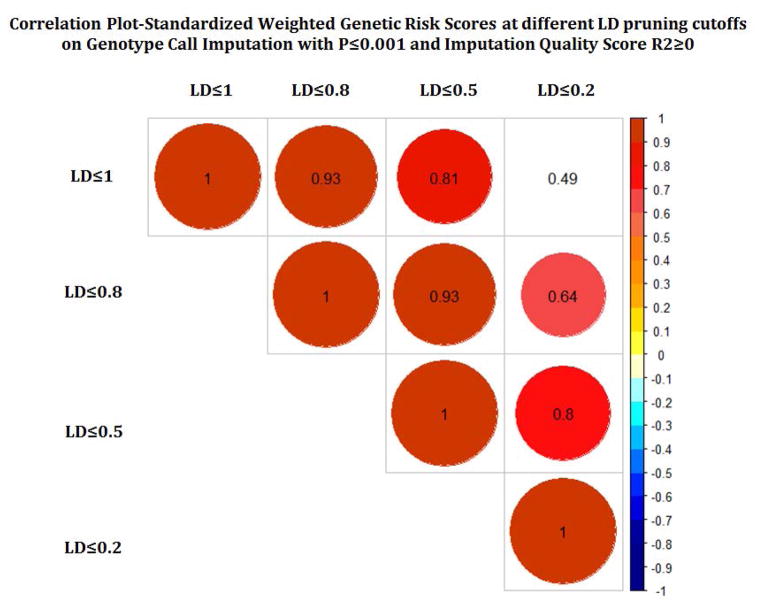
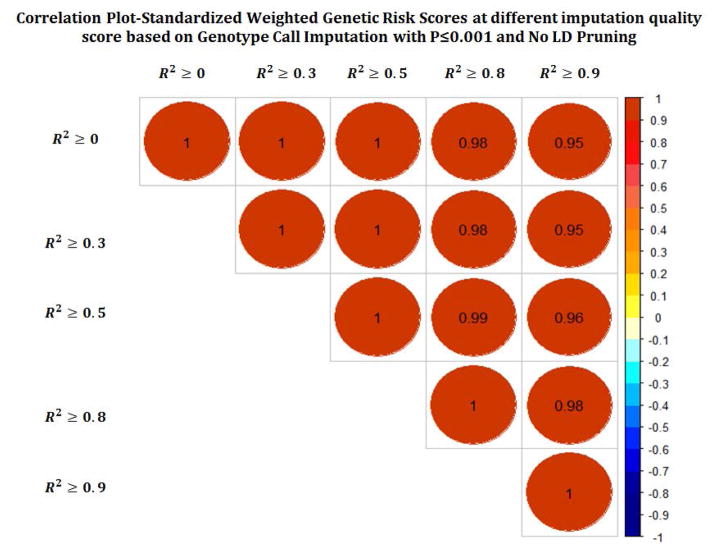
We created and compared a total of 256 GRS scores. Figures 2a–c show the correlation between different GRS scores based on different p-value, LD and imputation quality thresholds. While the correlation across different LD thresholds and different imputation quality scores were quite high, we observed more variability in correlations across the different p-value thresholds.
Figure 2.
Pairwise correlation across different GRS across while varying p-value threshhold (a), linkage disequilibrium (b), and imputation quality (c).
The predicted 10-year risk of developing CHD based on the FRS in this subcohort is 7.4% (interquartile range 4.3% to 12.3%), which coincides very well with the observed proportion that actually developed CHD (7.3%). The c-statistic and calibration slope for the prediction of CHD within 10-years using clinical factors alone was 0.774 (0.758, 0.791) and 3.12 (2.55, 3.69), respectively, suggesting moderately strong discrimination but suboptimal calibration. Table 2 shows the RR of CHD for a 1-standard deviation change in GRS for select models that we tested. Results for all models tested can be found in the supplement (Table S1). The RR for CHD varied from 1.08 to 1.28 for each standard deviation in GRS, with better performing GRSs having higher estimated RRs. Using 10-fold cross validation, we calculated the c-statistic for predicting CHD. The optimal GRSs improved the c-statistic by 0.009, while the worst did not improve the statistic at all. Most GRS constructions led to a significant improvement in discrimination compared to the baseline model with just clinical factors. All of the GRSs led to improved calibration, with a maximal improvement observed of 0.96 units translating to a 56% improvement of calibration. Not surprisingly there was some discrepancy between the optimal calibration and discrimination (Cook, 2007).
Table 2.
Select Results of GRS quality across different tuning parameters
| Parameters | #SNPs | Relative Risk | AUC | Calibration Score | |
|---|---|---|---|---|---|
| Pvaluea | |||||
| ≤ 5E-08 | 166 | 1.180(1.096, 1.270) | 0.777(0.761, 0.794)* | 2.684(2.067, 3.302) | |
| ≤ 0.00001 | 525 | 1.231(1.143, 1.325) | 0.780(0.763, 0.797)** | 2.563(1.905, 3.222) | |
| ≤ 0.001 | 7387 | 1.277(1.186, 1.375) | 0.783(0.766, 0.800)*** | 2.888(2.311, 3.465) | |
| ≤ 0.01 | 41017 | 1.213(1.126, 1.306) | 0.780(0.763, 0.796)** | 2.369(1.710, 3.028) | |
| ≤ 0.05 | 156193 | 1.215(1.127, 1.309) | 0.780(0.763, 0.796)** | 2.257(1.623, 2.891) | |
| ≤ 0.1 | 287398 | 1.201(1.114, 1.296) | 0.779(0.763, 0.796)** | 2.367(1.734, 3.001) | |
| ≤ 0.2 | 535188 | 1.179(1.090, 1.275) | 0.778(0.761, 0.794)* | 2.350(1.702, 2.998) | |
| ≤ 0.5 | 1244367 | 1.157(1.068, 1.253) | 0.777(0.760, 0.793)* | 2.473(1.806, 3.141) | |
|
| |||||
| Imputation Quality-R2b | |||||
| ≥ 0 | 7387 | 1.277(1.186, 1.375) | 0.783(0.766, 0.800)*** | 2.888(2.311, 3.465) | |
| ≥ 0.3 | 7205 | 1.272(1.181, 1.370) | 0.783(0.766, 0.799)*** | 2.915(2.346, 3.484) | |
| ≥ 0.5 | 6959 | 1.273(1.183, 1.371) | 0.783(0.766, 0.799)*** | 2.921(2.342, 3.501) | |
| ≥ 0.8 | 6015 | 1.264(1.174, 1.361) | 0.782(0.766, 0.799)** | 2.921(2.322, 3.520) | |
| ≥ 0.9 | 5055 | 1.252(1.162, 1.348) | 0.782(0.765, 0.798)** | 2.990(2.379, 3.602) | |
|
| |||||
| Ldc | |||||
| No LD prunning | 7387 | 1.277(1.186, 1.375) | 0.783(0.766, 0.800)*** | 2.888(2.311, 3.465) | |
| Prunned with LD R2≥ 0.8 | 2267 | 1.267(1.175, 1.368) | 0.782(0.765, 0.798)** | 2.350(1.781, 2.919) | |
| Prunned with LD R2≥ 0.5 | 1711 | 1.227(1.137, 1.324) | 0.780(0.763, 0.796)** | 2.340(1.746, 2.935) | |
| Prunned with LD R2≥ 0.2 | 1406 | 1.173(1.089, 1.264) | 0.777(0.761, 0.794)* | 2.377(1.825, 2.930) | |
Baseline c-stat 0.774(0.758, 0.791) and calibration score is 3.121(2.548, 3.693)
p < 0.05,
p < 0.01,
p < 0.001 compared to base model
R2 ≥ 0; no LD pruning
p-value ≤ 0.001; no LD pruning
p-value ≤ 0.001; R2 ≥ 0
We observed the greatest impact in GRS performance across different p-values. For discrimination, the optimal performance was observed at a threshold of 0.001. This threshold also corresponded to the largest estimated RR. A p-value cut point of 0.001, would correspond to a Benjamini-Hochberg false discovery rate (FDR)of 0.323 (Yoav Benjamini and Yosef Hochberg), beyond typical standards for statistical significance. On the other hand, calibration was optimized at a p threshold of around 0.01 – 0.05. Both showed decreased performance at very stringent and liberal thresholds. The least amount of variability in GRS quality was observed across different imputation metrics, with also little difference between dosage and genotype based calls. For LD, increasing the pruning threshold led to a slight deterioration in quality, with the relative risk and discrimination dropping with more pruning, but the calibration being slightly higher with some LD pruning.
Discussion
We illustrate an approach to empirically determining the optimal set of SNPs for a GRS. The focus is on performing a grid-search across the various tuning parameters (p-value, LD, imputation quality, and imputation type) and using cross-validation to select the optimal combination. The primary conclusions are that the addition of a GRS improves the overall model and that the GRSs are relatively robust to different specifications. Aside from extreme values, the results were relatively similar.
For 10 year risk prediction of CHD in the ARIC cohort using SNPs derived from the CARDIoGRAM consortium GWAS of ~2.5 M HapMap SNPs, we found optimal discrimination was achieved using a GRS that included all SNPs in CARDIoGRAM with a p < 0.0001 without a need to remove SNPs in high LD and without a need to filter by low imputation quality. Since each disease is likely to have a varying degree of evidence supporting the set of SNPs useful for risk prediction, we emphasize that our optimal parameterization is not generalizable to other outcomes. Moreover, even within a given a disease (such as CHD) it is possible that results will be different among different cohorts since they will each have different case mixes. However, our findings suggest, instead of arbitrarily selecting different cut-points an empirical evaluation is warranted to select the optimal cut-points.
Investigators have carefully considered how best to select a set of SNPs for a GRS. Wu et al.(Wu et al., 2013) examined this question in the context of a single study, where one wants to perform both discovery and prediction and found that estimating the effect size of SNPs was a critical part of the process, an issue we did not need to struggle with because we simply applied the weights from the CARDIoGRAM GWAS. Others (Purcell et al., 2009; Simonson et al., 2011) have suggested that the typical approach of using genome wide significance may not be sufficient, a finding that is somewhat confirmed in our analysis, though not to the extent that they report. In our case we found the optimal p-value threshold when considering RR to be 0.001. This corresponded to a Benjamin-Hochberg FDR of 0.323, suggesting there is value in incorporating what are still likely false associations. We anticipate that the optimal p-value threshold will strongly depend on the supporting data to create the GRS and the underlying genetic architecture of the trait of interest with a more polygenic trait benefiting from the addition of a larger fraction of the top SNP associations. In studies based on smaller studies, where the effects are less stable, it may be necessary to go further down the list to capture more of the genetic variability, where larger studies will be able to better estimate the true relationships. Ultimately the question of how far “down-the-list” is an important one that has implications beyond simply the optimal GRS. Our optimal score incorporated 7,387 SNPs. For a GRS computed off of a full genome chip, this is not a challenge. However, we can also envision a scenario where hospital systems may choose to create custom chips for their patients to capture a variety of GRSs. In this case, the slight loss in prediction quality may be worth creating a smaller, custom chip.
Imputation quality and LD level were less important determinants of GRS quality. The LD finding is counter to the findings of Wu et al, who noted LD pruning was beneficial. Theoretically, one would expect LD pruning to improve a GRS since LD violates one of the assumptions of the GRS construction: the independence of marginal effects. An increasingly important consideration is the role of imputation. In our data, 60% of the SNPs eligible for the GRS were imputed using 1000 genomes data. It is possible that as the percentage of the dataset that needs to be imputed increases, the imputation quality will matter more. Overall, the results suggested that we did not need to filter on imputation quality, as measured by r2. Similarly using genotype calls or dosages did not make a noticeable in quality. We note that over the long term, the need for imputation will diminish as more individuals undergo whole genome sequencing.
A very challenging matter is the evaluation of the clinical utility of a GRS. We considered only three potential evaluation metrics: relative risk, c-statistic, and calibration slope. These three metrics are interconnected. It has been noted that the Wald test on an odds ratio is a more powerful test than testing the increase in AUC (Demler et al., 2012; Pepe et al., 2013). While comparing GRSs are not technically nested tests, we did observe high correlation between the AUC and RR, with the RR showing more variability across constructions, Moreover, as noted previously, an increase in discrimination can often lead to a decrease in calibration (Cook, 2007), observed in our own results. Ultimately the choice of evaluation metric is an important consideration and partially depends on how the GRS will be used. If the goal is simply to estimate the strength of association of the GRS then RR is probably the best metric. However if one will be using the GRS as part of a clinical risk tool to group patients then discrimination should be considered. Calibration is most useful if one wants to give an individual accurate information about his or her risk of disease. Of course these metrics do not speak specifically to clinical utility, something that will ultimately need to be shown prospectively.
The process we suggest has implication both within analytic and clinical settings. As more genetic data becomes readily available researchers are using GRS in different ways. This includes assessing heritability (Chatterjee et al., 2013; Purcell et al., 2009), Mendelian Randomization studies (Palmer et al., 2012), and risk prediction (Weijmans et al., 2015). Having a means to optimally develop a GRS within a specific research context is important. In addition to the research context, the clinical environment is increasingly becoming “a learning environment.” With the proliferation of electronic health records and declining cost of genotyping, the incorporation of genetic data into clinical practice becomes more likely (Kannry and Williams, 2013). In such a setting it becomes important for the clinical system to properly validate the optimal scoring system to use, for their patient population.
Our analysis has several strengths and weaknesses. We took advantage of large GWAS data to get robust estimates of SNP association with CHD. Using an independent cohort we calculated a GRS and evaluated with cross-validation to minimize overfitting. While our results point out important considerations for creating a GRS it is important to note that this is only in one cohort and one disease. Our results will not necessarily directly translate to other diseases or even other cohorts. Instead, we suggest that others can be guided by our approach when constructing and testing their own GRS. Moreover, we have only considered GRS construction based on marginal associations. While this was necessary based on the use of outside source data, it is possible that approaches that used multivariable associations would provide additional insights (Goldstein et al., 2010).
Overall, our work helps to add an empirical approach to the construction and evaluation of a GRS. We suggest different criterion that one can assess and means to perform that assessment. As GRSs become more commonplace there will be more of a need for each user to determine the optimal set of parameters for their specific analysis.
Supplementary Material
Acknowledgments
Funding Sources
Benjamin A. Goldstein is supported by an NIH career development award K25DK097279. Themistocles L. Assimes is supported by an NIH career development award K23DK088942.
Contributor Information
Benjamin A. Goldstein, Department of Biostatistics and Bioinformatics, Duke University, Durham, NC.
Lingyao Yang, Quantitative Sciences Unit, Stanford School of Medicine, Palo Alto, CA.
Elias Salfati, Division of Cardiovascular Medicine, Stanford School of Medicine, Palo Alto, CA.
Themistoclies L. Assimes, Division of Cardiovascular Medicine, Stanford School of Medicine, Palo Alto, CA.
References
- 1.Amin N, van Duijn CM, Janssens ACJW. Genetic scoring analysis: a way forward in genome wide association studies? Eur J Epidemiol. 2009;24:585–587. doi: 10.1007/s10654-009-9387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45:400–405. 405e1–e3. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 4.Crowson CS, Atkinson EJ, Therneau TM. Assessing calibration of prognostic risk scores. Stat Methods Med Res. 2014 doi: 10.1177/0962280213497434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demler OV, Pencina MJ, D’Agostino RB. Misuse of DeLong test to compare AUCs for nested models. Stat Med. 2012;31:2577–2587. doi: 10.1002/sim.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein BA, Hubbard AE, Cutler A, Barcellos LF. An application of Random Forests to a genome-wide association dataset: Methodological considerations & new findings. BMC Genetics. 2010;11:49. doi: 10.1186/1471-2156-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein BA, Knowles JW, Salfati E, Ioannidis JPA, Assimes TL. Simple, standardized incorporation of genetic risk into non-genetic risk prediction tools for complex traits: coronary heart disease as an example. Front Genet. 2014;5:254. doi: 10.3389/fgene.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemani G, Shakhbazov K, Westra HJ, Esko T, Henders AK, McRae AF, Yang J, Gibson G, Martin NG, Metspalu A, et al. Detection and replication of epistasis influencing transcription in humans. Nature. 2014;508:249–253. doi: 10.1038/nature13005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, Aubin C, Bauer D, Heubach JF, Sandbrink R, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Hum Mol Genet. 2011;20:R182–R188. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannry JL, Williams MS. Integration of genomics into the electronic health record: mapping terra incognita. Genet Med. 2013;15:757–760. doi: 10.1038/gim.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, Davey Smith G, Sterne JAC. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21:223–242. doi: 10.1177/0962280210394459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepe MS, Kerr KF, Longton G, Wang Z. Testing for improvement in prediction model performance. Stat Med. 2013;32:1467–1482. doi: 10.1002/sim.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obuchowski, Lieber Confidence intervals for the receiver operating characteristic area in studies with small samples. Academic Radiology. 1998;5:561–571. doi: 10.1016/s1076-6332(98)80208-0. [DOI] [PubMed] [Google Scholar]
- 21.R Core Team. R: A language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 22.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonson MA, Wills AG, Keller MC, McQueen MB. Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Med Genet. 2011;12:146. doi: 10.1186/1471-2350-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, Kraft P, Chen R, Kallberg HJ, Kurreeman FAS, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamler J, Stamler R, Neaton JD, Wentworth D, Daviglus ML, Garside D, Dyer AR, Liu K, Greenland P. Low risk-factor profile and long-term cardiovascular and noncardiovascular mortality and life expectancy: findings for 5 large cohorts of young adult and middle-aged men and women. JAMA. 1999;282:2012–2018. doi: 10.1001/jama.282.21.2012. [DOI] [PubMed] [Google Scholar]
- 26.Thanassoulis G, Vasan RS. Genetic cardiovascular risk prediction: will we get there? Circulation. 2010;122:2323–2334. doi: 10.1161/CIRCULATIONAHA.109.909309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D’Agostino RB, Hwang SJ, O’Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113–121. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weijmans M, de Bakker PIW, van der Graaf Y, Asselbergs FW, Algra A, Jan de Borst G, Spiering W, Visseren FLJ SMART Study Group. Incremental value of a genetic risk score for the prediction of new vascular events in patients with clinically manifest vascular disease. Atherosclerosis. 2015;239:451–458. doi: 10.1016/j.atherosclerosis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White AD, Folsom AR, Chambless LE, Sharret AR, Yang K, Conwill D, Higgins M, Williams OD, Tyroler HA. Community surveillance of coronary heart disease in the Atherosclerosis Risk in Communities (ARIC) Study: methods and initial two years’ experience. J Clin Epidemiol. 1996;49:223–233. doi: 10.1016/0895-4356(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 32.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 33.Wray NR, Purcell SM, Visscher PM. Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biol. 2011;9:e1000579. doi: 10.1371/journal.pbio.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Pfeiffer RM, Gail MH. Strategies for developing prediction models from genome-wide association studies. Genet Epidemiol. 2013;37:768–777. doi: 10.1002/gepi.21762. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Yoav, Hochberg Yosef. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 57:289–300. [Google Scholar]
- 36.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 37.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 129:687–702. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.