Abstract
The excessive metastatic propensity of melanoma makes it the most deadly form of skin cancer, yet the underlying mechanism of metastasis remains elusive. Here, mining of cancer genome datasets discovered a frequent loss of chromosome 19p13.3 and associated down-regulation of the zinc finger transcription factor ZBTB7A in metastatic melanoma. Functional assessment of ZBTB7A-regulated genes identified MCAM, which encodes an adhesion protein key to melanoma metastasis. Using an integrated approach, it is demonstrated that ZBTB7A directly binds to the promoter and transcriptionally represses the expression of MCAM, establishing ZBTB7A as a bona fide transcriptional repressor of MCAM. Consistently, down-regulation of ZBTB7A results in marked upregulation of MCAM and enhanced melanoma cell invasion and metastasis. An inverse correlation of ZBTB7A and MCAM expression in association with melanoma metastasis is further validated with data from analysis of human melanoma specimens.
Implications
Together these results uncover a previously unrecognized role of ZBTB7A in negative regulation of melanoma metastasis and have important clinical implications.
Keywords: melanoma, metastasis, 19p13.3, ZBTB7A, MCAM
Introduction
Melanoma is a malignant tumor of melanocytes and is classified by stage, based primarily on degree of local invasion or metastatic status of the malignant melanocytes. The prognosis of melanoma drops sharply as the stage progresses, and metastasis is primarily responsible for the poor prognosis of cutaneous melanoma (1). Melanoma is thought to progress through multiple steps prior to distant metastatic spread to organs such as the liver and lung (2). The molecular changes associated with the transition of melanoma cells through to the most advanced phases are not fully understood.
In order to metastasize from primary sites, tumor cells must attach and invade through endothelial cells, implicating the critical roles cell adhesion molecules play in the metastatic progression of melanoma (3). One such cell adhesion molecule, Melanoma Cell Adhesion Molecule (MCAM) is widely known to play a pivotal role in the metastatic progression of melanoma (4). Reported as a valuable prognostic marker for melanoma, MCAM is highly expressed in advanced stage melanoma but not in benign melanocytes (5–7). Furthermore, forced expression of MCAM in non-metastatic melanoma cells strongly enhances metastatic potential while MCAM antibody treatment significantly suppresses metastatic progression of human melanoma in vivo (8,9). However, how MCAM expression is upregulated during human melanoma progression is not well understood.
Genetic alterations, such as recurrent chromosomal alterations, can be primary causes for many human cancers (10). Chromosomal focal amplifications or deletions often produce copy number variation (CNV) of genes, which may contribute to tumor progression. These chromosomal alterations can lead to deregulation of gene structure, function, and expression that functionally contribute to the pathogenesis of cancer (10). A recent study by The Cancer Genome Atlas (TCGA) Pan-Cancer Analysis Working Group performed an integrative analysis of somatic copy number alterations across 12 tumor types and provided a public resource of highly curated data and information (11). We performed data mining of the TCGA database and identified 19p13.3 as a region of frequent chromosomal deletion in metastatic melanoma. To investigate the biological significance of this chromosomal loss to melanoma, we analyzed genes localized within this region and discovered that ZBTB7A, a transcriptional repressor with Zinc-finger and BTB/POZ domains, was significantly down-regulated during progression from normal skin and benign nevus to late stage melanoma. We demonstrate that ZBTB7A suppresses melanoma metastasis by transcriptionally repressing the critical melanoma progression molecule MCAM in vitro and in vivo, implicating ZBTB7A as a novel tumor suppressor against melanoma metastasis.
Materials and Methods
Promoter mutation
ZBTB7A binding sites in human MCAM promoter region were mutated with Q5® Site-Directed Mutagenesis Kit (New England Biolabs). Mutation primers were designed using NEBaseChanger software provided by the Q5 Mutagenesis Kit. Mutation PCR, Kinase-Ligase-DpnI (KLD) enzyme treatment and transformation were performed according manufacturer’s instruction. All the mutations have been confirmed via sequencing.
Luciferase assay
293 or SK-MEL-28 cells were cultured in 48 well-plate, transfection were started when cell density was approximately 50% confluency. 20 ng of pGL3-MCAM vector, 10ng of pRL-CMV Renillar luciferase reporter and pcDNA3.1 control or increasing dose (5–30ng) of pcDNA3.1-ZBTB7A expression plasmid were co-transfected into 293 cells. After 36 hours, the luciferase activity was measured with the dual luciferase reporter assay system (Promega) according to manufacturer’s instruction.
Tissue Microarray analysis
The Tissue Microarray ME2082 and ME1003 used in the study was purchased from US Biomax, stained and analyzed at the Pathology and Molecular Cytology Core facilities. ME2082 contains 128 cases of primary malignant melanoma, 64 metastatic malignant melanoma, and 16 cases of adjacent normal tissue or normal tissue from autopsy. ME1003 contains 56 cases of malignant melanoma, 20 cases of metastatic malignant melanoma and 24 cases of benign nevi.
Lentiviral shRNA mediated knockdown
293T cells were seeded in 10-cm tissue culture plates, plasmid transfection was performed when the cell reach 60% confluence. Prior to transfection, 293T cells culture medium was changed to antibiotics free DMEM medium with 10% FBS. 10μg of shZBTB7A plasmid (Sigma, TRCN0000137332), shMCAM plasmid (Qiagen, KH00651N) or control non-targeting shRNA plasmid, 7.5μg of psPAX2 packaging plasmid and 2.5μg of pMD2.G envelope plasmid were co-transfected using 20ul Lipofectamine® 2000 Transfection Reagent (Invitrogen). The next day after transfection, the transfection medium were changed to fresh complete culture medium. The virus supernatant was collected every 12 hours, filtered through a 0.45μm filter, and frozen in −80°C until further usage. The virus titer was determined by serial dilution assay using 3T3 cells. For melanoma cell lentiviral transfection, 5×105 cells were seeded in 10-cm dish, incubated with virus at MOI=1 for 12 hours in the presence of 8 μg/ml polybrene (Sigma). 72 hours later, the cells were selected with 1μg/ml of puromycin (for shZBTB7A) for 3 days or 500 μg/ml of G418 (for shMCAM) for 1 week. The polyclonal cells after selection are used for downstream experiments.
Melanoma cell nude mice lung seeding assay
All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Harvard School of Public Health. 1×106 melanocytes of different genotype were injected through tail vein into nude mice (nu/nu, female, 6–8 weeks old, Charlies River Laboratories). 60 days later, nude mice lung were dissected, fixed with 4% formalin, embedded in paraffin blocks. Sections of lung were stained with hematoxylin and eosin. Degree of lung seeding was determined by histopathological scoring of infiltrating melanoma cells percentage among normal lung cells.
Statistical analysis
Log-rank tests were used for Kaplan Meier survival analysis. Student’s t-test was used for the comparisons of the means. Correlations of gene expression were determined using Pearson coefficient. All statistical tests were two-sided. P values of less than 0.05 were considered statistically significant.
Results
19p13.3 is frequently deleted in human melanoma, and ZBTB7A is the top down-regulated gene within this region
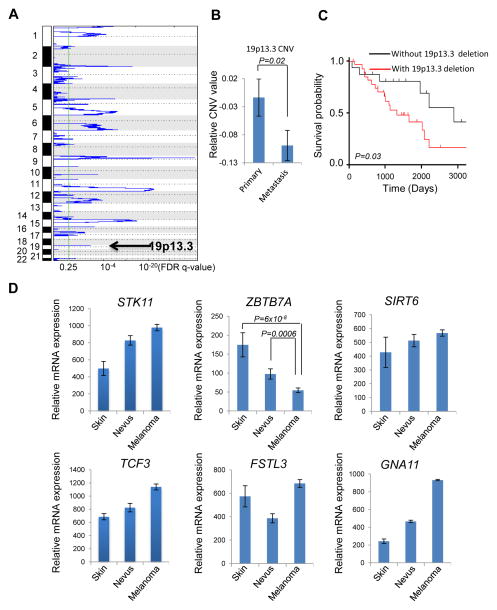
Genome wide SNP array analysis has uncovered several recurrent chromosomal alterations (12). We sought to understand the genetics of human melanoma metastasis by mining several human genomic databases and discovered 19p13.3 as one of the frequently deleted chromosomal regions, which is down-regulated in approximately 30% (47 out of total 158) of melanoma cases (Fig. 1A). Of note, chromosomal 19p13.3 is gradually lost along the progression of melanoma, as reflected by a significant drop from primary in situ melanoma to metastatic melanoma (Fig. 1B). Consistent with the close association of metastasis with mortality, data derived from TCGA Skin Cutaneous Melanoma datasets reveal that decreased chromosome 19p13.3 copy number correlates with poor prognoses in melanoma patients (Fig. 1C).
Figure 1.
19p13.3 deletion associated ZBTB7A down-regulation contributes to melanoma metastasis and poor prognosis. A, list of significant chromosomal focal deletions in human melanoma based on The Cancer Genome Atlas (TCGA) skin cutaneous melanoma SNP array dataset. GISTIC q-values (x-axis) are plotted across different chromosome (y-axis). B, 19p13.3 CNV status in primary and metastatic melanoma based on TCGA SNP array database, error bars represent mean ± s.e.m. C, kaplan–Meier overall survival curve of patients with and without 19p13.3 deletion based on TCGA melanoma dataset. D, expression of 19p13.3 cancer genes STK11, TCF3, FSTL3, GNA11, SIRT6 and ZBTB7A during the progression of human melanoma, error bars represent mean ± s.e.m.
To explore the critical genetic events associated with 19p13.3 loss in melanoma metastasis, we examined the expression of genes within this region relative to disease progression. Among the genes found within the 19p13.3 region (Supplementary Fig. S1), only a few were found to be significantly altered during disease progression according to two different datasets (13, 14) (Supplementary Fig. S2). ZBTB7A was one of the top down-regulated genes in both datasets (Supplementary Fig. S2), and importantly, its expression significantly decreases progressively from normal skin to nevus to melanoma (Fig. 1D). Known cancer genes as defined by The Sanger Institute’s Cancer Gene Census in the 19p13.3 region include STK11, GNA11, TCF3, FSTL3 and SIRT6 (15), none of which exhibited the same correlation as ZBTB7A, and no discernable pattern of regulation was detected during disease progression (Fig. 1D). In addition, no significant somatic mutations have been found in any of these known cancer genes in the TCGA skin cutaneous melanoma (SKCM) somatic mutation database (16), diminishing the likelihood that any of these genes functionally link to 19p13.3 loss during melanoma metastasis. The data together suggest ZBTB7A as a possible cancer gene candidate in 19p13.3 loss-associated melanoma.
ZBTB7A is down-regulated in human melanoma metastases
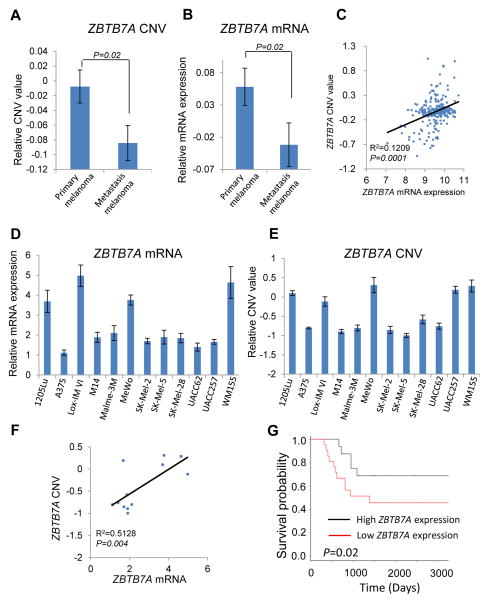
ZBTB7A (also known as POKEMON (17), FBI (18), LRF (19) and OCZF (20)) is a member of the POZ/BTB and Krüppel (POK) family of transcription factors. POK family transcription factors can bind DNA through Zinc finger domain and repress transcription by recruiting co-repressor complexes through the BTB (for BR-C, ttk and bab) domain (21). POK transcription factors have been recognized to be critical developmental regulators and have been directly implicated in human cancers (22–24). Analysis of copy number variation (CNV) values indicates that ZBTB7A copy number was significantly reduced in metastatic melanoma when compared with primary melanoma (Fig. 2A). In line with these CNV data, the mRNA expression levels of ZBTB7A also declined significantly upon disease progression (Fig. 2B), exhibiting a significant positive correlation between genomic CNV and mRNA expression of ZBTB7A (Fig. 2C). To corroborate these data derived from the human melanoma datasets, we analyzed a panel of 12 human melanoma cell lines. The result confirmed the correlation between the CNV and mRNA of ZBTB7A (Fig. 2D–2F), implicating CNV as a recurrent alteration at the ZBTB7A locus that contributes to down-regulation of its expression in human melanoma. Significantly, lower expression of ZBTB7A mRNA also correlates with poor prognosis (Fig. 2G).
Figure 2.
ZBTB7A is frequently down-regulated during the disease progression of human melanoma, and ZBTB7A expression correlates with metastatic propensity of melanoma. A and B, ZBTB7A copy number variation (CNV) values (A) and mRNA (B) in primary and metastatic melanoma based on TCGA dataset. Error bars represent mean ± s.e.m. C, correlation between ZBTB7A CNV and mRNA value in human melanoma samples based on TCGA dataset. D and E, relative values of ZBTB7A mRNA (D) and CNV (E) in 12 human melanoma cell lines (1205Lu, A375, Lox-IM VI, M14, Malme-3M, MeWo, SK-MEL-2, SK-MEL-5, SK-MEL-28, UACC62, UACC257, WM155). Average values of three independent experiments are shown with error bars represent mean ± s.e.m. F, correlation between ZBTB7A CNV and mRNA value in 12 human melanoma cell lines. G, kaplan–Meier overall survival curve of patients with high and low ZBTB7A expression level.
An inverse correlation between ZBTB7A and MCAM expression in human melanoma samples and cell lines
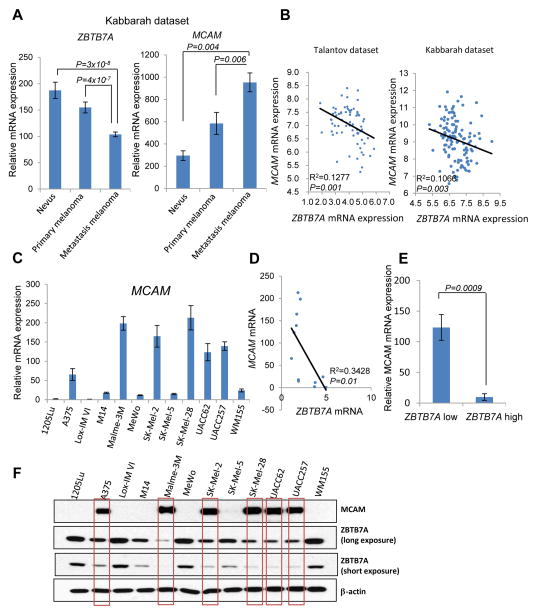
Given that ZBTB7A is a transcriptional repressor, we began to explore its role in melanoma development by identifying its potential target genes. Microarray analysis was performed to uncover differentially expressed genes between ZBTB7A proficient and deficient cells. Gene set enrichment analysis revealed cell adhesion molecules as a top gene signature enriched in differentially expressed genes (Supplementary Fig. S3A). Significantly, MCAM (Melanoma Cell Adhesion Molecule), a critical regulator of melanoma metastasis and progression, was found to be the top significantly up-regulated cell adhesion molecule in ZBTB7A deficient cells (Supplementary Fig. S3B and S4). MCAM, also known as MUC18, CD146, Mel-CAM, A32 antigen and S-Endo-1 Antigen, is a membrane glycoprotein, belonging to the immunoglobulin superfamily. It was previously identified as a marker of melanoma progression and metastasis (25).
The highly elevated MCAM expression in ZBTB7A-deficient cells prompted us to examine its expression levels in human clinical melanocytic lesions. In sharp contrast to the association of decreased ZBTB7A expression with melanoma metastasis, MCAM expression was seen to progressively increase from nevus to primary cutaneous melanoma to metastatic melanoma (Fig. 3A), and higher levels of MCAM expression were associated with poorer patient survival (Supplementary Fig. S5A), consistent with published findings (26). Indeed, mRNA transcripts of ZBTB7A and MCAM showed a clear inverse correlation based on data from two different databases (Fig. 3B). To complement these data, we analyzed the correlation between ZBTB7A and MCAM expression in a collection of melanoma cell lines (A374, Lox-IM VI, M14, Malme-3M, MeWo, SK-MEL-2, SK-MEL-5, SK-MEL-28, UACC62, WM155,1205Lu, UACC257). We observed that cell lines with low levels of ZBTB7A (A375, SK-Mel-2, UACC62 and UACC257) express approximately 20 fold more MCAM mRNA than cells with high ZBTB7A expression levels (1205Lu, Lox-IM VI, MeWo and WM155) (Fig. 3C–3E). This inverse correlation of ZBTB7A and MCAM expression was also apparent at the protein level (Fig. 3F). The data together suggested that ZBTB7A might suppress MCAM expression and thus modulate invasive behavior during melanoma progression.
Figure 3.
ZBTB7A expression negatively correlates with MCAM expression in both human melanoma patient samples and melanoma cell lines. A, the levels of ZBTB7A mRNA and MCAM mRNA in different stages of human melanoma were analyzed based on the values derived from Kabbarah dataset. Error bars represent mean ± s.e.m. B, the correlation between ZBTB7A and MCAM mRNA expression derived from two different human melanoma datasets was shown. C, the mRNA levels of 12 human melanoma cell lines were assessed by real time PCR, 18s served as internal control for the analysis. Average values of three independent experiments are shown as mean ± s.e.m. D and E, the correlation of ZBTB7A mRNA with MCAM mRNA in 12 human melanoma cell lines was analyzed. ZBTB7A low expression cell lines (A375, SK-Mel-2, UACC62 and UACC257) express an average 12-fold more MCAM than ZBTB7A high expression cell lines (1205Lu, Lox-IM VI, MeWo and WM155). F, ZBTB7A and MCAM protein expression detected by Western blot in 12 different human melanoma cell lines, β-actin served as loading control.
ZBTB7A transcriptionally represses MCAM expression in vitro and in vivo
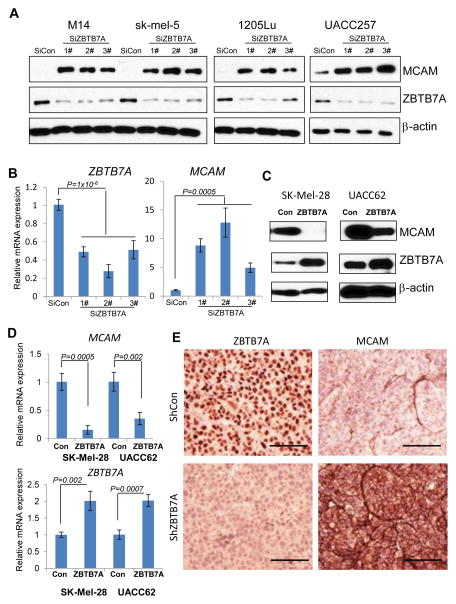
To test the hypothesis that ZBTB7A transcriptionally represses MCAM expression, we knocked down the expression of ZBTB7A with 3 independent siRNA sequences in multiple melanoma cell lines M14, SK-Mel-5, 1205Lu and UACC257. ZBTB7A knockdown was associated with a robust induction of the expression of MCAM at both protein and mRNA levels in all 4 melanoma cell lines (Fig. 4A and B), supporting a ZBTB7A-dependent regulation of MCAM expression. To complement the knockdown approach, we overexpressed ZBTB7A in two melanoma cell lines SK-Mel-28 and UACC62 that express a relatively low endogenous level of ZBTB7A. The expression of MCAM protein and mRNA was strongly suppressed upon introduction of exogenous ZBTB7A (Fig. 4C and D). These results are consistent with the hypothesis that ZBTB7A suppresses the transcription of MCAM. To further validate the ZBTB7A-dependent regulation of MCAM expression, we generated ZBTB7A stable knockdown and control M14 melanoma cells using shRNA and implanted the cells into nude mice. Immunohistochemical staining of tumor sections derived from implanted cells confirmed that knockdown of ZBTB7A was associated with a robust increase in MCAM expression (Fig. 4E), confirming that ZBTB7A can regulate the expression of MCAM in vivo.
Figure 4.
ZBTB7A regulates the expression of MCAM in vitro and in vivo. A, the indicated melanoma cell lines were transfected with control (SiCon) or 3 independent ZBTB7A targeting siRNAs. The cells were harvested 48h after transfection and analyzed by immunoblot with MCAM and ZBTB7A specific antibodies. β-actin served as loading control. B, the mRNA levels of ZBTB7A and MCAM were assessed by real time PCR in M14 cells after ZBTB7A knockdown with three different siRNA. Data shown represent mean ± s.d of three experiments. C, retrovirus mediated ZBTB7A overexpression in melanoma cell lines SK-MEL-28 and UACC62. Immunoblot was performed with MCAM and ZBTB7A specific antibodies. β-actin served as internal control for loading. D, the mRNA levels of ZBTB7A and MCAM were assessed by real time PCR in SK-MEL-28 and UACC62 cells after retrovirus mediated ZBTB7A overexpression. Data shown are mean ± s.d of three experiments. E, M14 cells stably expressing lentiviral shZBTB7A and shcontrol vector were subcutaneous injected into nude mice. The developed tumors were dissected, fixed, and stained with ZBTB7A or MCAM specific antibody. Bar, 50μm.
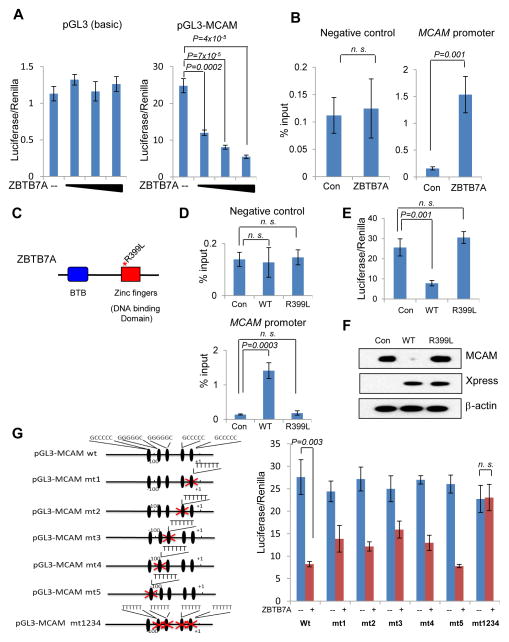
We next investigated the mechanism by which ZBTB7A represses the transcription of MCAM. It has been reported that POK family proteins can bind to DNA via the Zinc finger domain and repress the transcription by recruiting a co-repressor complex to the promoter of target genes (27). We asked whether ZBTB7A might repress expression of MCAM by binding to its promoter. To address this question, we created a human MCAM promoter-driven luciferase plasmid and co-transfected it with an increasing amount of ZBTB7A. Luciferase assays showed that ZBTB7A efficiently repressed human MCAM reporter activity in a dose-dependent manner in different cell types including melanoma cells (Fig. 5A and Supplementary Fig. S6A). We then performed chromatin immunoprecipitation (ChIP) to test whether ZBTB7A binds directly to the MCAM promoter. FLAG-ZBTB7A plasmids were stably expressed in HeLa cells at levels similar to that of endogenous ZBTB7A (Supplementary Fig. S6B) and ChIP was performed using an anti-FLAG antibody conjugated to agarose beads. Three different primer sets were designed to amplify the MCAM promoter region. The FLAG antibody specifically precipitated MCAM promoter sequences but not the control genomic sequence from cell nuclear extracts (Fig. 5B and Supplementary Fig. S6C). To additionally prove that ZBTB7A suppressed the expression of MCAM via directly binding to its promoter, we generated a Zinc finger (R399L) mutant of ZBTB7A (Fig. 5C), which is defective in binding to the MCAM promoter, as confirmed by ChIP assay (Fig. 5D). Indeed, unlike its wild type counterpart, ZBTB7A (R399L) failed to repress the expression of MCAM, as shown in the luciferase-based assay (Fig. 5E) as well as Western blot analysis (Fig. 5F).
Figure 5.
ZBTB7A binds to human MCAM promoter and directly represses the transcription of MCAM. A, human MCAM promoter was cloned into pGL3-Basic vector. Luciferase assay was carried out to assess the effects of increasing amount of ZBTB7A on the luciferase activity of pGL3-Basic and pGL3-MCAM vector in 293 cells. Data shown are mean ± s.d. of three independent experiments. B, chromatin immunoprecipitation assay were performed with anti-FLAG M2 beads in FLAG-ZBTB7A or FLAG control expressing HeLa cells. The abundance of DNA with MCAM promoter region and minimum protein binding intra-genic DNA were measured by quantitative real time PCR with each specific primer. Data shown are mean ± s.d. from three independent experiments. C, domain architecture of ZBTB7A protein and the design of R399L mutant. D, Xpress tagged wild type ZBTB7A (WT), R399L mutated ZBTB7A (R399L) and control Xpress vector were transfected in SK-Mel-28 melanoma cells, chromatin immunoprecipitation was performed with Xpress Ab, Data are mean ± s.d. of three independent experiments. E, luciferase assay as described in A was performed to assess the effect of R399L mutant on the activity of pGL3-MCAM reporter. Data are mean ± s.d. of three independent experiments. F, the expressions of MCAM and transfected Xpress-tagged WT or R399L ZBTB7A in SK-Mel-28 melanoma cells were analyzed by Western blot. G, human MCAM promoter has five conserved putative ZBTB7A binding sites, each of these ZBTB7A binding sites was mutated individually or in combination as shown in left panel. Luciferase assay as described in A was performed to assess each mutant. Data are mean ± s.d. of three independent experiments.
To further define the direct binding of ZBTB7A to the MCAM promoter, we made an attempt to identify the specific binding site. ZBTB7A binds to GC rich DNA sequence (17). Both human and mouse MCAM promoters are highly GC rich, and contain multiple putative ZBTB7A binding sites, five of which are shared by human and mouse MCAM (Supplementary Fig. S6D and S6E), these binding sites were named site 1 to site 5 based on their distance from the transcription start sites. To identify the functionally important binding site(s), we mutated each of the five putative binding sites in the reporter construct and performed luciferase assays (Fig. 5G). As shown in Figure 5G, mutation of putative ZBTB7A binding sites 1,2,3 and 4 significantly but incompletely attenuated the ability of ZBTB7A to bind to the MCAM promoter and repress its activity, whereas mutation of the putative ZBTB7A binding site 5 had little effects. Furthermore, when putative ZBTB7A binding sites 1 through 4 are mutated simultaneously, the ability of ZBTB7A to repress MCAM promoter was completely abolished (Fig. 5G). The results together establish ZBTB7A as a bona fide transcription repressor of MCAM.
ZBTB7A regulates melanoma cell adhesion, invasion and metastasis via repression of MCAM expression
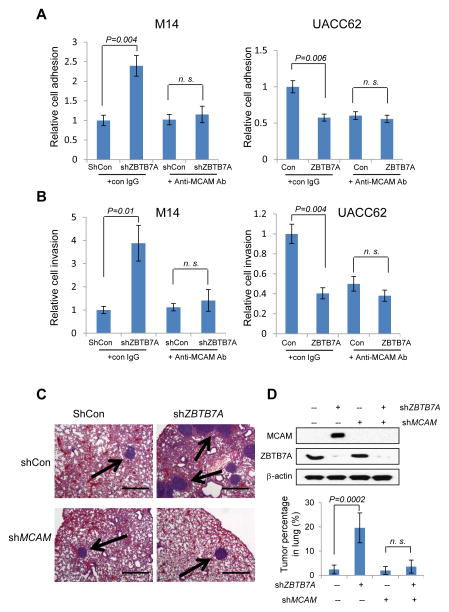
Having shown that ZBTB7A is a transcriptional repressor of MCAM, we asked whether ZBTB7A inhibits melanoma metastasis through inhibition of the expression of MCAM. MCAM is expressed not only in melanoma cells but also in the endothelia cells of melanoma associated blood vessels, and is reported to mediate melanoma cell endothelia cell adhesion and metastasis (28). We thus tested the functional consequence of ZBTB7A-mediated regulation of MCAM expression by examining melanoma cell adhesion onto Human umbilical vein endothelial cells (HUVEC). Two melanoma cell lines, M14 and UACC62 that express relatively low and high levels of MCAM respectively, were used to experimentally test the effects of ZBTB7A knockdown or overexpression. Lentiviral-mediated expression of shZBTB7A in M14 cells resulted in considerable increase in M14 cell adhesion to HUVEC cells (Fig. 6A, left). This increased adhesion was completely abolished by anti-MCAM antibody (relative to control antibody), demonstrating that the effect is mediated by MCAM (Fig. 6A, left). Overexpression of ZBTB7A in UACC62 cells, on the other hand, was associated with a considerable reduction of cell adhesion to HUVEC (Fig. 6A, right). These data together indicate that by controlling MCAM expression, ZBTB7A plays a critical role in regulation of melanoma cell adhesion to HUVEC cells. A matrigel-based invasion assay was also carried out to assess the role for ZBTB7A-dependent regulation of MCAM expression. The results indicate that ZBTB7A knockdown stimulated cell invasion and this effect of ZBTB7A-deficiency was mitigated by the use of the anti-MCAM antibody. Conversely, ZBTB7A overexpression suppressed cell invasion (Fig. 6B). Finally, we tested the role of ZBTB7A in melanoma metastasis using a nude mouse lung seeding assay. When compared to control shRNA expressing M14 cells, the shZBTB7A expressing melanoma cells developed much more lung metastasis, which was completely diminished by MCAM knockdown (Fig. 6C). Quantitative analysis of the lung tumor nodules revealed an approximate 5-fold increase in lung metastasis in ZBTB7A-deficient melanoma cells and remarkably, this increase was nearly completely eliminated when MCAM expression was knocked down (Fig. 6D). Together, our data demonstrate a critical role of ZBTB7A in control of melanoma metastasis and this transcription repressor does so via regulation of the expression of MCAM.
Figure 6.
ZBTB7A suppresses melanoma cell adhesion, invasion and in vivo metastasis by repressing MCAM. A, ZBTB7A was stably knocked down using lentiviral shRNA in M14 melanoma cell line that expresses a relatively high level of ZBTB7A. The cells were subject to a HUVEC cell adhesion assay in the absence or presence of anti-MCAM antibody (Left panel). Retrovirus mediated ZBTB7A overexpression in UACC62 melanoma cells that express a relatively low level of ZBTB7A. The cells were similarly subject to HUVEC cell adhesion assay. Data are mean ± s.d. of three independent experiments. B, the ZBTB7A stable knockdown M14 cells as in A were assessed for cell invasion through matrigel, again in the absence or presence of anti-MCAM antibody (Left panel). ZBTB7A overexpressing UACC62 cells were similarly assessed for cell invasion through matrigel (Right panel). Data are mean ± s.d. of three independent experiments. C, M14 melanoma cells stably expressing ShCon, shZBTB7A or shMCAM were generated and the expression levels of MCAM and ZBTB7A were analyzed by Western blot (D). The cells were injected into nude mice through tail vein. The animals were harvested 60 days later and lungs were collected for H&E staining. Representative H&E images of nude mice lung are shown. Bar, 500μm. Arrows indicate the lung nodules formed by melanoma cells. The tumor nodules were quantified and presented as percentage in each lung. The numbers represent mean ± s.d from 5 mice/group (D).
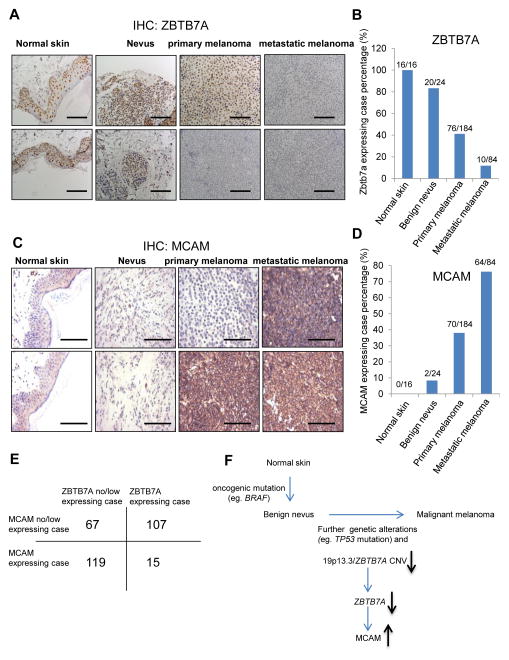
To further confirm the association of ZBTB7A and human melanoma, we examined the expression of ZBTB7A protein in a cohort of human melanoma samples by immunohistochemistry staining. The results indicate a gradual loss of ZBTB7A protein expression during progression of melanoma. The expression of ZBTB7A is readily detectable in normal skin and benign nevi, considerably reduced in melanoma in situ, and almost undetectable in metastatic melanoma (Fig. 7A), displaying an inverse correlation with ZBTB7A expression and melanoma progression (Fig. 7B). The expression of MCAM, on the other hand, exhibited a positive correlation with disease progression. The MCAM level was barely detectable in normal skin but increased progressively from nevus to primary melanoma to metastatic melanoma (Fig. 7C–E). The inverse correlation of ZBTB7A and MCAM expression in association with melanoma progression is in agreement with that uncovered from the TCGA database.
Figure 7.
An inverse correlation between ZBTB7A and MCAM in human melanoma. A, detection of ZBTB7A protein by immunohistochemistry in tissue microarrays of human normal skin, benign nevus, primary and metastatic melanoma samples. Bar, 50μm. B, statistical analysis of ZBTB7A expression levels in normal skin, benign nevus, primary and metastatic melanoma tissues. ZBTB7A is expressed in 100% (16 of 16) normal skin, 83% (20 of 24) benign nevus, 41% (76 of 184) primary melanoma and 12% (10 of 84) metastatic melanoma. C, detection of MCAM protein by immunohistochemistry in tissue microarrays of human normal skin, benign nevus, primary and metastatic melanoma samples. Bar, 50μm. D, statistical analysis of MCAM expression levels in normal skin, benign nevus, primary and metastatic melanoma samples. MCAM is expressed in 0% (0 of 16) normal skin, 8% (2 of 24) benign nevus, 38% (70 of 184) primary melanoma and 76% (64 of 84) metastatic melanoma. E, an inverse-correlation between ZBTB7A and MCAM expression in tissue microarray samples. F, model for the function of ZBTB7A in the regulation of melanoma progression and metastasis.
Discussion
The cancer genomic projects have produced rich data serving as valuable resources for cancer research (12). We conducted data mining of the TCGA database and identified 19p13.3 as a recurrently deleted region in melanoma. We discovered decreased copy number of ZBTB7A, a gene localized within 19p13.3, as a major genetic event with a measurable biological significance. Consistent with its function as a transcription repressor, we identified melanoma cell adhesion molecule (MCAM) as a target gene under the control of ZBTB7A. Indeed, in vitro and in vivo evidence showed that down-regulation of ZBTB7A resulted in robust induction of MCAM, promoting melanoma metastasis.
ZBTB7A is a POK family transcription repressor that is best known for its proto-oncogenic role through repressing the expression of tumor suppressor ARF (17). The finding that 19p13.3 loci containing ZBTB7A is frequently lost in human melanoma seems to be inconsistent with an oncogenic role of ZBTB7A in skin tumorigenesis. Analysis of several melanoma genomics databases revealed that the expression levels of ZBTB7A decrease progressively among normal skin, nevi, primary melanoma and metastatic melanoma, largely due to a reduction of copy number caused by 19p13.3 loss. Significantly, among the genes localized within 19p13.3, ZBTB7A was found to be the gene whose expression exhibits a reverse correlation with melanoma progression, arguing against an oncogenic role of ZBTB7A in human melanoma. Consistent with this notion is the finding that, similar to 19p13.3 loss, reduced expression of ZBTB7A is closely associated with poor prognosis of melanoma patients.
ZBTB7A is a known transcriptional repressor (17). Gene set enrichment analysis of differentially expressed genes between ZBTB7A-proficient and deficient cells revealed cell adhesion molecules as the most enriched gene signature. MCAM, a critical regulator of melanoma metastasis and progression, was among the significantly up-regulated genes in ZBTB7A deficient cells. Using a combination of loss- and gain-of-function approaches, we confirmed ZBTB7A-dependent control of MCAM expression. The multiple GC rich sequences matching consensus-binding sites for ZBTB7A were found in the MCAM promoter, and provide a structural basis for ZBTB7A-mediated regulation. Indeed, data obtained from experiments using complementary approaches including ChIP and site-directly mutagenesis demonstrated direct binding of ZBTB7A to the MCAM promoter, thereby establishing ZBTB7A as a bona fide transcriptional repressor of MCAM. Interestingly, the MCAM genomic locus is rarely amplified (Supplementary Fig. S5B), supporting transcriptional regulation as the major mechanism of MCAM control. The importance of ZBTB7A-mediated suppression of MCAM expression in human melanoma is further supported by data derived from our analysis of a cohort of human melanoma patient samples, which showed a clear inverse correlation of ZBTB7A and MCAM expression in association with melanoma progression. Recently we identified a tumor suppressive function of ZBTB7A by repressing the transcription of glycolysis genes (29). In human colon cancers, we observed significantly reverse correlation between ZBTB7A and glycolysis genes (29), while in melanoma ZBTB7A do not show significant reverse correlation with glycolysis genes, and similarly we do not observe the reverse correlation between ZBTB7A and MCAM in human colon cancers. Most importantly depletion of MCAM nearly completely rescued the melanoma metastasis phenotype of ZBTB7A deficient melanoma cells in vitro and in vivo. Our study indicates cancer type specific mechanisms for ZBTB7A in cancer suppression, and MCAM play a major function in ZBTB7A regulated melanoma metastasis.
MCAM has been identified as a major contributor to melanoma progression and metastasis. This cell adhesion protein is highly expressed in a wide range of advanced and metastatic melanoma, but is rarely expressed in benign nevus cells or normal melanocytes (26), implicating that the upregulation of MCAM is late event during melanoma progression. We also analyzed the genetic information of the cell lines used in this study for the association. Among the melanoma lines, MeWo harbors wild type BRAF, NRAS and PTEN; A375, Lox-IM VI, M14, SK-MEL-2, SK-MEL-5, WM155, and UACC257 have mutations in BRAF or NRAS, but wild type for PTEN; SK-MEL-28 and UACC62 have mutations in BRAF or NRAS, and homozygous or heterozygous loss of PTEN, representing more advanced stage of melanoma. Interestingly, MeWo cells express a relatively high level of ZBTB7A, and a low level of MCAM. SK-MEL-28 and UACC62, on the other hand, express a low level of ZBTB7A that correlates with a high MCAM level. The correlation derived from melanoma cell lines is consistent with the notion that ZBTB7A down-regulation and MCAM up-regulation underscores a late genetic event in melanoma progression. Chromosome 19p13.3 loss and its associated reduction of ZBTB7A expression are likely the result of genomic instability. The resulting upregulation of MCAM may represent an important consequence to these genetic alterations during melanoma progression. MCAM expression correlates with tumor metastatic potential, and has been used as a prognostic marker for melanoma (5–7). In agreement with published work (30), we showed that ZBTB7A overexpression repressed the expression of MCAM resulting in diminished ability of melanoma cells to adhere to endothelial cells and to invade. ZBTB7A underexpression in melanoma cells was associated with enhanced cell adhesion and invasion, both of which were completely blocked by the use of anti-MCAM antibody. The importance of ZBTB7A-dependent transcriptional repression of MCAM in the suppression of melanoma metastasis was additionally corroborated by animal studies. Our data collectively suggest that reduced expression of ZBTB7A due to chromosome 19p13.3 loss is an important genetic event contributing to the progression of melanoma.
In summary, our study has uncovered a novel and common genetic event associated with human melanoma metastasis, i.e. a frequent loss of chromosome 19p13.3. Biological significance of this chromosome loss is highlighted by decreased expression of ZBTB7A, leading to enhanced melanoma progression and metastasis due to compromised repression of MCAM expression (Fig. 7F). The ZBTB7A-mediated transcriptional control of MCAM expression not only represents a previously unrecognized mechanism of regulation but also carries therapeutic implications.
Supplementary Material
Acknowledgments
We are grateful to current and former members of the Yuan lab for experimental support, advice and helpful discussions. We thank The Nikon Imaging Center at Harvard Medical School for help in H&E and IHC image acquisition. This work was supported in part by the Morningside Foundation and grants from NIH/NCI (R01CA085679, RO1CA167814 and RO1CA125144).
Footnotes
All authors declare no conflict of interest.
References
- 1.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer. 2003;3:559–70. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen DX, Massagué J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–52. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 4.Luca M, et al. Direct correlation between MUC18 expression and metastatic potential of human melanoma cells. Melanoma Res. 1993;3:35–41. doi: 10.1097/00008390-199304000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Pacifico MD, et al. Development of a tissue array for primary melanoma with long-term follow-up: discovering melanoma cell adhesion molecule as an important prognostic marker. Plast Reconstr Surg. 2005;115:367–75. doi: 10.1097/01.prs.0000148417.86768.c9. [DOI] [PubMed] [Google Scholar]
- 6.Gould Rothberg BE, Bracken MB, Rimm DL. Tissue biomarkers for prognosis in cutaneous melanoma: a systematic review and meta-analysis. J Natl Cancer Inst. 2009;101:452–474. doi: 10.1093/jnci/djp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rapanotti MC, Bianchi L, Ricozzi I, Campione E, Pierantozzi A, Orlandi A, et al. Melanoma-associated markers expression in blood: MUC-18 is associated with advanced stages in melanoma patients. Br J Dermatol. 2009;160:338–344. doi: 10.1111/j.1365-2133.2008.08929.x. [DOI] [PubMed] [Google Scholar]
- 8.Mills L, Tellez C, Huang S, Baker C, McCarty M, Green L, et al. Fully human antibodies to MCAM/MUC18 inhibit tumor growth and metastasis of human melanoma. Cancer Res. 2002;62:5106–14. [PubMed] [Google Scholar]
- 9.Melnikova VO, Bar-Eli M. Bioimmunotherapy for melanoma using fully human antibodies targeting MCAM/MUC18 and IL-8. Pigment Cell Res. 2006;19:395–405. doi: 10.1111/j.1600-0749.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. The hallmarks of cancer. The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garraway LA, Lander ES. Lessons from the Cancer Genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 14.Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 18.Pessler F, Pendergrast PS, Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol. 1997;17:3786–98. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J Biol Chem. 2004;279:47081–91. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- 20.Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–97. [PubMed] [Google Scholar]
- 21.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic. 2007;6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–7. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–87. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–6. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann JM, Holzmann B, Breitbart EW, Schmiegelow P, Riethmüller G, Johnson JP. Discrimination between benign and malignant cells of melanocytic lineage by two novel antigens, a glycoprotein with a molecular weight of 113,000 and a protein with molecular weight of 76,000. Cancer Res. 1987;47:841–5. [PubMed] [Google Scholar]
- 26.Shih IM, Elder DE, Speicher D, Johnson JP, Herlyn M. Isolation and functional characterization of the A32 melanoma-associated antigen. Cancer Res. 1994;54:2514–20. [PubMed] [Google Scholar]
- 27.Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–18. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sers C, Riethmüller G, Johnson JP. MUC18, a melanoma-progression associated molecule, and its potential role in tumor vascularization and hematogenous spread. Cancer Res. 1994;54:5689–94. [PubMed] [Google Scholar]
- 29.Liu XS, Haines JE, Mehanna EK, Genet MD, Ben-Sahra I, Asara JM, et al. ZBTB7A acts as a tumor suppressor through the transcriptional repression of glycolysis. Genes Dev. 2014;28:1917–1928. doi: 10.1101/gad.245910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie S, Luca M, Huang S, Gutman M, Reich R, Johnson JP, et al. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 1997;57:2295–303. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.